library(readxl)
library(tidyverse)
library(stringr)
library(zoo)
library(hrbrthemes)
library(ggthemes)
library(ggsci)
# ---------------
# N Cases
# ---------------
y <- usa.sf %>%
st_set_geometry(NULL) %>%
inner_join(data, by="County.Code") %>%
group_by(loc) %>%
summarise(y = mean(synthetic_opioid_deaths, na.rm=T)) %>%
mutate(Strata = str_sub(loc,1,1))
# ---------------
# PLOT
# ---------------
p <- read_excel("./_data/Spat_EMMERG_tables_v3.xlsx", sheet=2) %>%
mutate(label = str_remove_all(model,pattern = "spatial"),
label = str_remove_all(label,pattern = "spatio-temporal"),
label = str_trim(label),
Strata = na.locf(Strata)
) %>%
inner_join(y, by="Strata") %>%
mutate(NRMSE = RMSE/y,
NMAE = MAE/y,
NMSLE = MSLE/y)
start <- p %>% filter(Type=="baseline")
end <- p %>% filter(Type=="spatial")
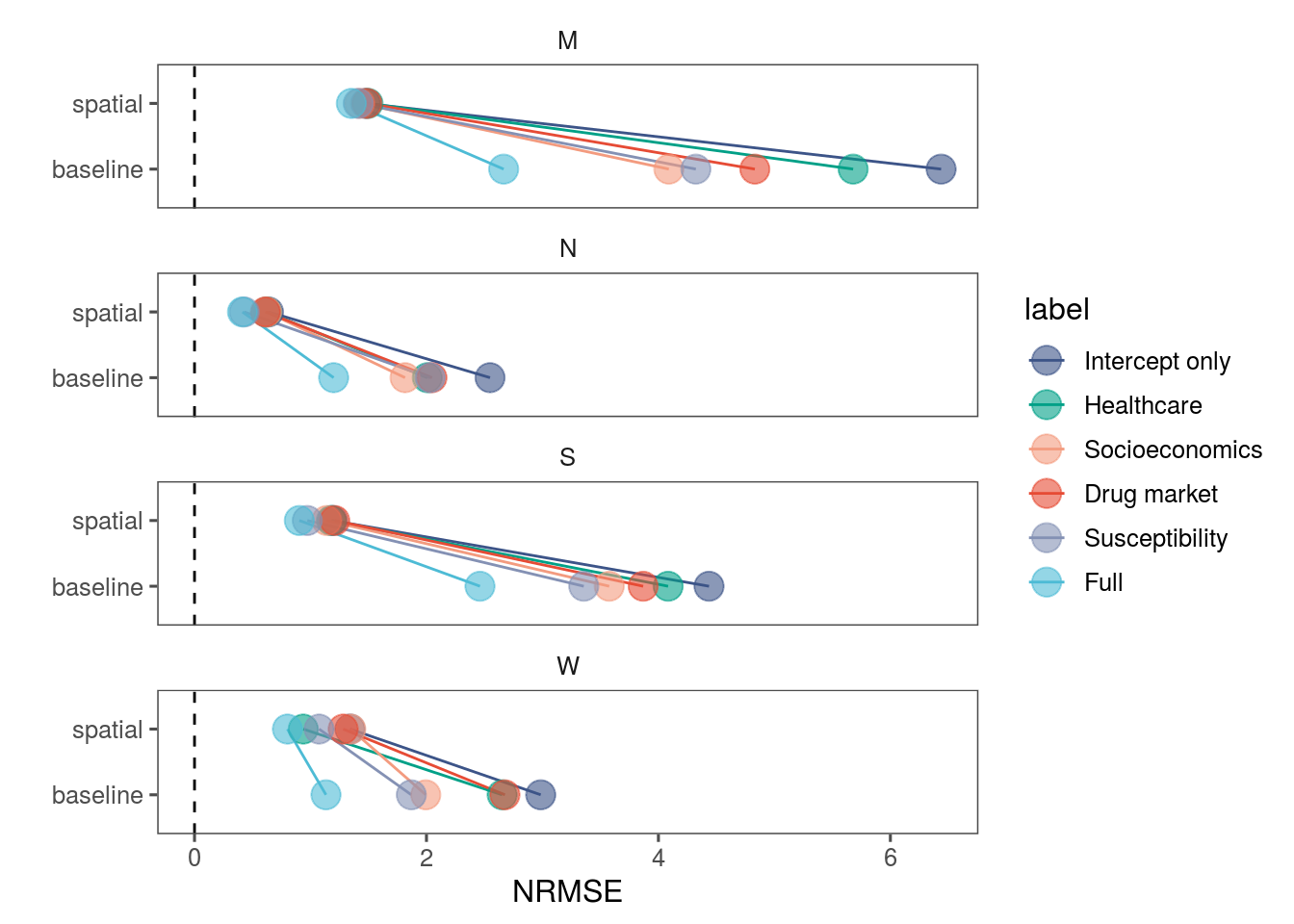
p %>%
ggplot(aes(y=Type, x=NRMSE, col=label)) +
geom_segment(data=start, aes(x = NRMSE, y= Type, xend=end$NRMSE, yend=end$Type)) +
geom_point(size = 5, alpha=.6) +
geom_vline(xintercept = 0, linetype="dashed") +
labs(y="") +
scale_color_npg(breaks = c("Intercept only", "Healthcare", "Socioeconomics", "Drug market", "Susceptibility", "Full")) +
theme_few() +
#theme(legend.position = "top") +
facet_wrap(.~Strata, ncol = 1)
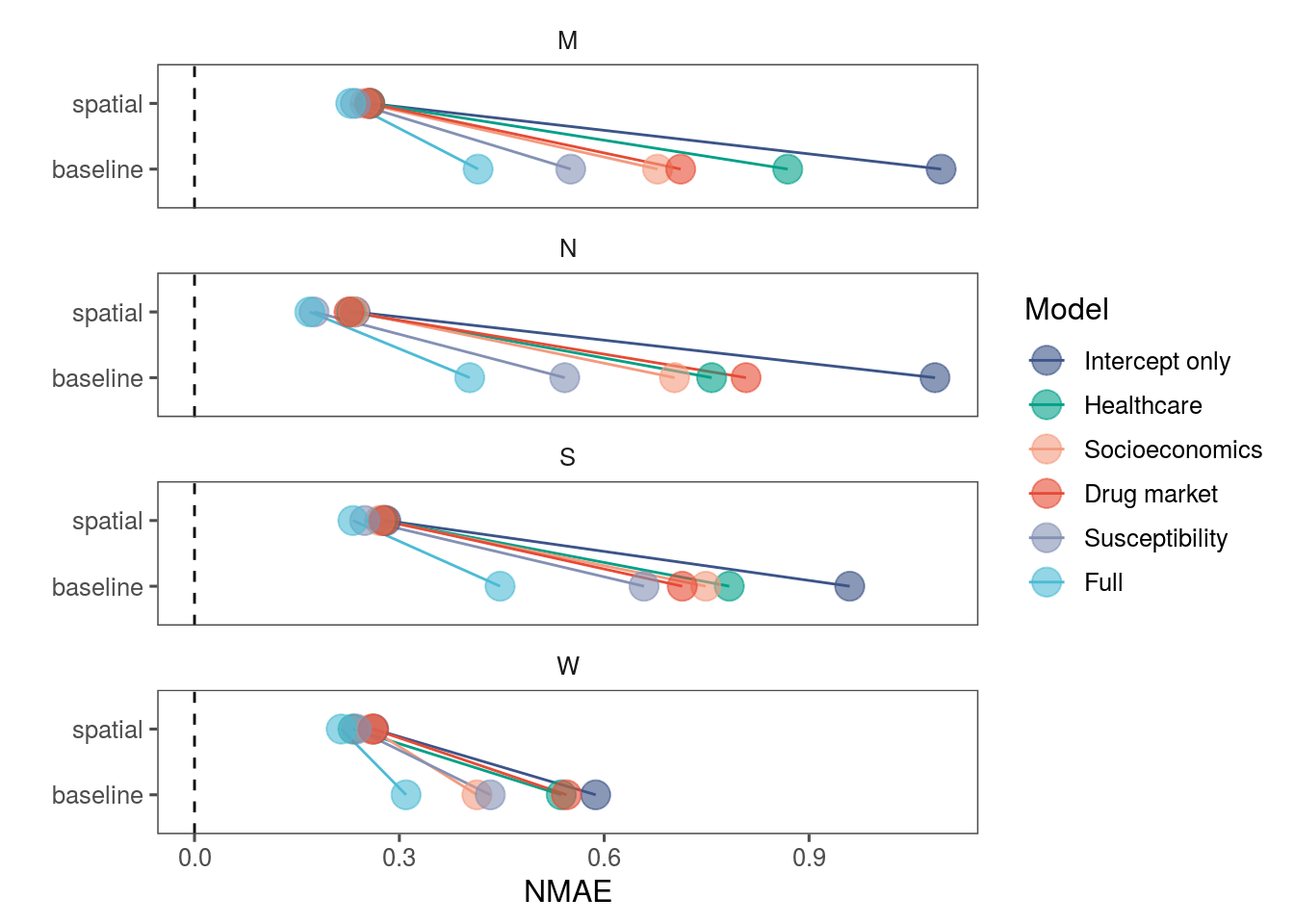
p %>%
filter(model!="Spatio-temporal only") %>%
ggplot(aes(y=Type, x=NMAE, col=label)) +
geom_segment(data=start, aes(x = NMAE, y= Type, xend=end$NMAE, yend=end$Type)) +
geom_point(size = 5, alpha=.6) +
geom_vline(xintercept = 0, linetype="dashed") +
labs(y="", color = "Model") +
scale_color_npg(breaks = c("Intercept only", "Healthcare", "Socioeconomics", "Drug market", "Susceptibility", "Full")) +
theme_few() +
#theme(legend.position = "top") +
facet_wrap(.~Strata, ncol = 1)
#ggsave("./_out/f1.png", height = 7, width = 8, dpi = "retina")