Chapter 2 Data
2.1 LIBRARIES
2.2 ADMINISTRATIVE AND SPATIAL DATA
data <- read.csv("./_data/Cleaned_Final_Dataset_2018_7-2-20.csv", stringsAsFactors = F)
usa.sf <- st_read("./_data/GIS/tl_2017_us_county/tl_2017_us_county.shp", stringsAsFactors = F) %>%
#filter(STATEFP=="01") %>%
filter(STATEFP!="02", STATEFP!="15", STATEFP!="60", STATEFP!="66", STATEFP!="69", STATEFP!="72", STATEFP!="78") %>%
mutate(county_code = as.numeric(GEOID),
STATEFP = as.numeric(STATEFP)) %>%
left_join(read_excel("./_data/fp_codes.xlsx") %>%
dplyr::rename(STATEFP = `Numeric code`), by = "STATEFP") ## Reading layer `tl_2017_us_county' from data source `/research-home/gcarrasco/EMMERG_MAP2/_data/GIS/tl_2017_us_county/tl_2017_us_county.shp' using driver `ESRI Shapefile'
## Simple feature collection with 3233 features and 17 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -179.2311 ymin: -14.60181 xmax: 179.8597 ymax: 71.43979
## CRS: 4269usa.sf %>%
group_by(loc) %>%
summarise(c = mean(county_code)) %>%
ggplot() +
geom_sf(aes(fill = loc), size = 0.5, col = "black") +
theme_bw()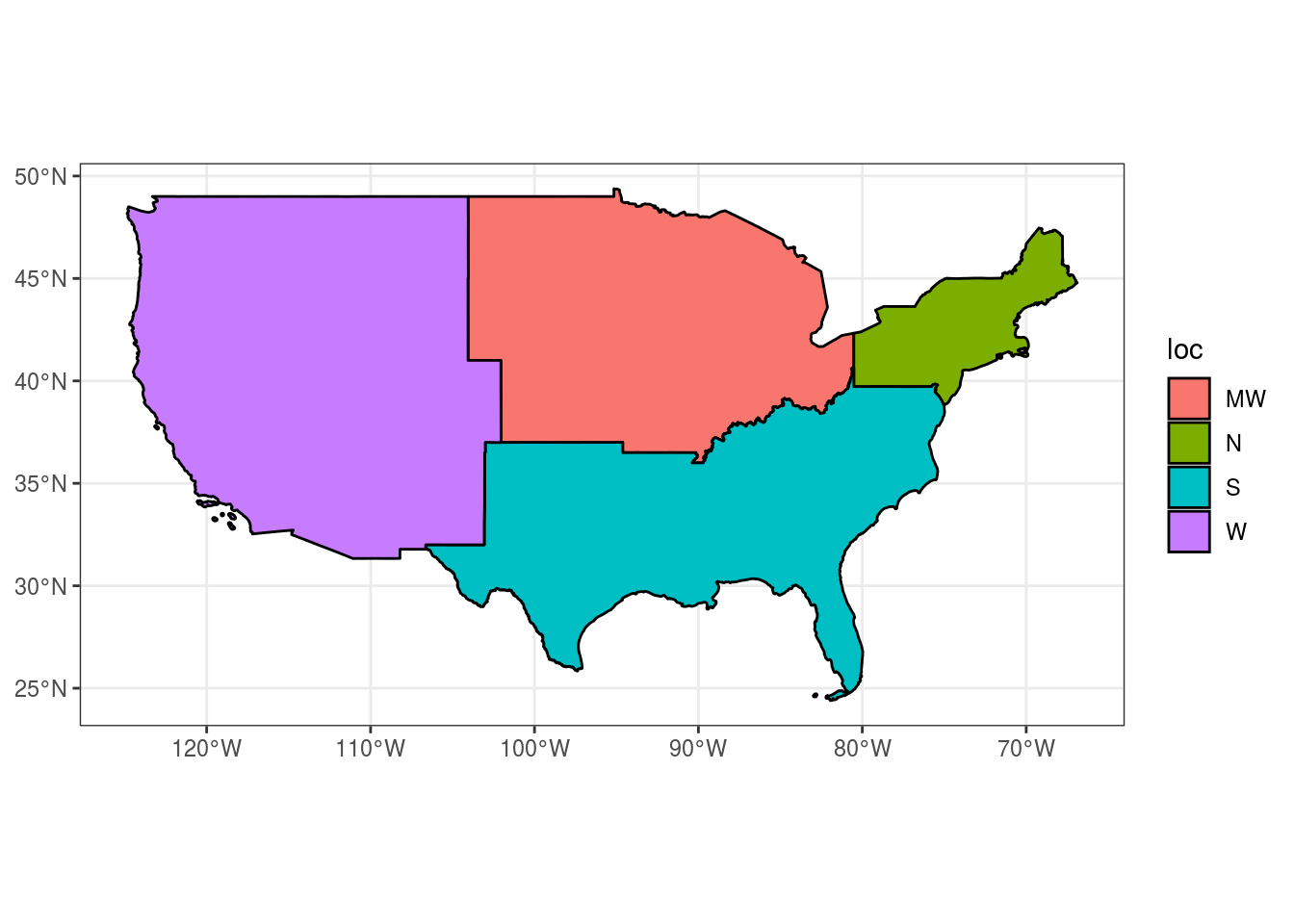
2.3 Synthetic Opioid Deaths
library(colorspace)
library(ggsci)
library(ggthemes)
library(cowplot)
sd_m <- dat.sf %>%
ggplot() +
geom_sf(aes(fill=synthetic_opioid_deaths), size = 0.3, col = NA) +
#scale_fill_continuous_sequential("BurgYl", trans = "log10", na.value = "#CDCDC1") +
scale_fill_continuous_sequential("BurgYl", trans = "pseudo_log", na.value = "#CDCDC1", breaks = c(1,3,10,30,100,300,3000)) +
labs(fill = "Synthetic Opioid Deaths") +
theme_void() +
theme(panel.grid.major = element_line(color = "white"),
strip.text = element_text(size=15),
legend.position = "bottom") +
facet_wrap(.~year, ncol = 2)
sd_t <- dat.sf %>%
st_set_geometry(NULL) %>%
group_by(year, loc) %>%
summarise(synthetic_opioid_deaths = sum(synthetic_opioid_deaths, na.rm = T)) %>%
rbind(dat.sf %>%
st_set_geometry(NULL) %>%
mutate(loc = "US") %>%
group_by(year, loc) %>%
summarise(synthetic_opioid_deaths = sum(synthetic_opioid_deaths, na.rm = T))) %>%
ggplot(aes(x = year, y = synthetic_opioid_deaths, col = loc)) +
geom_line() +
labs(y = "Synthetic Opioid Deaths", color = "Area") +
scale_color_npg() +
theme_few() +
theme(legend.position = "bottom")
plot_grid(sd_t, sd_m, labels = c("A)", "B)"), nrow = 1)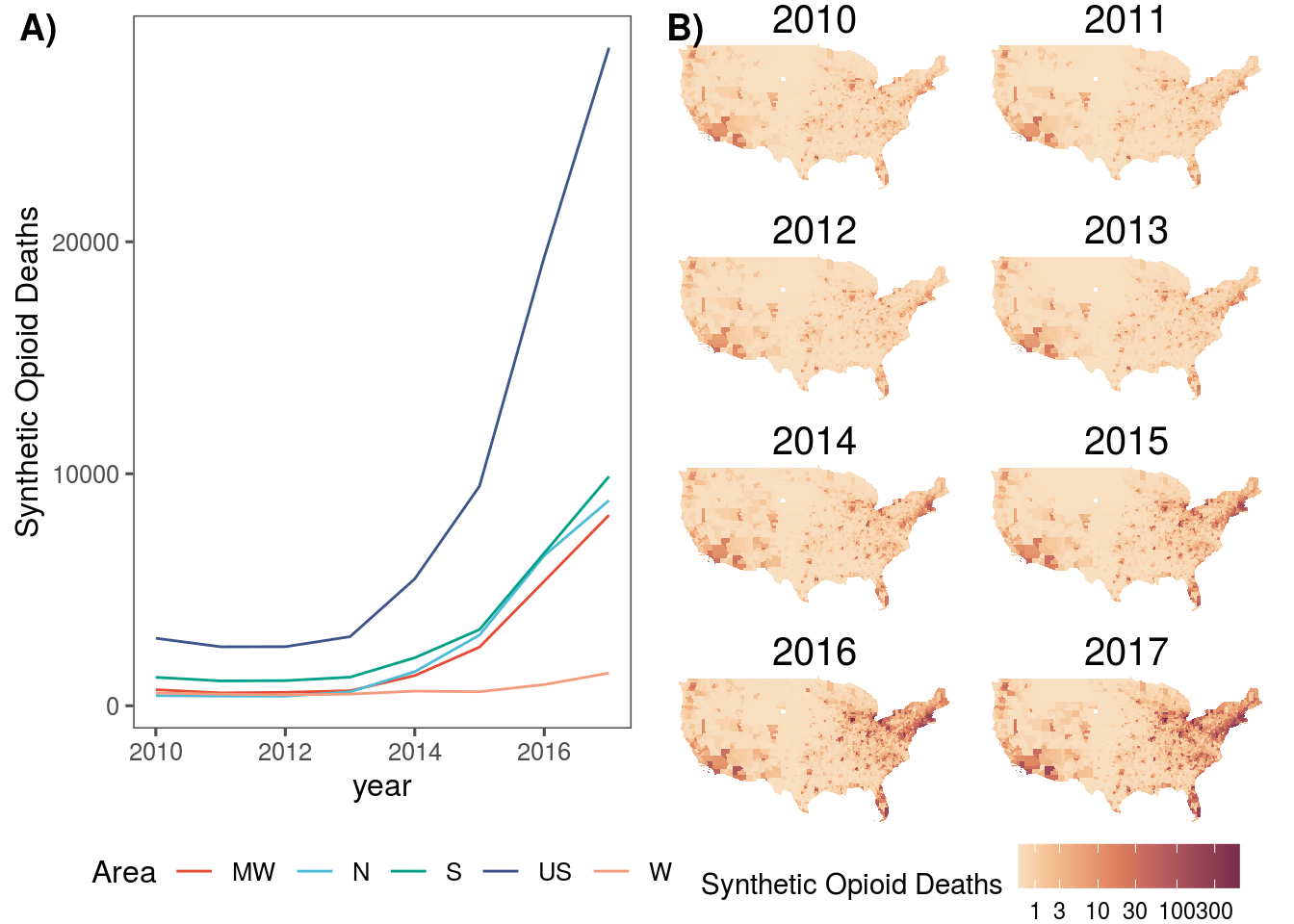
# IN TEXT
dat.sf %>%
st_set_geometry(NULL) %>%
ungroup() %>%
filter(synthetic_opioid_deaths>=10) %>%
summarise(synthetic_opioid_deaths = sum(synthetic_opioid_deaths, nana.rm = T))## synthetic_opioid_deaths
## 1 53532dat.sf %>%
st_set_geometry(NULL) %>%
ungroup() %>%
filter(synthetic_opioid_deaths>=10) %>%
distinct(GEOID) %>%
count()## n
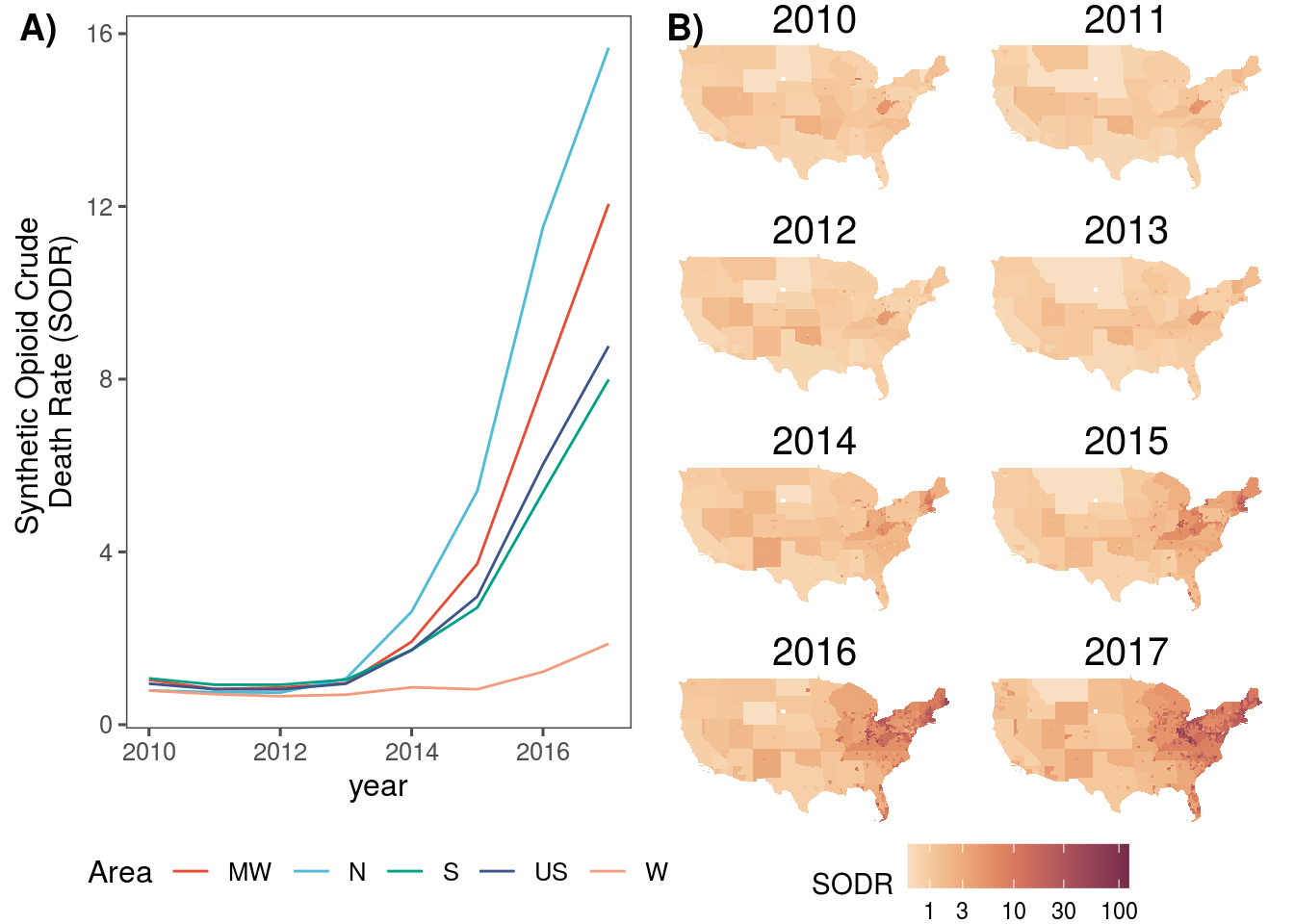
## 1 5022.4 Synthetic Opioid Deaths Rate
library(colorspace)
library(ggsci)
library(ggthemes)
library(cowplot)
sr_m <- dat.sf %>%
ggplot() +
geom_sf(aes(fill=synthetic_opioid_crude_death_rate), size = 0.3, col = NA) +
#scale_fill_continuous_sequential("BurgYl", trans = "log10", na.value = "#CDCDC1") +
scale_fill_continuous_sequential("BurgYl", trans = "pseudo_log", na.value = "#CDCDC1", breaks = c(1,3,10,30,100,300,3000)) +
labs(fill = "SODR") +
theme_void() +
theme(panel.grid.major = element_line(color = "white"),
strip.text = element_text(size=15),
legend.position = "bottom") +
facet_wrap(.~year, ncol = 2)
sr_t <- dat.sf %>%
st_set_geometry(NULL) %>%
group_by(year, loc) %>%
summarise(synthetic_opioid_deaths = sum(synthetic_opioid_deaths, na.rm = T),
population = sum(population, na.rm=T)) %>%
rbind(dat.sf %>%
st_set_geometry(NULL) %>%
mutate(loc = "US") %>%
group_by(year, loc) %>%
summarise(synthetic_opioid_deaths = sum(synthetic_opioid_deaths, na.rm = T),
population = sum(population, na.rm=T))) %>%
mutate(synthetic_opioid_crude_death_rate = 100000*synthetic_opioid_deaths/population) %>%
ggplot(aes(x = year, y = synthetic_opioid_crude_death_rate, col = loc)) +
geom_line() +
labs(y = "Synthetic Opioid Crude \nDeath Rate (SODR)", color = "Area") +
scale_color_npg() +
theme_few() +
theme(legend.position = "bottom")
plot_grid(sr_t, sr_m, labels = c("A)", "B)"), nrow = 1)