3 Estrutura da comunidade
3.0.1 Organização básica
dev.off() #apaga os graficos, se houver algum
rm(list=ls(all=TRUE)) #limpa a memória
cat("\014") #limpa o console Instalando os pacotes necessários
install.packages("vegan")
install.packages("moments")
install.packages("ggplot2")
install.packages("dplyr")
install.packages("tidyr")
install.packages("tibble")
install.packages("tidyverse") #atente para alguma msg de erro qdo executar essa linha
install.packages("forcats")
install.packages("iNEXT")
install.packages("openxlsx")
install.packages('Rcpp')Depois de instalado, carregue os pacotes no seu computador.
library(tibble); library(tidyverse); library(forcats); library(openxlsx); library(Rcpp)3.1 Importar a base de dados
#dir <- getwd()
#shell.exec(dir) #abre o diretorio de trabalho no Windows Explorer
m_bruta <- read.csv("D:/Elvio/OneDrive/MSS/_Zoo-Rebio/R_ZooRebio.rmd/zoorebio.csv",
sep = ";", dec = ",",
header = T,
row.names = 1,
na.strings = NA)
str(m_bruta)
#View(m_bruta)## 'data.frame': 49 obs. of 85 variables:
## $ Bra.angularis : num 0 0.533 0.533 0 0 ...
## $ Lepadella.sp : num 0 0.178 0.178 0.533 0.178 ...
## $ Lecane.sp : num 0 0 0 0 0 ...
## $ Lec.leontina : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.bulla : num 1.067 0 0.356 0.889 0.178 ...
## $ Lec.cornuta : num 0 0 0 0 0 ...
## $ Lec.curvicornis : num 0 0 0 0 0 ...
## $ Notholca.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bra.urceolaris : num 0.178 0 0 0 0 ...
## $ Trichocerca.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.quadridentata : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.kluchor : num 0.178 0 0 0 0 ...
## $ Lec.lunaris : num 0.356 0.711 1.067 0 0.178 ...
## $ Rotaria.sp : num 0 1.07 0 0 0 ...
## $ Aspelta.sp : num 0.178 0 0 0 0 ...
## $ Lec.furcata : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Col.geophila : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lep.dactyliseta : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bra.calyciflorus : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bra.caudatus : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.crepida : num 0 0 0 0 0 ...
## $ Pla.patulus : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.ovalis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.elasma : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Pla.quadricornis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.aculeata : num 1.6 0.889 0 0.178 0 ...
## $ Ker.tropica : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bdelloidea : num 0 0 0 0 0 ...
## $ Ker.lenzi : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Con.unicornis : num 0 0 0 0 0 ...
## $ Alo.dadayi : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Macrothrix.sp : num 0.178 0 0 0 0 ...
## $ Alonella.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Nauplii : num 0.711 0.889 0.178 0.889 0.711 ...
## $ Chy.eurynotus : num 0.356 0 0 0 0 ...
## $ Cyclopoida : num 0 0 0 0 0 ...
## $ Mac.collinsi : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Copepodite : num 0 0 0 0.178 0 ...
## $ Paracyclops.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Asc.ecaudis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lep.ovalis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Polyarthra.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bra.havanaensis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Hexarthra.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.hastata : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Tri.tetractis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Ker.serrulata : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Notodiaptomus.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bra.falcatus : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Fil.longiseta : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Myt.crassipes : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lep.patella : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Euchlanis.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Colurella.sp : num 0 0 0.178 0 0 ...
## $ Mytilina.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.monostyla : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.ligona : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Ascomorpha.sp : num 0.178 0 0 0 0 ...
## $ Pom.sulcata : num 0 0 0 0.356 0.356 ...
## $ Harpacticoida : num 0 0 0 0 0 ...
## $ Epi.senta : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Rot.neptunia : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Dis.aculeata : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.luna : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Euc.dilatata : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bea.eudactylota : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Chy.sphaericus : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Lec.hornemanni : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Dia.birgei : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Mac.subquadratus : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Bra.quadridentatus: num 0 0 0 0 0 0 0 0 0 0 ...
## $ Pol.vulgaris : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Pol.bicerca : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Dicranop.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Asc.saltans : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Moi.minuta : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Alo.hamulata : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Mac.laticornis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Mac.mira : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Pro.similis : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Alo.pulchella : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Dia.spinulosum : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Chydorus.sp : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Ily.spinifer : num 0 0 0 0 0 0 0 0 0 0 ...
## $ Dun.odontoplax : num 0 0 0 0 0 0 0 0 0 0 ...3.2 Reset point
m_trab <- m_brutaAqui substitui-se uma nova matriz de dados, relativizada e/ou transformada, pela matriz bruta.
3.3 Transpor a matriz para trabalhar com as espécies
A função t transpõe a matriz. Só deve ser usada uma vez, pois se repetida com Ctrl+Enter continua “girando” a matriz. As espécies como colunas representam uma matriz comunitária e as espécies como linhas representam uma matriz (comunitária) transposta.
m_trab <- t(m_trab)
str(m_trab)
#View(m_trab)
m_trab## num [1:85, 1:49] 0 0 0 0 1.07 ...
## - attr(*, "dimnames")=List of 2
## ..$ : chr [1:85] "Bra.angularis" "Lepadella.sp" "Lecane.sp" "Lec.leontina" ...
## ..$ : chr [1:49] "BB1-P01" "BB1-P02" "BB1-P03" "BB2-P01" ...
## BB1-P01 BB1-P02 BB1-P03 BB2-P01 BB2-P02 BB2-P03 BB3-P01 BB3-P02 BB3-P03 BB4-P01
## Bra.angularis 0.0000000 0.5333333 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lepadella.sp 0.0000000 0.1777778 0.1777778 0.5333333 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.bulla 1.0666667 0.0000000 0.3555556 0.8888889 0.1777778 0.0000000 0.5333333 1.6000000 1.4222222 1.4222222
## Lec.cornuta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.1777778 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.kluchor 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.lunaris 0.3555556 0.7111111 1.0666667 0.0000000 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000
## Rotaria.sp 0.0000000 1.0666667 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Aspelta.sp 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## BB4-P02 BB4-P03 BB5-P01 BB5-P02 BB5-P03 BB6-P01 BB6-P02 BB6-P03 CA1-P04 CA1-P05
## Bra.angularis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Lepadella.sp 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.1777778 0.3555556 0.7111111 0.7111111
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 0.0000000 0.3555556 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.bulla 0.3555556 1.0666667 2.1333333 0.7111111 0.5333333 0.8888889 0.5333333 0.3555556 0.8888889 1.2444444
## Lec.cornuta 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Notholca.sp 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.1777778
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000
## Lec.kluchor 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.3555556 0.1777778
## Lec.lunaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.3555556 0.1777778
## Rotaria.sp 0.3555556 0.1777778 0.0000000 0.1777778 0.1777778 0.1777778 0.7111111 0.1777778 0.3555556 0.0000000
## Aspelta.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 1.0666667
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.3555556
## CA1-P06 CA2-P04 CA2-P05 CA2-P06 CA3-P04 CA3-P05 CA3-P06 CA4-P04 CA4-P05 CA4-P06
## Bra.angularis 0.0000000 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lepadella.sp 0.0000000 0.1777778 0.3555556 0.5333333 0.3555556 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 0.0000000 0.3555556 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.bulla 0.5333333 0.5333333 0.7111111 1.6000000 1.6000000 1.7777778 0.7111111 0.5333333 0.1777778 0.0000000
## Lec.cornuta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.1777778 0.0000000 0.1777778 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.kluchor 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.lunaris 0.7111111 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Rotaria.sp 0.8888889 0.0000000 0.5333333 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Aspelta.sp 0.0000000 0.0000000 0.1777778 0.7111111 0.7111111 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## CA5-P04 CA5-P05 CA5-P06 CA6-P04 CA6-P05 CA6-P06 EN08 EN09 EN10 EN11
## Bra.angularis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lepadella.sp 0.0000000 0.1777778 0.0000000 6.4000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.3555556 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 1.0666667 0.0000000
## Lec.bulla 0.7111111 0.1777778 0.3555556 4.2666667 2.1333333 0.0000000 0.0000000 2.1333333 1.0666667 72.0000000
## Lec.cornuta 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.0000000 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 1.6000000 1.6000000
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 2.6666667 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.quadridentata 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.kluchor 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.lunaris 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.1777778 0.5333333 0.0000000 0.5333333 0.0000000
## Rotaria.sp 0.3555556 0.0000000 0.1777778 2.6666667 0.5333333 0.0000000 1.0666667 0.5333333 1.0666667 0.5333333
## Aspelta.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 1.0666667 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 10.6666667
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 5.3333333 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## EN12 EN13 EN15 EN16 EN14 EN17 RE19 RE18 RE07
## Bra.angularis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lepadella.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 1.6000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.bulla 0.5333333 1.0666667 12.8000000 0.0000000 0.0000000 0.0000000 1.0666667 2.133333 0.0000000
## Lec.cornuta 0.5333333 0.0000000 2.6666667 0.5333333 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 1.6000000
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.5333333
## Lec.kluchor 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.lunaris 0.0000000 0.0000000 0.0000000 1.6000000 0.5333333 0.0000000 0.0000000 0.000000 2.6666667
## Rotaria.sp 0.0000000 0.0000000 2.6666667 10.1333333 9.0666667 10.1333333 2.1333333 3.733333 24.5333333
## Aspelta.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 1.6000000 0.0000000 0.0000000 0.0000000 0.5333333 0.000000 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## [ reached getOption("max.print") -- omitted 65 rows ]3.3.1 Informações básicas da matriz
range(m_trab) #menor e maior valores
length(m_trab) #no. de colunas
ncol(m_trab) #no. de N colunas
nrow(m_trab) #no. de M linhas
sum(lengths(m_trab)) #soma os nos. de colunas
length(as.matrix(m_trab)) #tamanho da matriz m x n
sum(m_trab == 0) #número de observações igual a zero
sum(m_trab > 0) #número de observações maiores que zero
zeros <- (sum(m_trab == 0)/length(as.matrix(m_trab)))*100 #proporção de zeros na matriz
zeros## [1] 0 72
## [1] 4165
## [1] 49
## [1] 85
## [1] 4165
## [1] 4165
## [1] 3763
## [1] 402
## [1] 90.34814Tabela que resume as informações geradas (Tabela 3.1).
## Função Resultado
## 1 range 0 - 72
## 2 lenght 4165
## 3 n cols 49
## 4 m linhas 85
## 5 Tamanho 4165
## 6 Tamanho 4165
## 7 Zeros 3763
## 8 Nao zeros 402
## 9 % Zeros 90.3| Função | Resultado |
|---|---|
| range | 0 - 72 |
| lenght | 4165 |
| n cols | 49 |
| m linhas | 85 |
| Tamanho | 4165 |
| Tamanho | 4165 |
| Zeros | 3763 |
| Nao zeros | 402 |
| % Zeros | 90.3 |
Ou seja, temos uma matriz de tamanho m x n igual a 85 objetos por 49 atributos, onde 90.35% dos valores da matriz são iguais a zero!
3.4 Calculando os descritores da comunidade
#?apply
Sum <- rowSums(m_trab)
#ou
Sum <- apply(m_trab,1,sum)
Sum
### Media
Mean <- rowMeans(m_trab)
Mean
##Ou
Mean <- apply(m_trab,1,mean)
Mean
### Desvio padrao
DP <- apply(m_trab,1,sd)
DP
### Máximo
Max <- apply(m_trab,1,max)
Max
### Minimo
Min <- apply(m_trab,1,min)
Min## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 1.7777778 11.7333333 0.7111111 3.9111111 124.8000000 5.6888889
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 3.3777778 0.1777778 0.5333333 4.9777778 1.6000000 1.2444444
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 10.4888889 74.3111111 1.9555556 14.0444444 0.3555556 6.5777778
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.1777778 0.3555556 1.6000000 4.8000000 0.5333333 0.1777778
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.7111111 3.9111111 0.1777778 60.5333333 0.7111111 1.0666667
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 1.6000000 1.9555556 4.8000000 234.0444444 1.4222222 7.4666667
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.3555556 23.2888889 0.3555556 0.1777778 2.3111111 0.1777778
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.1777778 0.1777778 1.7777778 2.3111111 0.1777778 0.3555556
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 25.7777778 0.1777778 0.7111111 65.6000000 2.6666667 7.8222222
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 1.9555556 1.7777778 1.6000000 0.1777778 0.7111111 0.1777778
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.1777778 1.6000000 5.8666667 2.6666667 1.0666667 0.5333333
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 3.7333333 3.7333333 2.1333333 0.5333333 10.1333333 19.2000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 58.1333333 68.8000000 1.0666667 2.6666667 1.0666667 2.6666667
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 2.1333333 0.5333333 3.7333333 1.6000000 1.0666667 3.7333333
## Dun.odontoplax
## 0.5333333
## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.036281179 0.239455782 0.014512472 0.079818594 2.546938776 0.116099773
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.068934240 0.003628118 0.010884354 0.101587302 0.032653061 0.025396825
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0.214058957 1.516553288 0.039909297 0.286621315 0.007256236 0.134240363
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.003628118 0.007256236 0.032653061 0.097959184 0.010884354 0.003628118
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.014512472 0.079818594 0.003628118 1.235374150 0.014512472 0.021768707
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.032653061 0.039909297 0.097959184 4.776417234 0.029024943 0.152380952
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.007256236 0.475283447 0.007256236 0.003628118 0.047165533 0.003628118
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.003628118 0.003628118 0.036281179 0.047165533 0.003628118 0.007256236
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.526077098 0.003628118 0.014512472 1.338775510 0.054421769 0.159637188
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.039909297 0.036281179 0.032653061 0.003628118 0.014512472 0.003628118
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.003628118 0.032653061 0.119727891 0.054421769 0.021768707 0.010884354
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.076190476 0.076190476 0.043537415 0.010884354 0.206802721 0.391836735
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 1.186394558 1.404081633 0.021768707 0.054421769 0.021768707 0.054421769
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.043537415 0.010884354 0.076190476 0.032653061 0.021768707 0.076190476
## Dun.odontoplax
## 0.010884354
## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.036281179 0.239455782 0.014512472 0.079818594 2.546938776 0.116099773
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.068934240 0.003628118 0.010884354 0.101587302 0.032653061 0.025396825
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0.214058957 1.516553288 0.039909297 0.286621315 0.007256236 0.134240363
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.003628118 0.007256236 0.032653061 0.097959184 0.010884354 0.003628118
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.014512472 0.079818594 0.003628118 1.235374150 0.014512472 0.021768707
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.032653061 0.039909297 0.097959184 4.776417234 0.029024943 0.152380952
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.007256236 0.475283447 0.007256236 0.003628118 0.047165533 0.003628118
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.003628118 0.003628118 0.036281179 0.047165533 0.003628118 0.007256236
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.526077098 0.003628118 0.014512472 1.338775510 0.054421769 0.159637188
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.039909297 0.036281179 0.032653061 0.003628118 0.014512472 0.003628118
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.003628118 0.032653061 0.119727891 0.054421769 0.021768707 0.010884354
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.076190476 0.076190476 0.043537415 0.010884354 0.206802721 0.391836735
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 1.186394558 1.404081633 0.021768707 0.054421769 0.021768707 0.054421769
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.043537415 0.010884354 0.076190476 0.032653061 0.021768707 0.076190476
## Dun.odontoplax
## 0.010884354
## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.13073816 0.91771842 0.07980829 0.28123472 10.30321459 0.39677883
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.32011588 0.02539683 0.04306241 0.43997755 0.11286596 0.09601097
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0.48139659 4.12290234 0.14635345 1.53952216 0.03554044 0.77366584
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.02539683 0.05079365 0.11855631 0.60945767 0.04306241 0.02539683
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.07980829 0.27651261 0.02539683 2.55779257 0.06111970 0.09345779
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.11855631 0.23027000 0.32873114 13.24363377 0.08391207 0.38404389
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.05079365 0.98407919 0.05079365 0.02539683 0.13949001 0.02539683
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.02539683 0.02539683 0.22945157 0.24009740 0.02539683 0.03554044
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 3.43095743 0.02539683 0.07980829 7.69424793 0.38095238 1.06620045
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.23027000 0.13073816 0.22857143 0.02539683 0.07108087 0.02539683
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.02539683 0.16894139 0.46527164 0.38095238 0.10662131 0.07619048
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.34426519 0.53333333 0.23942486 0.07619048 1.44761905 2.74285714
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 5.83147557 9.82857143 0.15238095 0.31259768 0.10662131 0.27205442
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.30476190 0.07619048 0.53333333 0.22857143 0.15238095 0.53333333
## Dun.odontoplax
## 0.07619048
## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.5333333 6.4000000 0.5333333 1.6000000 72.0000000 2.6666667
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 1.6000000 0.1777778 0.1777778 2.6666667 0.5333333 0.5333333
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 2.6666667 24.5333333 0.7111111 10.6666667 0.1777778 5.3333333
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.1777778 0.3555556 0.5333333 4.2666667 0.1777778 0.1777778
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.5333333 1.6000000 0.1777778 13.3333333 0.3555556 0.5333333
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.7111111 1.6000000 1.6000000 70.9333333 0.3555556 1.6000000
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.3555556 4.8000000 0.3555556 0.1777778 0.5333333 0.1777778
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.1777778 0.1777778 1.6000000 1.6000000 0.1777778 0.1777778
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 24.0000000 0.1777778 0.5333333 53.8666667 2.6666667 7.4666667
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 1.6000000 0.5333333 1.6000000 0.1777778 0.3555556 0.1777778
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.1777778 1.0666667 2.6666667 2.6666667 0.5333333 0.5333333
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 2.1333333 3.7333333 1.6000000 0.5333333 10.1333333 19.2000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 31.4666667 68.8000000 1.0666667 2.1333333 0.5333333 1.6000000
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 2.1333333 0.5333333 3.7333333 1.6000000 1.0666667 3.7333333
## Dun.odontoplax
## 0.5333333
## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0 0 0 0 0 0
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0 0 0 0 0 0
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0 0 0 0 0 0
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0 0 0 0 0 0
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0 0 0 0 0 0
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0 0 0 0 0 0
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0 0 0 0 0 0
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0 0 0 0 0 0
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0 0 0 0 0 0
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0 0 0 0 0 0
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0 0 0 0 0 0
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0 0 0 0 0 0
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 0 0 0 0 0 0
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0 0 0 0 0 0
## Dun.odontoplax
## 03.5 Riqueza
Ou frequência de ocorrência (no caso da matrix transposta). Converte-se primeiro para matriz binaria
library(vegan)
bin <- decostand(m_trab,"pa")
bin
S <- apply(bin,1,sum)
S
#OU
Riqueza <- specnumber(m_trab)
Riqueza
Riqueza_total <- specnumber(colSums(m_trab))
Riqueza_total## BB1-P01 BB1-P02 BB1-P03 BB2-P01 BB2-P02 BB2-P03 BB3-P01 BB3-P02 BB3-P03 BB4-P01 BB4-P02 BB4-P03
## Bra.angularis 0 1 1 0 0 0 0 0 0 0 0 0
## Lepadella.sp 0 1 1 1 1 1 0 0 0 0 0 0
## Lecane.sp 0 0 0 0 0 0 1 0 0 0 0 0
## Lec.leontina 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.bulla 1 0 1 1 1 0 1 1 1 1 1 1
## Lec.cornuta 0 0 0 0 0 0 1 0 1 0 0 0
## Lec.curvicornis 0 0 0 0 0 0 0 0 0 1 0 0
## Notholca.sp 0 0 0 0 0 0 0 0 0 0 1 0
## Bra.urceolaris 1 0 0 0 0 0 0 0 0 0 0 0
## Trichocerca.sp 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.quadridentata 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.kluchor 1 0 0 0 0 0 0 0 0 0 0 0
## Lec.lunaris 1 1 1 0 1 1 0 0 0 0 0 0
## Rotaria.sp 0 1 0 0 0 0 0 0 0 0 1 1
## Aspelta.sp 1 0 0 0 0 0 0 0 0 0 0 0
## Lec.furcata 0 0 0 0 0 0 0 0 0 0 0 0
## Col.geophila 0 0 0 0 0 0 0 0 0 0 0 0
## Lep.dactyliseta 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.calyciflorus 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.caudatus 0 0 0 0 0 0 0 0 0 0 0 0
## BB5-P01 BB5-P02 BB5-P03 BB6-P01 BB6-P02 BB6-P03 CA1-P04 CA1-P05 CA1-P06 CA2-P04 CA2-P05 CA2-P06
## Bra.angularis 0 0 0 0 0 0 1 0 0 1 0 0
## Lepadella.sp 0 1 0 0 1 1 1 1 0 1 1 1
## Lecane.sp 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.leontina 0 1 0 0 0 0 0 0 0 0 0 1
## Lec.bulla 1 1 1 1 1 1 1 1 1 1 1 1
## Lec.cornuta 0 1 0 0 0 0 0 0 0 0 0 0
## Lec.curvicornis 0 0 0 0 0 0 0 0 0 0 0 0
## Notholca.sp 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.urceolaris 0 0 0 0 0 0 1 0 0 0 0 1
## Trichocerca.sp 0 0 0 0 0 0 1 1 0 0 0 0
## Lec.quadridentata 0 1 0 0 0 0 1 0 0 0 0 1
## Lec.kluchor 0 0 0 0 0 0 1 1 1 0 0 0
## Lec.lunaris 0 0 0 0 1 0 1 1 1 0 0 0
## Rotaria.sp 0 1 1 1 1 1 1 0 1 0 1 1
## Aspelta.sp 0 0 0 0 0 0 1 0 0 0 1 1
## Lec.furcata 0 0 0 0 0 0 1 0 0 0 0 0
## Col.geophila 0 0 0 0 0 0 0 1 0 0 0 0
## Lep.dactyliseta 0 0 0 0 0 0 0 1 0 0 0 0
## Bra.calyciflorus 0 0 0 0 0 0 0 1 0 0 0 0
## Bra.caudatus 0 0 0 0 0 0 0 1 0 0 0 0
## CA3-P04 CA3-P05 CA3-P06 CA4-P04 CA4-P05 CA4-P06 CA5-P04 CA5-P05 CA5-P06 CA6-P04 CA6-P05 CA6-P06 EN08
## Bra.angularis 0 0 0 0 0 0 0 0 0 0 0 0 0
## Lepadella.sp 1 1 1 0 0 0 0 1 0 1 0 0 0
## Lecane.sp 0 1 0 0 0 0 0 0 0 0 0 0 0
## Lec.leontina 0 0 0 0 0 0 0 1 1 0 0 0 0
## Lec.bulla 1 1 1 1 1 0 1 1 1 1 1 0 0
## Lec.cornuta 0 1 1 0 1 0 1 1 0 0 0 0 1
## Lec.curvicornis 0 0 0 0 0 0 0 0 0 0 0 0 0
## Notholca.sp 0 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.urceolaris 0 0 0 0 0 0 0 0 0 0 0 0 0
## Trichocerca.sp 1 0 0 1 0 0 0 0 0 1 0 0 0
## Lec.quadridentata 0 0 0 0 0 0 0 1 0 0 0 0 0
## Lec.kluchor 0 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.lunaris 0 0 0 0 0 0 0 0 0 1 0 1 1
## Rotaria.sp 0 0 0 0 0 0 1 0 1 1 1 0 1
## Aspelta.sp 1 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.furcata 0 0 0 0 0 0 0 0 0 1 0 0 0
## Col.geophila 1 0 0 0 0 0 0 0 0 0 0 0 0
## Lep.dactyliseta 0 0 0 0 0 0 0 0 0 1 0 1 0
## Bra.calyciflorus 0 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.caudatus 0 0 0 0 0 0 0 0 0 0 0 0 0
## EN09 EN10 EN11 EN12 EN13 EN15 EN16 EN14 EN17 RE19 RE18 RE07
## Bra.angularis 0 0 0 0 0 0 0 0 0 0 0 0
## Lepadella.sp 0 0 0 0 0 0 0 0 0 0 0 0
## Lecane.sp 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.leontina 0 1 0 0 0 1 0 0 0 0 0 0
## Lec.bulla 1 1 1 1 1 1 0 0 0 1 1 0
## Lec.cornuta 0 0 0 1 0 1 1 0 0 0 0 0
## Lec.curvicornis 0 1 1 0 0 0 0 0 0 0 0 0
## Notholca.sp 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.urceolaris 0 0 0 0 0 0 0 0 0 0 0 0
## Trichocerca.sp 0 0 0 0 0 0 0 0 0 0 0 1
## Lec.quadridentata 0 0 0 0 0 0 0 0 0 0 0 1
## Lec.kluchor 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.lunaris 0 1 0 0 0 0 1 1 0 0 0 1
## Rotaria.sp 1 1 1 0 0 1 1 1 1 1 1 1
## Aspelta.sp 0 0 0 0 0 0 0 0 0 0 0 0
## Lec.furcata 0 0 1 0 0 1 0 0 0 1 0 0
## Col.geophila 0 0 0 0 0 0 0 0 0 0 0 0
## Lep.dactyliseta 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.calyciflorus 0 0 0 0 0 0 0 0 0 0 0 0
## Bra.caudatus 0 0 0 0 0 0 0 0 0 0 0 0
## [ reached getOption("max.print") -- omitted 65 rows ]
## attr(,"decostand")
## [1] "pa"
## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 4 18 2 6 40 12
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 3 1 3 6 5 4
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 16 27 5 5 2 3
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 1 1 4 4 3 1
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 2 6 1 24 3 3
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 5 3 8 44 6 11
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 1 23 1 1 6 1
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 1 1 2 3 1 2
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 3 1 2 8 1 3
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 3 4 1 1 2 1
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 1 2 4 1 2 1
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 3 1 2 1 1 1
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 2 1 1 2 2 2
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 1 1 1 1 1 1
## Dun.odontoplax
## 1
## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 4 18 2 6 40 12
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 3 1 3 6 5 4
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 16 27 5 5 2 3
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 1 1 4 4 3 1
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 2 6 1 24 3 3
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 5 3 8 44 6 11
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 1 23 1 1 6 1
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 1 1 2 3 1 2
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 3 1 2 8 1 3
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 3 4 1 1 2 1
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 1 2 4 1 2 1
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 3 1 2 1 1 1
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 2 1 1 2 2 2
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 1 1 1 1 1 1
## Dun.odontoplax
## 1
## [1] 493.6 Índices de Diversidade
3.6.1 Shannon
O índice de diversidade de Shannon ou Shannon–Weaver (or Shannon–Wiener) é uma medida de diversidade que leva em consideração tanto a riqueza de espécies quanto a uniformidade na distribuição dessas espécies (Dixon 2003; Oksanen et al. 2017). Este índice é definido por:
\[ H' = -\sum_{i} p_i\log_{b} p_i \] Onde:
- \(H'\) é a entropia de Shannon.
- \(p_i\) é a abundância proporcional da espécie \(i\).
- \(b\) é a base do logaritmo.
É mais comum usar-se o logarítmo natural, embora pode-se argumentar que para o logarítmo de base = 2 (o que faz sentido mas nenhuma diferença)
Outra fórmula para calcular o índice de diversidade de Shannon é:
\[ H' = -\sum_{i=1}^{S} \left( \frac{n_i}{N} \times \ln\frac{n_i}{N} \right) \]
Onde:
- \(H'\) é a entropia de Shannon (ou a diversidade de Shannon).
- \(n_i\) é o número de indivíduos da espécie \(i\).
- \(N\) é o número total de indivíduos na comunidade.
- \(S\) é o número total de espécies na comunidade.
Esta fórmula mede a incerteza na identificação de uma espécie selecionada aleatoriamente da comunidade. Quanto maior o valor de \(H'\), maior a diversidade da comunidade.
H <- diversity(m_trab, index = "shannon")
H## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 1.3138340 1.9040851 0.5623351 1.5144760 2.0055324 1.8873527
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.8628578 0.0000000 1.0986123 1.1752126 1.4648164 1.2770343
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 2.4132557 2.2565973 1.3896812 0.8316999 0.6931472 0.5626347
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.0000000 0.0000000 1.3107837 0.4709001 1.0986123 0.0000000
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.5623351 1.5267750 0.0000000 2.5728447 1.0397208 1.0114043
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 1.4270610 0.6001661 1.6576082 2.2897987 1.7328680 2.0941190
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.0000000 2.5817736 0.0000000 0.0000000 1.6977336 0.0000000
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.0000000 0.0000000 0.3250830 0.7902680 0.0000000 0.6931472
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.2733743 0.0000000 0.5623351 0.7815039 0.0000000 0.2164141
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.6001661 1.3138340 0.0000000 0.0000000 0.6931472 0.0000000
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.0000000 0.6365142 1.2406843 0.0000000 0.6931472 0.0000000
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.9556999 0.0000000 0.5623351 0.0000000 0.0000000 0.0000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 0.6897345 0.0000000 0.0000000 0.5004024 0.6931472 0.6730117
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Dun.odontoplax
## 0.00000003.6.2 Simpson
O índice de diversidade de Simpson é uma medida de diversidade que leva em consideração a riqueza de espécies e a abundância relativa de cada espécie em uma comunidade. A fórmula para calcular o índice de diversidade de Simpson é:
\[ D = 1 - \sum_{i=1}^{S} \left( \frac{n_i (n_i - 1)}{N (N - 1)} \right) \]
Onde:
- \(D\) é o índice de diversidade de Simpson.
- \(n_i\) é o número de indivíduos da espécie \(i\).
- \(N\) é o número total de indivíduos na comunidade.
- \(S\) é o número total de espécies na comunidade.
Esta fórmula fornece um valor entre 0 e 1, onde 0 indica uma comunidade com apenas uma espécie presente e 1 indica uma comunidade com uma distribuição uniforme de espécies.
Existem duas variantes do índice de Simpson baseado em \(D = \sum p_i^2\) (Dixon 2003; Oksanen et al. 2017; Hurlbert 1971):
- Ao escolhermos
simpsono R retorna \(1 - D\), e - ao escolhermos
invsimpsono R retorna \(1/D\) .
D <- diversity(m_trab, "simpson")
D## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.72000000 0.68595041 0.37500000 0.73140496 0.65243383 0.74609375
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.54847645 0.00000000 0.66666667 0.60459184 0.74074074 0.69387755
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0.87848319 0.83183764 0.71074380 0.40282006 0.50000000 0.31555880
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.00000000 0.00000000 0.71604938 0.20576132 0.66666667 0.00000000
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.37500000 0.73966942 0.00000000 0.89389147 0.62500000 0.61111111
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.71604938 0.31404959 0.75445816 0.82589714 0.81250000 0.85260771
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.00000000 0.89388730 0.00000000 0.00000000 0.80473373 0.00000000
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.00000000 0.00000000 0.18000000 0.46153846 0.00000000 0.50000000
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.12927467 0.00000000 0.37500000 0.31925441 0.00000000 0.08780992
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.31404959 0.72000000 0.00000000 0.00000000 0.50000000 0.00000000
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.00000000 0.44444444 0.67768595 0.00000000 0.50000000 0.00000000
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.57142857 0.00000000 0.37500000 0.00000000 0.00000000 0.00000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 0.49659120 0.00000000 0.00000000 0.32000000 0.50000000 0.48000000
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.00000000 0.00000000 0.00000000 0.00000000 0.00000000 0.00000000
## Dun.odontoplax
## 0.00000000D[is.na(D)] <- 0 #substitui NA ou NaN por 0
D## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.72000000 0.68595041 0.37500000 0.73140496 0.65243383 0.74609375
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.54847645 0.00000000 0.66666667 0.60459184 0.74074074 0.69387755
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0.87848319 0.83183764 0.71074380 0.40282006 0.50000000 0.31555880
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.00000000 0.00000000 0.71604938 0.20576132 0.66666667 0.00000000
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.37500000 0.73966942 0.00000000 0.89389147 0.62500000 0.61111111
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.71604938 0.31404959 0.75445816 0.82589714 0.81250000 0.85260771
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.00000000 0.89388730 0.00000000 0.00000000 0.80473373 0.00000000
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.00000000 0.00000000 0.18000000 0.46153846 0.00000000 0.50000000
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.12927467 0.00000000 0.37500000 0.31925441 0.00000000 0.08780992
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.31404959 0.72000000 0.00000000 0.00000000 0.50000000 0.00000000
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.00000000 0.44444444 0.67768595 0.00000000 0.50000000 0.00000000
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.57142857 0.00000000 0.37500000 0.00000000 0.00000000 0.00000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 0.49659120 0.00000000 0.00000000 0.32000000 0.50000000 0.48000000
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.00000000 0.00000000 0.00000000 0.00000000 0.00000000 0.00000000
## Dun.odontoplax
## 0.000000003.6.3 Equitabilidade de Pielou
O índice de equitabilidade de Pielou é uma medida de uniformidade em uma distribuição de espécies (Pielou 1975). Sua fórmula é dada por:
\[ J' = \frac{H'}{\ln(S)} \]
Onde:
- \(J'\) é o índice de equitabilidade de Pielou.
- \(H'\) é a entropia de Shannon (ou a diversidade de Shannon), que é calculada como \(-\sum_{i=1}^{S} p_i \cdot \ln(p_i)\), onde \(S\) é o número total de espécies e \(p_i\) é a proporção da espécie \(i\).
- \(\ln(S)\) é o logarítmo natural do número total de espécies na comunidade.
Esta fórmula fornece um valor entre 0 e 1, onde 0 indica uma distribuição completamente desigual onde uma comunidade apresenta uma única espécie dominante e 1 indica uma distribuição completamente equitativa onde todas as espécies têm a mesma abundância.
E <- H/log(specnumber(m_trab))
E## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.9477309 0.6587682 0.8112781 0.8452452 0.5436698 0.7595266
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.7854070 NaN 1.0000000 0.6558986 0.9101416 0.9211855
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0.8703980 0.6846811 0.8634575 0.5167642 1.0000000 0.5121322
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## NaN NaN 0.9455306 0.3396826 1.0000000 NaN
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.8112781 0.8521093 NaN 0.8095661 0.9463946 0.9206198
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.8866829 0.5462947 0.7971410 0.6050962 0.9671320 0.8733155
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## NaN 0.8234024 NaN NaN 0.9475232 NaN
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## NaN NaN 0.4689956 0.7193329 NaN 1.0000000
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.2488360 NaN 0.8112781 0.3758239 NaN 0.1969886
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.5462947 0.9477309 NaN NaN 1.0000000 NaN
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## NaN 0.9182958 0.8949645 NaN 1.0000000 NaN
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.8699155 NaN 0.8112781 NaN NaN NaN
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 0.9950765 NaN NaN 0.7219281 1.0000000 0.9709506
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## NaN NaN NaN NaN NaN NaN
## Dun.odontoplax
## NaNE[is.na(E)] <- 0 #substitui NA ou NaN por 0
E## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.9477309 0.6587682 0.8112781 0.8452452 0.5436698 0.7595266
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.7854070 0.0000000 1.0000000 0.6558986 0.9101416 0.9211855
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 0.8703980 0.6846811 0.8634575 0.5167642 1.0000000 0.5121322
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.0000000 0.0000000 0.9455306 0.3396826 1.0000000 0.0000000
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.8112781 0.8521093 0.0000000 0.8095661 0.9463946 0.9206198
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.8866829 0.5462947 0.7971410 0.6050962 0.9671320 0.8733155
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.0000000 0.8234024 0.0000000 0.0000000 0.9475232 0.0000000
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.0000000 0.0000000 0.4689956 0.7193329 0.0000000 1.0000000
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.2488360 0.0000000 0.8112781 0.3758239 0.0000000 0.1969886
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.5462947 0.9477309 0.0000000 0.0000000 1.0000000 0.0000000
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.0000000 0.9182958 0.8949645 0.0000000 1.0000000 0.0000000
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.8699155 0.0000000 0.8112781 0.0000000 0.0000000 0.0000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 0.9950765 0.0000000 0.0000000 0.7219281 1.0000000 0.9709506
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Dun.odontoplax
## 0.00000003.6.3.1 Assimetria e curtose
library(moments)
Assimetria <- apply(m_trab,1,skewness)
Assimetria## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 3.476655 6.363013 5.957144 4.273584 6.461601 5.645693
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 4.597181 6.783866 3.660403 4.991817 3.786828 4.123459
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 3.413726 4.149242 4.094856 6.461638 4.641396 6.419702
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 6.783866 6.783866 3.614935 6.732329 3.660403 6.783866
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 5.957144 4.190788 6.783866 2.978950 4.446423 4.496792
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 4.466614 6.549959 4.053351 3.797043 2.912236 2.816981
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 6.783866 2.886668 6.783866 6.783866 2.900572 6.783866
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 6.783866 6.783866 6.663554 5.865222 6.783866 4.641396
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 6.738488 6.783866 5.957144 6.693569 6.783866 6.772263
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 6.549959 3.476655 6.783866 6.783866 4.641396 6.783866
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 6.783866 5.366983 4.306765 6.783866 4.641396 6.783866
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 5.031153 6.783866 5.957144 6.783866 6.783866 6.783866
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 4.694623 6.783866 6.783866 6.261844 4.641396 4.935917
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 6.783866 6.783866 6.783866 6.783866 6.783866 6.783866
## Dun.odontoplax
## 6.783866Curtose <- apply(m_trab,1,kurtosis)
Curtose## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 13.364950 43.193400 38.356754 21.445502 43.934851 36.397244
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 22.260592 47.020833 14.398551 27.433083 16.652473 19.908163
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 16.091866 21.721303 18.872150 43.964610 22.542553 43.417838
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 47.020833 47.020833 14.753227 46.553383 14.398551 47.020833
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 38.356754 21.301967 47.020833 12.576162 22.785803 22.720535
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 24.127985 44.815176 18.620513 17.005265 10.606273 10.016103
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 47.020833 11.289662 47.020833 47.020833 9.967459 47.020833
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 47.020833 47.020833 45.879552 37.571252 47.020833 22.542553
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 46.601720 47.020833 38.356754 46.197490 47.020833 46.916558
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 44.815176 13.364950 47.020833 47.020833 22.542553 47.020833
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 47.020833 31.487216 21.658190 47.020833 22.542553 47.020833
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 28.640000 47.020833 38.356754 47.020833 47.020833 47.020833
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 23.214798 47.020833 47.020833 41.720125 22.542553 26.231706
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 47.020833 47.020833 47.020833 47.020833 47.020833 47.020833
## Dun.odontoplax
## 47.0208333.7 Descritores da estrutura da comunidade
Descritores1 <- cbind(Sum, Mean, DP, Max, Min, S, E, H, D)
Descritores1 <- as.data.frame(Descritores1)
Descritores1
#Descritores1 <- Descritores1 %>% rownames_to_column(var="Espécies") #da nome a primeira coluna
SomaTotalD <- apply(Descritores1,2,sum)
SomaTotalD
MediaTotalD <- apply(Descritores1,2,mean)
MediaTotalD
DPTotalD <- apply(Descritores1,2,sd)
DPTotalD
Descritores2 <- cbind(SomaTotalD, MediaTotalD, DPTotalD)
Descritores2 <- as.data.frame(Descritores2)
Descritores2 <- t(Descritores2)
Descritores2
DescritoresFinal <- rbind(Descritores1, Descritores2)
DescritoresFinal
DescritoresFinal <- round (DescritoresFinal, 2)
DescritoresFinal## Sum Mean DP Max Min S E H D
## Bra.angularis 1.7777778 0.036281179 0.13073816 0.5333333 0 4 0.9477309 1.3138340 0.72000000
## Lepadella.sp 11.7333333 0.239455782 0.91771842 6.4000000 0 18 0.6587682 1.9040851 0.68595041
## Lecane.sp 0.7111111 0.014512472 0.07980829 0.5333333 0 2 0.8112781 0.5623351 0.37500000
## Lec.leontina 3.9111111 0.079818594 0.28123472 1.6000000 0 6 0.8452452 1.5144760 0.73140496
## Lec.bulla 124.8000000 2.546938776 10.30321459 72.0000000 0 40 0.5436698 2.0055324 0.65243383
## Lec.cornuta 5.6888889 0.116099773 0.39677883 2.6666667 0 12 0.7595266 1.8873527 0.74609375
## Lec.curvicornis 3.3777778 0.068934240 0.32011588 1.6000000 0 3 0.7854070 0.8628578 0.54847645
## Notholca.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Bra.urceolaris 0.5333333 0.010884354 0.04306241 0.1777778 0 3 1.0000000 1.0986123 0.66666667
## Trichocerca.sp 4.9777778 0.101587302 0.43997755 2.6666667 0 6 0.6558986 1.1752126 0.60459184
## Lec.quadridentata 1.6000000 0.032653061 0.11286596 0.5333333 0 5 0.9101416 1.4648164 0.74074074
## Lec.kluchor 1.2444444 0.025396825 0.09601097 0.5333333 0 4 0.9211855 1.2770343 0.69387755
## Lec.lunaris 10.4888889 0.214058957 0.48139659 2.6666667 0 16 0.8703980 2.4132557 0.87848319
## Rotaria.sp 74.3111111 1.516553288 4.12290234 24.5333333 0 27 0.6846811 2.2565973 0.83183764
## Aspelta.sp 1.9555556 0.039909297 0.14635345 0.7111111 0 5 0.8634575 1.3896812 0.71074380
## Lec.furcata 14.0444444 0.286621315 1.53952216 10.6666667 0 5 0.5167642 0.8316999 0.40282006
## Col.geophila 0.3555556 0.007256236 0.03554044 0.1777778 0 2 1.0000000 0.6931472 0.50000000
## Lep.dactyliseta 6.5777778 0.134240363 0.77366584 5.3333333 0 3 0.5121322 0.5626347 0.31555880
## Bra.calyciflorus 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Bra.caudatus 0.3555556 0.007256236 0.05079365 0.3555556 0 1 0.0000000 0.0000000 0.00000000
## Lec.crepida 1.6000000 0.032653061 0.11855631 0.5333333 0 4 0.9455306 1.3107837 0.71604938
## Pla.patulus 4.8000000 0.097959184 0.60945767 4.2666667 0 4 0.3396826 0.4709001 0.20576132
## Lec.ovalis 0.5333333 0.010884354 0.04306241 0.1777778 0 3 1.0000000 1.0986123 0.66666667
## Lec.elasma 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Pla.quadricornis 0.7111111 0.014512472 0.07980829 0.5333333 0 2 0.8112781 0.5623351 0.37500000
## Lec.aculeata 3.9111111 0.079818594 0.27651261 1.6000000 0 6 0.8521093 1.5267750 0.73966942
## Ker.tropica 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Bdelloidea 60.5333333 1.235374150 2.55779257 13.3333333 0 24 0.8095661 2.5728447 0.89389147
## Ker.lenzi 0.7111111 0.014512472 0.06111970 0.3555556 0 3 0.9463946 1.0397208 0.62500000
## Con.unicornis 1.0666667 0.021768707 0.09345779 0.5333333 0 3 0.9206198 1.0114043 0.61111111
## Alo.dadayi 1.6000000 0.032653061 0.11855631 0.7111111 0 5 0.8866829 1.4270610 0.71604938
## Macrothrix.sp 1.9555556 0.039909297 0.23027000 1.6000000 0 3 0.5462947 0.6001661 0.31404959
## Alonella.sp 4.8000000 0.097959184 0.32873114 1.6000000 0 8 0.7971410 1.6576082 0.75445816
## Nauplii 234.0444444 4.776417234 13.24363377 70.9333333 0 44 0.6050962 2.2897987 0.82589714
## Chy.eurynotus 1.4222222 0.029024943 0.08391207 0.3555556 0 6 0.9671320 1.7328680 0.81250000
## Cyclopoida 7.4666667 0.152380952 0.38404389 1.6000000 0 11 0.8733155 2.0941190 0.85260771
## Mac.collinsi 0.3555556 0.007256236 0.05079365 0.3555556 0 1 0.0000000 0.0000000 0.00000000
## Copepodite 23.2888889 0.475283447 0.98407919 4.8000000 0 23 0.8234024 2.5817736 0.89388730
## Paracyclops.sp 0.3555556 0.007256236 0.05079365 0.3555556 0 1 0.0000000 0.0000000 0.00000000
## Asc.ecaudis 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Lep.ovalis 2.3111111 0.047165533 0.13949001 0.5333333 0 6 0.9475232 1.6977336 0.80473373
## Polyarthra.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Bra.havanaensis 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Hexarthra.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Lec.hastata 1.7777778 0.036281179 0.22945157 1.6000000 0 2 0.4689956 0.3250830 0.18000000
## Tri.tetractis 2.3111111 0.047165533 0.24009740 1.6000000 0 3 0.7193329 0.7902680 0.46153846
## Ker.serrulata 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Notodiaptomus.sp 0.3555556 0.007256236 0.03554044 0.1777778 0 2 1.0000000 0.6931472 0.50000000
## Bra.falcatus 25.7777778 0.526077098 3.43095743 24.0000000 0 3 0.2488360 0.2733743 0.12927467
## Fil.longiseta 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Myt.crassipes 0.7111111 0.014512472 0.07980829 0.5333333 0 2 0.8112781 0.5623351 0.37500000
## Lep.patella 65.6000000 1.338775510 7.69424793 53.8666667 0 8 0.3758239 0.7815039 0.31925441
## Euchlanis.sp 2.6666667 0.054421769 0.38095238 2.6666667 0 1 0.0000000 0.0000000 0.00000000
## Colurella.sp 7.8222222 0.159637188 1.06620045 7.4666667 0 3 0.1969886 0.2164141 0.08780992
## Mytilina.sp 1.9555556 0.039909297 0.23027000 1.6000000 0 3 0.5462947 0.6001661 0.31404959
## Lec.monostyla 1.7777778 0.036281179 0.13073816 0.5333333 0 4 0.9477309 1.3138340 0.72000000
## Lec.ligona 1.6000000 0.032653061 0.22857143 1.6000000 0 1 0.0000000 0.0000000 0.00000000
## Ascomorpha.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Pom.sulcata 0.7111111 0.014512472 0.07108087 0.3555556 0 2 1.0000000 0.6931472 0.50000000
## Harpacticoida 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Epi.senta 0.1777778 0.003628118 0.02539683 0.1777778 0 1 0.0000000 0.0000000 0.00000000
## Rot.neptunia 1.6000000 0.032653061 0.16894139 1.0666667 0 2 0.9182958 0.6365142 0.44444444
## Dis.aculeata 5.8666667 0.119727891 0.46527164 2.6666667 0 4 0.8949645 1.2406843 0.67768595
## Lec.luna 2.6666667 0.054421769 0.38095238 2.6666667 0 1 0.0000000 0.0000000 0.00000000
## Euc.dilatata 1.0666667 0.021768707 0.10662131 0.5333333 0 2 1.0000000 0.6931472 0.50000000
## Bea.eudactylota 0.5333333 0.010884354 0.07619048 0.5333333 0 1 0.0000000 0.0000000 0.00000000
## Chy.sphaericus 3.7333333 0.076190476 0.34426519 2.1333333 0 3 0.8699155 0.9556999 0.57142857
## Lec.hornemanni 3.7333333 0.076190476 0.53333333 3.7333333 0 1 0.0000000 0.0000000 0.00000000
## Dia.birgei 2.1333333 0.043537415 0.23942486 1.6000000 0 2 0.8112781 0.5623351 0.37500000
## Mac.subquadratus 0.5333333 0.010884354 0.07619048 0.5333333 0 1 0.0000000 0.0000000 0.00000000
## Bra.quadridentatus 10.1333333 0.206802721 1.44761905 10.1333333 0 1 0.0000000 0.0000000 0.00000000
## Pol.vulgaris 19.2000000 0.391836735 2.74285714 19.2000000 0 1 0.0000000 0.0000000 0.00000000
## Pol.bicerca 58.1333333 1.186394558 5.83147557 31.4666667 0 2 0.9950765 0.6897345 0.49659120
## Dicranop.sp 68.8000000 1.404081633 9.82857143 68.8000000 0 1 0.0000000 0.0000000 0.00000000
## Asc.saltans 1.0666667 0.021768707 0.15238095 1.0666667 0 1 0.0000000 0.0000000 0.00000000
## Moi.minuta 2.6666667 0.054421769 0.31259768 2.1333333 0 2 0.7219281 0.5004024 0.32000000
## Alo.hamulata 1.0666667 0.021768707 0.10662131 0.5333333 0 2 1.0000000 0.6931472 0.50000000
## Mac.laticornis 2.6666667 0.054421769 0.27205442 1.6000000 0 2 0.9709506 0.6730117 0.48000000
## Mac.mira 2.1333333 0.043537415 0.30476190 2.1333333 0 1 0.0000000 0.0000000 0.00000000
## Pro.similis 0.5333333 0.010884354 0.07619048 0.5333333 0 1 0.0000000 0.0000000 0.00000000
## Alo.pulchella 3.7333333 0.076190476 0.53333333 3.7333333 0 1 0.0000000 0.0000000 0.00000000
## Dia.spinulosum 1.6000000 0.032653061 0.22857143 1.6000000 0 1 0.0000000 0.0000000 0.00000000
## Chydorus.sp 1.0666667 0.021768707 0.15238095 1.0666667 0 1 0.0000000 0.0000000 0.00000000
## Ily.spinifer 3.7333333 0.076190476 0.53333333 3.7333333 0 1 0.0000000 0.0000000 0.00000000
## Dun.odontoplax 0.5333333 0.010884354 0.07619048 0.5333333 0 1 0.0000000 0.0000000 0.00000000
## Sum Mean DP Max Min S E H D
## 934.22222 19.06576 78.86398 499.91111 0.00000 402.00000 40.85574 59.78162 29.56409
## Sum Mean DP Max Min S E H D
## 10.9908497 0.2243031 0.9278115 5.8813072 0.0000000 4.7294118 0.4806558 0.7033132 0.3478128
## Sum Mean DP Max Min S E H D
## 31.6800338 0.6465313 2.3220881 14.6991518 0.0000000 7.7265594 0.4178274 0.7474313 0.3230295
## Sum Mean DP Max Min S E H D
## SomaTotalD 934.22222 19.0657596 78.8639769 499.911111 0 402.000000 40.8557437 59.7816180 29.5640893
## MediaTotalD 10.99085 0.2243031 0.9278115 5.881307 0 4.729412 0.4806558 0.7033132 0.3478128
## DPTotalD 31.68003 0.6465313 2.3220881 14.699152 0 7.726559 0.4178274 0.7474313 0.3230295
## Sum Mean DP Max Min S E H D
## Bra.angularis 1.7777778 0.036281179 0.13073816 0.5333333 0 4.000000 0.9477309 1.3138340 0.72000000
## Lepadella.sp 11.7333333 0.239455782 0.91771842 6.4000000 0 18.000000 0.6587682 1.9040851 0.68595041
## Lecane.sp 0.7111111 0.014512472 0.07980829 0.5333333 0 2.000000 0.8112781 0.5623351 0.37500000
## Lec.leontina 3.9111111 0.079818594 0.28123472 1.6000000 0 6.000000 0.8452452 1.5144760 0.73140496
## Lec.bulla 124.8000000 2.546938776 10.30321459 72.0000000 0 40.000000 0.5436698 2.0055324 0.65243383
## Lec.cornuta 5.6888889 0.116099773 0.39677883 2.6666667 0 12.000000 0.7595266 1.8873527 0.74609375
## Lec.curvicornis 3.3777778 0.068934240 0.32011588 1.6000000 0 3.000000 0.7854070 0.8628578 0.54847645
## Notholca.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Bra.urceolaris 0.5333333 0.010884354 0.04306241 0.1777778 0 3.000000 1.0000000 1.0986123 0.66666667
## Trichocerca.sp 4.9777778 0.101587302 0.43997755 2.6666667 0 6.000000 0.6558986 1.1752126 0.60459184
## Lec.quadridentata 1.6000000 0.032653061 0.11286596 0.5333333 0 5.000000 0.9101416 1.4648164 0.74074074
## Lec.kluchor 1.2444444 0.025396825 0.09601097 0.5333333 0 4.000000 0.9211855 1.2770343 0.69387755
## Lec.lunaris 10.4888889 0.214058957 0.48139659 2.6666667 0 16.000000 0.8703980 2.4132557 0.87848319
## Rotaria.sp 74.3111111 1.516553288 4.12290234 24.5333333 0 27.000000 0.6846811 2.2565973 0.83183764
## Aspelta.sp 1.9555556 0.039909297 0.14635345 0.7111111 0 5.000000 0.8634575 1.3896812 0.71074380
## Lec.furcata 14.0444444 0.286621315 1.53952216 10.6666667 0 5.000000 0.5167642 0.8316999 0.40282006
## Col.geophila 0.3555556 0.007256236 0.03554044 0.1777778 0 2.000000 1.0000000 0.6931472 0.50000000
## Lep.dactyliseta 6.5777778 0.134240363 0.77366584 5.3333333 0 3.000000 0.5121322 0.5626347 0.31555880
## Bra.calyciflorus 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Bra.caudatus 0.3555556 0.007256236 0.05079365 0.3555556 0 1.000000 0.0000000 0.0000000 0.00000000
## Lec.crepida 1.6000000 0.032653061 0.11855631 0.5333333 0 4.000000 0.9455306 1.3107837 0.71604938
## Pla.patulus 4.8000000 0.097959184 0.60945767 4.2666667 0 4.000000 0.3396826 0.4709001 0.20576132
## Lec.ovalis 0.5333333 0.010884354 0.04306241 0.1777778 0 3.000000 1.0000000 1.0986123 0.66666667
## Lec.elasma 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Pla.quadricornis 0.7111111 0.014512472 0.07980829 0.5333333 0 2.000000 0.8112781 0.5623351 0.37500000
## Lec.aculeata 3.9111111 0.079818594 0.27651261 1.6000000 0 6.000000 0.8521093 1.5267750 0.73966942
## Ker.tropica 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Bdelloidea 60.5333333 1.235374150 2.55779257 13.3333333 0 24.000000 0.8095661 2.5728447 0.89389147
## Ker.lenzi 0.7111111 0.014512472 0.06111970 0.3555556 0 3.000000 0.9463946 1.0397208 0.62500000
## Con.unicornis 1.0666667 0.021768707 0.09345779 0.5333333 0 3.000000 0.9206198 1.0114043 0.61111111
## Alo.dadayi 1.6000000 0.032653061 0.11855631 0.7111111 0 5.000000 0.8866829 1.4270610 0.71604938
## Macrothrix.sp 1.9555556 0.039909297 0.23027000 1.6000000 0 3.000000 0.5462947 0.6001661 0.31404959
## Alonella.sp 4.8000000 0.097959184 0.32873114 1.6000000 0 8.000000 0.7971410 1.6576082 0.75445816
## Nauplii 234.0444444 4.776417234 13.24363377 70.9333333 0 44.000000 0.6050962 2.2897987 0.82589714
## Chy.eurynotus 1.4222222 0.029024943 0.08391207 0.3555556 0 6.000000 0.9671320 1.7328680 0.81250000
## Cyclopoida 7.4666667 0.152380952 0.38404389 1.6000000 0 11.000000 0.8733155 2.0941190 0.85260771
## Mac.collinsi 0.3555556 0.007256236 0.05079365 0.3555556 0 1.000000 0.0000000 0.0000000 0.00000000
## Copepodite 23.2888889 0.475283447 0.98407919 4.8000000 0 23.000000 0.8234024 2.5817736 0.89388730
## Paracyclops.sp 0.3555556 0.007256236 0.05079365 0.3555556 0 1.000000 0.0000000 0.0000000 0.00000000
## Asc.ecaudis 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Lep.ovalis 2.3111111 0.047165533 0.13949001 0.5333333 0 6.000000 0.9475232 1.6977336 0.80473373
## Polyarthra.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Bra.havanaensis 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Hexarthra.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Lec.hastata 1.7777778 0.036281179 0.22945157 1.6000000 0 2.000000 0.4689956 0.3250830 0.18000000
## Tri.tetractis 2.3111111 0.047165533 0.24009740 1.6000000 0 3.000000 0.7193329 0.7902680 0.46153846
## Ker.serrulata 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Notodiaptomus.sp 0.3555556 0.007256236 0.03554044 0.1777778 0 2.000000 1.0000000 0.6931472 0.50000000
## Bra.falcatus 25.7777778 0.526077098 3.43095743 24.0000000 0 3.000000 0.2488360 0.2733743 0.12927467
## Fil.longiseta 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Myt.crassipes 0.7111111 0.014512472 0.07980829 0.5333333 0 2.000000 0.8112781 0.5623351 0.37500000
## Lep.patella 65.6000000 1.338775510 7.69424793 53.8666667 0 8.000000 0.3758239 0.7815039 0.31925441
## Euchlanis.sp 2.6666667 0.054421769 0.38095238 2.6666667 0 1.000000 0.0000000 0.0000000 0.00000000
## Colurella.sp 7.8222222 0.159637188 1.06620045 7.4666667 0 3.000000 0.1969886 0.2164141 0.08780992
## Mytilina.sp 1.9555556 0.039909297 0.23027000 1.6000000 0 3.000000 0.5462947 0.6001661 0.31404959
## Lec.monostyla 1.7777778 0.036281179 0.13073816 0.5333333 0 4.000000 0.9477309 1.3138340 0.72000000
## Lec.ligona 1.6000000 0.032653061 0.22857143 1.6000000 0 1.000000 0.0000000 0.0000000 0.00000000
## Ascomorpha.sp 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Pom.sulcata 0.7111111 0.014512472 0.07108087 0.3555556 0 2.000000 1.0000000 0.6931472 0.50000000
## Harpacticoida 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Epi.senta 0.1777778 0.003628118 0.02539683 0.1777778 0 1.000000 0.0000000 0.0000000 0.00000000
## Rot.neptunia 1.6000000 0.032653061 0.16894139 1.0666667 0 2.000000 0.9182958 0.6365142 0.44444444
## Dis.aculeata 5.8666667 0.119727891 0.46527164 2.6666667 0 4.000000 0.8949645 1.2406843 0.67768595
## Lec.luna 2.6666667 0.054421769 0.38095238 2.6666667 0 1.000000 0.0000000 0.0000000 0.00000000
## Euc.dilatata 1.0666667 0.021768707 0.10662131 0.5333333 0 2.000000 1.0000000 0.6931472 0.50000000
## Bea.eudactylota 0.5333333 0.010884354 0.07619048 0.5333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Chy.sphaericus 3.7333333 0.076190476 0.34426519 2.1333333 0 3.000000 0.8699155 0.9556999 0.57142857
## Lec.hornemanni 3.7333333 0.076190476 0.53333333 3.7333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Dia.birgei 2.1333333 0.043537415 0.23942486 1.6000000 0 2.000000 0.8112781 0.5623351 0.37500000
## Mac.subquadratus 0.5333333 0.010884354 0.07619048 0.5333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Bra.quadridentatus 10.1333333 0.206802721 1.44761905 10.1333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Pol.vulgaris 19.2000000 0.391836735 2.74285714 19.2000000 0 1.000000 0.0000000 0.0000000 0.00000000
## Pol.bicerca 58.1333333 1.186394558 5.83147557 31.4666667 0 2.000000 0.9950765 0.6897345 0.49659120
## Dicranop.sp 68.8000000 1.404081633 9.82857143 68.8000000 0 1.000000 0.0000000 0.0000000 0.00000000
## Asc.saltans 1.0666667 0.021768707 0.15238095 1.0666667 0 1.000000 0.0000000 0.0000000 0.00000000
## Moi.minuta 2.6666667 0.054421769 0.31259768 2.1333333 0 2.000000 0.7219281 0.5004024 0.32000000
## Alo.hamulata 1.0666667 0.021768707 0.10662131 0.5333333 0 2.000000 1.0000000 0.6931472 0.50000000
## Mac.laticornis 2.6666667 0.054421769 0.27205442 1.6000000 0 2.000000 0.9709506 0.6730117 0.48000000
## Mac.mira 2.1333333 0.043537415 0.30476190 2.1333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Pro.similis 0.5333333 0.010884354 0.07619048 0.5333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Alo.pulchella 3.7333333 0.076190476 0.53333333 3.7333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Dia.spinulosum 1.6000000 0.032653061 0.22857143 1.6000000 0 1.000000 0.0000000 0.0000000 0.00000000
## Chydorus.sp 1.0666667 0.021768707 0.15238095 1.0666667 0 1.000000 0.0000000 0.0000000 0.00000000
## Ily.spinifer 3.7333333 0.076190476 0.53333333 3.7333333 0 1.000000 0.0000000 0.0000000 0.00000000
## Dun.odontoplax 0.5333333 0.010884354 0.07619048 0.5333333 0 1.000000 0.0000000 0.0000000 0.00000000
## SomaTotalD 934.2222222 19.065759637 78.86397687 499.9111111 0 402.000000 40.8557437 59.7816180 29.56408929
## MediaTotalD 10.9908497 0.224303055 0.92781149 5.8813072 0 4.729412 0.4806558 0.7033132 0.34781282
## DPTotalD 31.6800338 0.646531302 2.32208806 14.6991518 0 7.726559 0.4178274 0.7474313 0.32302953
## Sum Mean DP Max Min S E H D
## Bra.angularis 1.78 0.04 0.13 0.53 0 4.00 0.95 1.31 0.72
## Lepadella.sp 11.73 0.24 0.92 6.40 0 18.00 0.66 1.90 0.69
## Lecane.sp 0.71 0.01 0.08 0.53 0 2.00 0.81 0.56 0.38
## Lec.leontina 3.91 0.08 0.28 1.60 0 6.00 0.85 1.51 0.73
## Lec.bulla 124.80 2.55 10.30 72.00 0 40.00 0.54 2.01 0.65
## Lec.cornuta 5.69 0.12 0.40 2.67 0 12.00 0.76 1.89 0.75
## Lec.curvicornis 3.38 0.07 0.32 1.60 0 3.00 0.79 0.86 0.55
## Notholca.sp 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Bra.urceolaris 0.53 0.01 0.04 0.18 0 3.00 1.00 1.10 0.67
## Trichocerca.sp 4.98 0.10 0.44 2.67 0 6.00 0.66 1.18 0.60
## Lec.quadridentata 1.60 0.03 0.11 0.53 0 5.00 0.91 1.46 0.74
## Lec.kluchor 1.24 0.03 0.10 0.53 0 4.00 0.92 1.28 0.69
## Lec.lunaris 10.49 0.21 0.48 2.67 0 16.00 0.87 2.41 0.88
## Rotaria.sp 74.31 1.52 4.12 24.53 0 27.00 0.68 2.26 0.83
## Aspelta.sp 1.96 0.04 0.15 0.71 0 5.00 0.86 1.39 0.71
## Lec.furcata 14.04 0.29 1.54 10.67 0 5.00 0.52 0.83 0.40
## Col.geophila 0.36 0.01 0.04 0.18 0 2.00 1.00 0.69 0.50
## Lep.dactyliseta 6.58 0.13 0.77 5.33 0 3.00 0.51 0.56 0.32
## Bra.calyciflorus 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Bra.caudatus 0.36 0.01 0.05 0.36 0 1.00 0.00 0.00 0.00
## Lec.crepida 1.60 0.03 0.12 0.53 0 4.00 0.95 1.31 0.72
## Pla.patulus 4.80 0.10 0.61 4.27 0 4.00 0.34 0.47 0.21
## Lec.ovalis 0.53 0.01 0.04 0.18 0 3.00 1.00 1.10 0.67
## Lec.elasma 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Pla.quadricornis 0.71 0.01 0.08 0.53 0 2.00 0.81 0.56 0.38
## Lec.aculeata 3.91 0.08 0.28 1.60 0 6.00 0.85 1.53 0.74
## Ker.tropica 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Bdelloidea 60.53 1.24 2.56 13.33 0 24.00 0.81 2.57 0.89
## Ker.lenzi 0.71 0.01 0.06 0.36 0 3.00 0.95 1.04 0.62
## Con.unicornis 1.07 0.02 0.09 0.53 0 3.00 0.92 1.01 0.61
## Alo.dadayi 1.60 0.03 0.12 0.71 0 5.00 0.89 1.43 0.72
## Macrothrix.sp 1.96 0.04 0.23 1.60 0 3.00 0.55 0.60 0.31
## Alonella.sp 4.80 0.10 0.33 1.60 0 8.00 0.80 1.66 0.75
## Nauplii 234.04 4.78 13.24 70.93 0 44.00 0.61 2.29 0.83
## Chy.eurynotus 1.42 0.03 0.08 0.36 0 6.00 0.97 1.73 0.81
## Cyclopoida 7.47 0.15 0.38 1.60 0 11.00 0.87 2.09 0.85
## Mac.collinsi 0.36 0.01 0.05 0.36 0 1.00 0.00 0.00 0.00
## Copepodite 23.29 0.48 0.98 4.80 0 23.00 0.82 2.58 0.89
## Paracyclops.sp 0.36 0.01 0.05 0.36 0 1.00 0.00 0.00 0.00
## Asc.ecaudis 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Lep.ovalis 2.31 0.05 0.14 0.53 0 6.00 0.95 1.70 0.80
## Polyarthra.sp 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Bra.havanaensis 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Hexarthra.sp 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Lec.hastata 1.78 0.04 0.23 1.60 0 2.00 0.47 0.33 0.18
## Tri.tetractis 2.31 0.05 0.24 1.60 0 3.00 0.72 0.79 0.46
## Ker.serrulata 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Notodiaptomus.sp 0.36 0.01 0.04 0.18 0 2.00 1.00 0.69 0.50
## Bra.falcatus 25.78 0.53 3.43 24.00 0 3.00 0.25 0.27 0.13
## Fil.longiseta 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Myt.crassipes 0.71 0.01 0.08 0.53 0 2.00 0.81 0.56 0.38
## Lep.patella 65.60 1.34 7.69 53.87 0 8.00 0.38 0.78 0.32
## Euchlanis.sp 2.67 0.05 0.38 2.67 0 1.00 0.00 0.00 0.00
## Colurella.sp 7.82 0.16 1.07 7.47 0 3.00 0.20 0.22 0.09
## Mytilina.sp 1.96 0.04 0.23 1.60 0 3.00 0.55 0.60 0.31
## Lec.monostyla 1.78 0.04 0.13 0.53 0 4.00 0.95 1.31 0.72
## Lec.ligona 1.60 0.03 0.23 1.60 0 1.00 0.00 0.00 0.00
## Ascomorpha.sp 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Pom.sulcata 0.71 0.01 0.07 0.36 0 2.00 1.00 0.69 0.50
## Harpacticoida 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Epi.senta 0.18 0.00 0.03 0.18 0 1.00 0.00 0.00 0.00
## Rot.neptunia 1.60 0.03 0.17 1.07 0 2.00 0.92 0.64 0.44
## Dis.aculeata 5.87 0.12 0.47 2.67 0 4.00 0.89 1.24 0.68
## Lec.luna 2.67 0.05 0.38 2.67 0 1.00 0.00 0.00 0.00
## Euc.dilatata 1.07 0.02 0.11 0.53 0 2.00 1.00 0.69 0.50
## Bea.eudactylota 0.53 0.01 0.08 0.53 0 1.00 0.00 0.00 0.00
## Chy.sphaericus 3.73 0.08 0.34 2.13 0 3.00 0.87 0.96 0.57
## Lec.hornemanni 3.73 0.08 0.53 3.73 0 1.00 0.00 0.00 0.00
## Dia.birgei 2.13 0.04 0.24 1.60 0 2.00 0.81 0.56 0.37
## Mac.subquadratus 0.53 0.01 0.08 0.53 0 1.00 0.00 0.00 0.00
## Bra.quadridentatus 10.13 0.21 1.45 10.13 0 1.00 0.00 0.00 0.00
## Pol.vulgaris 19.20 0.39 2.74 19.20 0 1.00 0.00 0.00 0.00
## Pol.bicerca 58.13 1.19 5.83 31.47 0 2.00 1.00 0.69 0.50
## Dicranop.sp 68.80 1.40 9.83 68.80 0 1.00 0.00 0.00 0.00
## Asc.saltans 1.07 0.02 0.15 1.07 0 1.00 0.00 0.00 0.00
## Moi.minuta 2.67 0.05 0.31 2.13 0 2.00 0.72 0.50 0.32
## Alo.hamulata 1.07 0.02 0.11 0.53 0 2.00 1.00 0.69 0.50
## Mac.laticornis 2.67 0.05 0.27 1.60 0 2.00 0.97 0.67 0.48
## Mac.mira 2.13 0.04 0.30 2.13 0 1.00 0.00 0.00 0.00
## Pro.similis 0.53 0.01 0.08 0.53 0 1.00 0.00 0.00 0.00
## Alo.pulchella 3.73 0.08 0.53 3.73 0 1.00 0.00 0.00 0.00
## Dia.spinulosum 1.60 0.03 0.23 1.60 0 1.00 0.00 0.00 0.00
## Chydorus.sp 1.07 0.02 0.15 1.07 0 1.00 0.00 0.00 0.00
## Ily.spinifer 3.73 0.08 0.53 3.73 0 1.00 0.00 0.00 0.00
## Dun.odontoplax 0.53 0.01 0.08 0.53 0 1.00 0.00 0.00 0.00
## SomaTotalD 934.22 19.07 78.86 499.91 0 402.00 40.86 59.78 29.56
## MediaTotalD 10.99 0.22 0.93 5.88 0 4.73 0.48 0.70 0.35
## DPTotalD 31.68 0.65 2.32 14.70 0 7.73 0.42 0.75 0.323.8 Descritores da normalidade
Normalidade1 <- cbind(Assimetria, Curtose)
Normalidade1 <- as.data.frame(Normalidade1)
Normalidade1
SomaTotalN <- apply(Normalidade1,2,sum)
SomaTotalN
MediaTotalN <- apply(Normalidade1,2,mean)
MediaTotalN
DPTotalN <- apply(Normalidade1,2,sd)
DPTotalN
Normalidade2<-cbind(SomaTotalN, MediaTotalN, DPTotalN)
Normalidade2<-as.data.frame(Normalidade2)
Normalidade2 <- t(Normalidade2) #"t" transpoe a matriz
Normalidade2
NormalidadeFinal <- rbind(Normalidade1, Normalidade2)
NormalidadeFinal
NormalidadeFinal <- round(NormalidadeFinal, 2)
NormalidadeFinal## Assimetria Curtose
## Bra.angularis 3.476655 13.364950
## Lepadella.sp 6.363013 43.193400
## Lecane.sp 5.957144 38.356754
## Lec.leontina 4.273584 21.445502
## Lec.bulla 6.461601 43.934851
## Lec.cornuta 5.645693 36.397244
## Lec.curvicornis 4.597181 22.260592
## Notholca.sp 6.783866 47.020833
## Bra.urceolaris 3.660403 14.398551
## Trichocerca.sp 4.991817 27.433083
## Lec.quadridentata 3.786828 16.652473
## Lec.kluchor 4.123459 19.908163
## Lec.lunaris 3.413726 16.091866
## Rotaria.sp 4.149242 21.721303
## Aspelta.sp 4.094856 18.872150
## Lec.furcata 6.461638 43.964610
## Col.geophila 4.641396 22.542553
## Lep.dactyliseta 6.419702 43.417838
## Bra.calyciflorus 6.783866 47.020833
## Bra.caudatus 6.783866 47.020833
## Lec.crepida 3.614935 14.753227
## Pla.patulus 6.732329 46.553383
## Lec.ovalis 3.660403 14.398551
## Lec.elasma 6.783866 47.020833
## Pla.quadricornis 5.957144 38.356754
## Lec.aculeata 4.190788 21.301967
## Ker.tropica 6.783866 47.020833
## Bdelloidea 2.978950 12.576162
## Ker.lenzi 4.446423 22.785803
## Con.unicornis 4.496792 22.720535
## Alo.dadayi 4.466614 24.127985
## Macrothrix.sp 6.549959 44.815176
## Alonella.sp 4.053351 18.620513
## Nauplii 3.797043 17.005265
## Chy.eurynotus 2.912236 10.606273
## Cyclopoida 2.816981 10.016103
## Mac.collinsi 6.783866 47.020833
## Copepodite 2.886668 11.289662
## Paracyclops.sp 6.783866 47.020833
## Asc.ecaudis 6.783866 47.020833
## Lep.ovalis 2.900572 9.967459
## Polyarthra.sp 6.783866 47.020833
## Bra.havanaensis 6.783866 47.020833
## Hexarthra.sp 6.783866 47.020833
## Lec.hastata 6.663554 45.879552
## Tri.tetractis 5.865222 37.571252
## Ker.serrulata 6.783866 47.020833
## Notodiaptomus.sp 4.641396 22.542553
## Bra.falcatus 6.738488 46.601720
## Fil.longiseta 6.783866 47.020833
## Myt.crassipes 5.957144 38.356754
## Lep.patella 6.693569 46.197490
## Euchlanis.sp 6.783866 47.020833
## Colurella.sp 6.772263 46.916558
## Mytilina.sp 6.549959 44.815176
## Lec.monostyla 3.476655 13.364950
## Lec.ligona 6.783866 47.020833
## Ascomorpha.sp 6.783866 47.020833
## Pom.sulcata 4.641396 22.542553
## Harpacticoida 6.783866 47.020833
## Epi.senta 6.783866 47.020833
## Rot.neptunia 5.366983 31.487216
## Dis.aculeata 4.306765 21.658190
## Lec.luna 6.783866 47.020833
## Euc.dilatata 4.641396 22.542553
## Bea.eudactylota 6.783866 47.020833
## Chy.sphaericus 5.031153 28.640000
## Lec.hornemanni 6.783866 47.020833
## Dia.birgei 5.957144 38.356754
## Mac.subquadratus 6.783866 47.020833
## Bra.quadridentatus 6.783866 47.020833
## Pol.vulgaris 6.783866 47.020833
## Pol.bicerca 4.694623 23.214798
## Dicranop.sp 6.783866 47.020833
## Asc.saltans 6.783866 47.020833
## Moi.minuta 6.261844 41.720125
## Alo.hamulata 4.641396 22.542553
## Mac.laticornis 4.935917 26.231706
## Mac.mira 6.783866 47.020833
## Pro.similis 6.783866 47.020833
## Alo.pulchella 6.783866 47.020833
## Dia.spinulosum 6.783866 47.020833
## Chydorus.sp 6.783866 47.020833
## Ily.spinifer 6.783866 47.020833
## Dun.odontoplax 6.783866 47.020833
## Assimetria Curtose
## 476.6836 2976.7207
## Assimetria Curtose
## 5.608042 35.020243
## Assimetria Curtose
## 1.345975 13.495948
## Assimetria Curtose
## SomaTotalN 476.683560 2976.72066
## MediaTotalN 5.608042 35.02024
## DPTotalN 1.345975 13.49595
## Assimetria Curtose
## Bra.angularis 3.476655 13.364950
## Lepadella.sp 6.363013 43.193400
## Lecane.sp 5.957144 38.356754
## Lec.leontina 4.273584 21.445502
## Lec.bulla 6.461601 43.934851
## Lec.cornuta 5.645693 36.397244
## Lec.curvicornis 4.597181 22.260592
## Notholca.sp 6.783866 47.020833
## Bra.urceolaris 3.660403 14.398551
## Trichocerca.sp 4.991817 27.433083
## Lec.quadridentata 3.786828 16.652473
## Lec.kluchor 4.123459 19.908163
## Lec.lunaris 3.413726 16.091866
## Rotaria.sp 4.149242 21.721303
## Aspelta.sp 4.094856 18.872150
## Lec.furcata 6.461638 43.964610
## Col.geophila 4.641396 22.542553
## Lep.dactyliseta 6.419702 43.417838
## Bra.calyciflorus 6.783866 47.020833
## Bra.caudatus 6.783866 47.020833
## Lec.crepida 3.614935 14.753227
## Pla.patulus 6.732329 46.553383
## Lec.ovalis 3.660403 14.398551
## Lec.elasma 6.783866 47.020833
## Pla.quadricornis 5.957144 38.356754
## Lec.aculeata 4.190788 21.301967
## Ker.tropica 6.783866 47.020833
## Bdelloidea 2.978950 12.576162
## Ker.lenzi 4.446423 22.785803
## Con.unicornis 4.496792 22.720535
## Alo.dadayi 4.466614 24.127985
## Macrothrix.sp 6.549959 44.815176
## Alonella.sp 4.053351 18.620513
## Nauplii 3.797043 17.005265
## Chy.eurynotus 2.912236 10.606273
## Cyclopoida 2.816981 10.016103
## Mac.collinsi 6.783866 47.020833
## Copepodite 2.886668 11.289662
## Paracyclops.sp 6.783866 47.020833
## Asc.ecaudis 6.783866 47.020833
## Lep.ovalis 2.900572 9.967459
## Polyarthra.sp 6.783866 47.020833
## Bra.havanaensis 6.783866 47.020833
## Hexarthra.sp 6.783866 47.020833
## Lec.hastata 6.663554 45.879552
## Tri.tetractis 5.865222 37.571252
## Ker.serrulata 6.783866 47.020833
## Notodiaptomus.sp 4.641396 22.542553
## Bra.falcatus 6.738488 46.601720
## Fil.longiseta 6.783866 47.020833
## Myt.crassipes 5.957144 38.356754
## Lep.patella 6.693569 46.197490
## Euchlanis.sp 6.783866 47.020833
## Colurella.sp 6.772263 46.916558
## Mytilina.sp 6.549959 44.815176
## Lec.monostyla 3.476655 13.364950
## Lec.ligona 6.783866 47.020833
## Ascomorpha.sp 6.783866 47.020833
## Pom.sulcata 4.641396 22.542553
## Harpacticoida 6.783866 47.020833
## Epi.senta 6.783866 47.020833
## Rot.neptunia 5.366983 31.487216
## Dis.aculeata 4.306765 21.658190
## Lec.luna 6.783866 47.020833
## Euc.dilatata 4.641396 22.542553
## Bea.eudactylota 6.783866 47.020833
## Chy.sphaericus 5.031153 28.640000
## Lec.hornemanni 6.783866 47.020833
## Dia.birgei 5.957144 38.356754
## Mac.subquadratus 6.783866 47.020833
## Bra.quadridentatus 6.783866 47.020833
## Pol.vulgaris 6.783866 47.020833
## Pol.bicerca 4.694623 23.214798
## Dicranop.sp 6.783866 47.020833
## Asc.saltans 6.783866 47.020833
## Moi.minuta 6.261844 41.720125
## Alo.hamulata 4.641396 22.542553
## Mac.laticornis 4.935917 26.231706
## Mac.mira 6.783866 47.020833
## Pro.similis 6.783866 47.020833
## Alo.pulchella 6.783866 47.020833
## Dia.spinulosum 6.783866 47.020833
## Chydorus.sp 6.783866 47.020833
## Ily.spinifer 6.783866 47.020833
## Dun.odontoplax 6.783866 47.020833
## SomaTotalN 476.683560 2976.720655
## MediaTotalN 5.608042 35.020243
## DPTotalN 1.345975 13.495948
## Assimetria Curtose
## Bra.angularis 3.48 13.36
## Lepadella.sp 6.36 43.19
## Lecane.sp 5.96 38.36
## Lec.leontina 4.27 21.45
## Lec.bulla 6.46 43.93
## Lec.cornuta 5.65 36.40
## Lec.curvicornis 4.60 22.26
## Notholca.sp 6.78 47.02
## Bra.urceolaris 3.66 14.40
## Trichocerca.sp 4.99 27.43
## Lec.quadridentata 3.79 16.65
## Lec.kluchor 4.12 19.91
## Lec.lunaris 3.41 16.09
## Rotaria.sp 4.15 21.72
## Aspelta.sp 4.09 18.87
## Lec.furcata 6.46 43.96
## Col.geophila 4.64 22.54
## Lep.dactyliseta 6.42 43.42
## Bra.calyciflorus 6.78 47.02
## Bra.caudatus 6.78 47.02
## Lec.crepida 3.61 14.75
## Pla.patulus 6.73 46.55
## Lec.ovalis 3.66 14.40
## Lec.elasma 6.78 47.02
## Pla.quadricornis 5.96 38.36
## Lec.aculeata 4.19 21.30
## Ker.tropica 6.78 47.02
## Bdelloidea 2.98 12.58
## Ker.lenzi 4.45 22.79
## Con.unicornis 4.50 22.72
## Alo.dadayi 4.47 24.13
## Macrothrix.sp 6.55 44.82
## Alonella.sp 4.05 18.62
## Nauplii 3.80 17.01
## Chy.eurynotus 2.91 10.61
## Cyclopoida 2.82 10.02
## Mac.collinsi 6.78 47.02
## Copepodite 2.89 11.29
## Paracyclops.sp 6.78 47.02
## Asc.ecaudis 6.78 47.02
## Lep.ovalis 2.90 9.97
## Polyarthra.sp 6.78 47.02
## Bra.havanaensis 6.78 47.02
## Hexarthra.sp 6.78 47.02
## Lec.hastata 6.66 45.88
## Tri.tetractis 5.87 37.57
## Ker.serrulata 6.78 47.02
## Notodiaptomus.sp 4.64 22.54
## Bra.falcatus 6.74 46.60
## Fil.longiseta 6.78 47.02
## Myt.crassipes 5.96 38.36
## Lep.patella 6.69 46.20
## Euchlanis.sp 6.78 47.02
## Colurella.sp 6.77 46.92
## Mytilina.sp 6.55 44.82
## Lec.monostyla 3.48 13.36
## Lec.ligona 6.78 47.02
## Ascomorpha.sp 6.78 47.02
## Pom.sulcata 4.64 22.54
## Harpacticoida 6.78 47.02
## Epi.senta 6.78 47.02
## Rot.neptunia 5.37 31.49
## Dis.aculeata 4.31 21.66
## Lec.luna 6.78 47.02
## Euc.dilatata 4.64 22.54
## Bea.eudactylota 6.78 47.02
## Chy.sphaericus 5.03 28.64
## Lec.hornemanni 6.78 47.02
## Dia.birgei 5.96 38.36
## Mac.subquadratus 6.78 47.02
## Bra.quadridentatus 6.78 47.02
## Pol.vulgaris 6.78 47.02
## Pol.bicerca 4.69 23.21
## Dicranop.sp 6.78 47.02
## Asc.saltans 6.78 47.02
## Moi.minuta 6.26 41.72
## Alo.hamulata 4.64 22.54
## Mac.laticornis 4.94 26.23
## Mac.mira 6.78 47.02
## Pro.similis 6.78 47.02
## Alo.pulchella 6.78 47.02
## Dia.spinulosum 6.78 47.02
## Chydorus.sp 6.78 47.02
## Ily.spinifer 6.78 47.02
## Dun.odontoplax 6.78 47.02
## SomaTotalN 476.68 2976.72
## MediaTotalN 5.61 35.02
## DPTotalN 1.35 13.50A função fix(nome da matriz) dá acesso ao grid da matriz criada para manipulação dos dados numéricos.
3.8.1 Salvando as tabelas criadas em txt direto no diretório de trabalho
write.table(data.frame("Spp"=rownames(DescritoresFinal),
DescritoresFinal),
"Descritores.csv",
row.names=FALSE,
sep="\t")
write.table(data.frame("Spp"=rownames(NormalidadeFinal),
NormalidadeFinal),
"Normalidade.txt",
row.names=FALSE,
sep="\t")Agora é necessário voltar à matriz comunitária (antes de ter sido transposta) antes de continuar a análise. As análises anteriores também precisam ser refeitas com a matriz bruta (não transposta). A seguir confere-se se está sendo usada a matriz comunitária com as espécies nas colunas.
m_trab## BB1-P01 BB1-P02 BB1-P03 BB2-P01 BB2-P02 BB2-P03 BB3-P01 BB3-P02 BB3-P03 BB4-P01
## Bra.angularis 0.0000000 0.5333333 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lepadella.sp 0.0000000 0.1777778 0.1777778 0.5333333 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.bulla 1.0666667 0.0000000 0.3555556 0.8888889 0.1777778 0.0000000 0.5333333 1.6000000 1.4222222 1.4222222
## Lec.cornuta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.1777778 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.kluchor 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.lunaris 0.3555556 0.7111111 1.0666667 0.0000000 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000
## Rotaria.sp 0.0000000 1.0666667 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Aspelta.sp 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## BB4-P02 BB4-P03 BB5-P01 BB5-P02 BB5-P03 BB6-P01 BB6-P02 BB6-P03 CA1-P04 CA1-P05
## Bra.angularis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Lepadella.sp 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.1777778 0.3555556 0.7111111 0.7111111
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 0.0000000 0.3555556 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.bulla 0.3555556 1.0666667 2.1333333 0.7111111 0.5333333 0.8888889 0.5333333 0.3555556 0.8888889 1.2444444
## Lec.cornuta 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Notholca.sp 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.1777778
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000
## Lec.kluchor 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.3555556 0.1777778
## Lec.lunaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.3555556 0.1777778
## Rotaria.sp 0.3555556 0.1777778 0.0000000 0.1777778 0.1777778 0.1777778 0.7111111 0.1777778 0.3555556 0.0000000
## Aspelta.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 1.0666667
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.3555556
## CA1-P06 CA2-P04 CA2-P05 CA2-P06 CA3-P04 CA3-P05 CA3-P06 CA4-P04 CA4-P05 CA4-P06
## Bra.angularis 0.0000000 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lepadella.sp 0.0000000 0.1777778 0.3555556 0.5333333 0.3555556 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 0.0000000 0.3555556 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.bulla 0.5333333 0.5333333 0.7111111 1.6000000 1.6000000 1.7777778 0.7111111 0.5333333 0.1777778 0.0000000
## Lec.cornuta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.1777778 0.0000000 0.1777778 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.kluchor 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.lunaris 0.7111111 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Rotaria.sp 0.8888889 0.0000000 0.5333333 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Aspelta.sp 0.0000000 0.0000000 0.1777778 0.7111111 0.7111111 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## CA5-P04 CA5-P05 CA5-P06 CA6-P04 CA6-P05 CA6-P06 EN08 EN09 EN10 EN11
## Bra.angularis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lepadella.sp 0.0000000 0.1777778 0.0000000 6.4000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.leontina 0.0000000 0.3555556 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 1.0666667 0.0000000
## Lec.bulla 0.7111111 0.1777778 0.3555556 4.2666667 2.1333333 0.0000000 0.0000000 2.1333333 1.0666667 72.0000000
## Lec.cornuta 0.1777778 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.0000000 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 1.6000000 1.6000000
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 2.6666667 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.quadridentata 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.kluchor 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.lunaris 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.1777778 0.5333333 0.0000000 0.5333333 0.0000000
## Rotaria.sp 0.3555556 0.0000000 0.1777778 2.6666667 0.5333333 0.0000000 1.0666667 0.5333333 1.0666667 0.5333333
## Aspelta.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 0.0000000 1.0666667 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 10.6666667
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 5.3333333 0.0000000 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## EN12 EN13 EN15 EN16 EN14 EN17 RE19 RE18 RE07
## Bra.angularis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lepadella.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lecane.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.leontina 0.0000000 0.0000000 1.6000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.bulla 0.5333333 1.0666667 12.8000000 0.0000000 0.0000000 0.0000000 1.0666667 2.133333 0.0000000
## Lec.cornuta 0.5333333 0.0000000 2.6666667 0.5333333 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.curvicornis 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Notholca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Bra.urceolaris 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Trichocerca.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 1.6000000
## Lec.quadridentata 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.5333333
## Lec.kluchor 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.lunaris 0.0000000 0.0000000 0.0000000 1.6000000 0.5333333 0.0000000 0.0000000 0.000000 2.6666667
## Rotaria.sp 0.0000000 0.0000000 2.6666667 10.1333333 9.0666667 10.1333333 2.1333333 3.733333 24.5333333
## Aspelta.sp 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lec.furcata 0.0000000 0.0000000 1.6000000 0.0000000 0.0000000 0.0000000 0.5333333 0.000000 0.0000000
## Col.geophila 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Lep.dactyliseta 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Bra.calyciflorus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## Bra.caudatus 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000
## [ reached getOption("max.print") -- omitted 65 rows ]m_trab <- m_bruta3.9 Gráficos descritivos
3.9.1 Série de Hill e pefil da diversidade
Pode-se utilizar uma série de Hill ao invés dos índices específicos de diversidade. Assim, quanto maior o valor de q (definido em scales), maior será o peso para a equabilidade. Quanto mais próximo de zero, maior o peso para riqueza. Quando q = 0, o valor de Hill é igual a riqueza de espécies.
Hill <- renyi(m_trab,
scales = c(0:5),
hill = TRUE)
Hill## 0 1 2 3 4 5
## BB1-P01 10 6.993228 5.369863 4.616559 4.224982 3.989802
## BB1-P02 6 5.425658 5.142857 4.977368 4.863511 4.776764
## BB1-P03 6 4.649327 3.769231 3.286815 3.019087 2.858625
## BB2-P01 6 5.008145 4.446154 4.137447 3.956045 3.840456
## BB2-P02 5 4.166436 3.521739 3.117691 2.878806 2.732619
## BB2-P03 4 3.789291 3.571429 3.370999 3.204120 3.073941
## BB3-P01 4 3.464102 3.000000 2.683282 2.489480 2.371130
## BB3-P02 7 4.875670 3.574257 2.974949 2.692430 2.541384
## BB3-P03 5 3.917181 3.247191 2.878366 2.670599 2.544773
## BB4-P01 5 3.271768 2.724528 2.465629 2.314415 2.219427
## BB4-P02 6 4.340807 3.266667 2.753200 2.507716 2.376050
## BB4-P03 4 3.061696 2.682540 2.530859 2.458091 2.415698
## BB5-P01 4 2.861364 2.337900 2.103454 1.982793 1.912301
## BB5-P02 12 6.663205 4.238390 3.393326 3.033681 2.845689
## BB5-P03 6 4.649327 3.769231 3.286815 3.019087 2.858625
## BB6-P01 6 2.318844 1.628406 1.467689 1.408026 1.378355
## BB6-P02 8 3.744164 2.369439 1.996998 1.856265 1.786608
## BB6-P03 7 3.632068 2.484472 2.122083 1.971082 1.892683
## CA1-P04 13 9.549730 7.163743 5.849605 5.143030 4.734564
## CA1-P05 15 6.749183 3.753906 2.935254 2.628108 2.477269
## CA1-P06 10 7.911397 6.760000 6.154617 5.798450 5.561875
## CA2-P04 13 6.928135 4.061281 3.139865 2.788817 2.618190
## CA2-P05 17 9.402285 5.141328 3.791271 3.298925 3.064739
## CA2-P06 16 7.639079 4.448399 3.517988 3.142456 2.946765
## CA3-P04 8 5.895652 4.839506 4.324172 4.040513 3.864580
## CA3-P05 9 5.920820 4.595745 4.046051 3.777578 3.624548
## CA3-P06 10 7.491776 5.730159 4.821913 4.357572 4.091111
## CA4-P04 4 3.369922 3.000000 2.799770 2.684871 2.611787
## CA4-P05 5 3.190527 2.327273 2.005885 1.869255 1.799195
## CA4-P06 2 2.000000 2.000000 2.000000 2.000000 2.000000
## CA5-P04 5 4.559014 4.235294 4.000000 3.826847 3.697009
## CA5-P05 9 6.399822 4.741935 3.938599 3.538647 3.313975
## CA5-P06 7 4.293598 2.892857 2.391160 2.186971 2.084572
## CA6-P04 18 11.394566 8.347826 6.932216 6.194479 5.761539
## CA6-P05 8 3.648667 2.442462 2.084490 1.937335 1.861519
## CA6-P06 6 5.298702 4.571429 4.000000 3.624826 3.390389
## EN08 6 5.656854 5.333333 5.059644 4.845655 4.685277
## EN09 7 5.743998 4.800000 4.242641 3.928556 3.739855
## EN10 10 8.402666 7.078431 6.155894 5.558073 5.168814
## EN11 10 3.354695 2.616329 2.437927 2.357712 2.308763
## EN12 16 6.530448 5.021704 4.412047 4.052348 3.818622
## EN13 7 6.335826 5.761905 5.321574 5.001823 4.770356
## EN15 19 4.783592 2.387139 1.977841 1.837953 1.769859
## EN16 9 4.092259 2.973698 2.620632 2.451970 2.352227
## EN14 9 5.680780 4.476821 3.998635 3.762505 3.625985
## EN17 5 3.040662 2.250000 1.961161 1.835225 1.769337
## RE19 8 6.915809 6.081081 5.514110 5.139080 4.883641
## RE18 4 3.736031 3.481928 3.264547 3.094332 2.967205
## RE07 6 3.236803 2.295993 1.980658 1.848619 1.780782Podemos mapear isso em um gráfico de perfil de diversidade.
library(ggplot2)
library(tidyr)
library(tidyverse)
grafico1 <- Hill %>%
rownames_to_column() %>%
pivot_longer(-rowname) %>%
mutate(name = factor(name, name[1:length(Hill)])) %>%
ggplot(aes(x = name, y = value, group = rowname,
col = rowname)) +
geom_point(size = 2) +
geom_line(size = 1) +
xlab("Parâmetro de ordem de diversidade (q)") +
ylab("Diversidade") +
labs(col = "Locais") +
theme_bw() +
theme(text = element_text(size = 5)) #ajustar a fonte caso nao caiba no output html
grafico1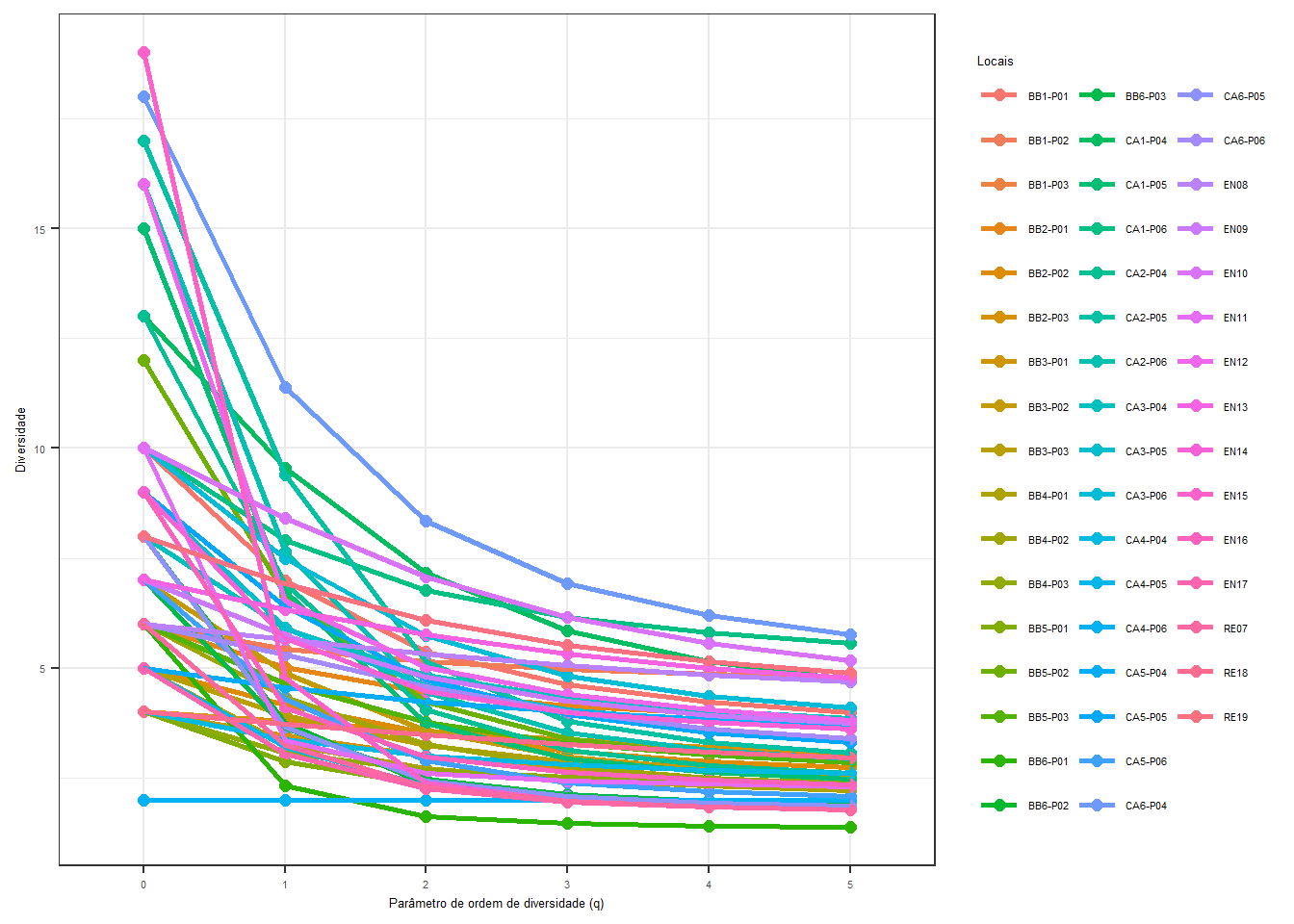
Figure 3.1: Mapeamento dos pontos de coleta em um gráfico de perfil de diversidade.
ggsave(grafico1, dpi = 300, filename = "fig-hill.png")## Saving 7 x 5 in image3.10 Distribuição de abundância
Uma maneira de observar a diversidade de espécies é usando um gráfico de distribuição de abundâncias de uma comunidade, como mostrado a seguir. Note que aqui utiliza-se as abundâncias totais das espécies, mas é possível fazer por linha, basta substituir o objeto abund pela abundância de uma linha. Como na linha marcada com o #.
abund <- colSums(m_trab)Na primeira linha do código abaixo, retira-se o # do início da linha para rodar a distribuição de abundância apenas para o local (linha) 1 comunidade[1,]. Pode-se mudar o número para o local de interesse.
#abund <- m_trab[1, ]
df <- data.frame(sp = colnames(m_trab),
abun = abund)
grafico2 <- ggplot(df, aes(fct_reorder(sp, -abun),
abun, group = 1)) +
geom_col() +
geom_line(col = "red", linetype = "dashed") +
geom_point(col = "red") +
xlab("Espécies") +
ylab("Abundância") +
theme_bw() +
theme(axis.text.x = element_text(
angle = 45,
hjust = 1,
face = "italic"))
grafico2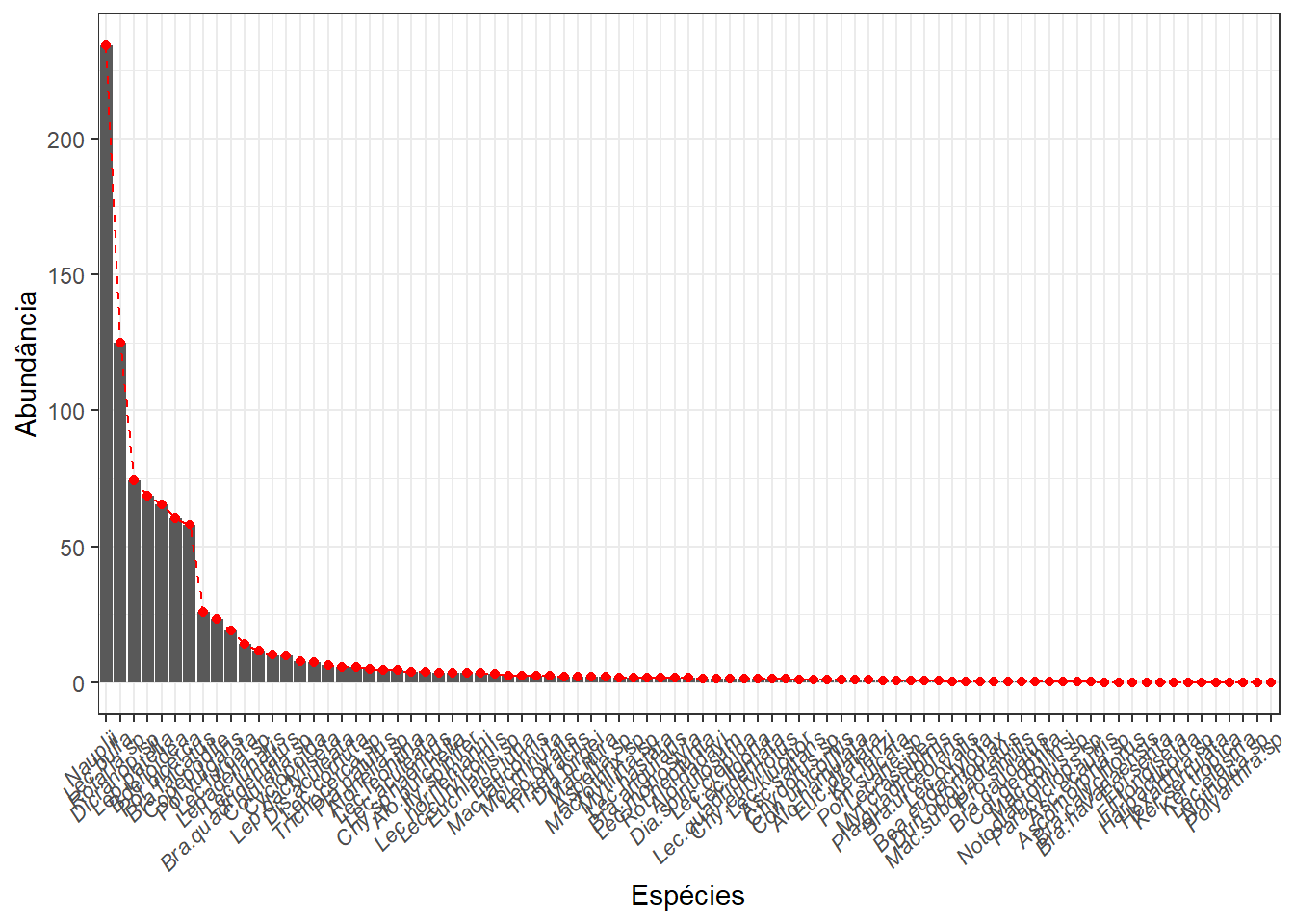
Figure 3.2: Distribuição da densidade de indivíduos.
ggsave(grafico2, dpi = 300, filename = "fig-abun.png")## Saving 7 x 5 in image3.11 Curva de rarefação
O código abaixo indica como usar o pacote iNEXT para extrapolar e interpolar curvas de rarefação visando comparar a riqueza de diferentes locais. A curva de raferação vai ser construida para os pontos demarcados em rarefa <- t(m_trab[c("BB1-P1","BB1-P2","BB1-P3"),]).
m_trab_r <- mutate(m_trab, across(everything(), ceiling)) #arredonda a matriz BRUTA para números inteiros
colnames(t(m_trab_r))## [1] "BB1-P01" "BB1-P02" "BB1-P03" "BB2-P01" "BB2-P02" "BB2-P03" "BB3-P01" "BB3-P02" "BB3-P03" "BB4-P01" "BB4-P02"
## [12] "BB4-P03" "BB5-P01" "BB5-P02" "BB5-P03" "BB6-P01" "BB6-P02" "BB6-P03" "CA1-P04" "CA1-P05" "CA1-P06" "CA2-P04"
## [23] "CA2-P05" "CA2-P06" "CA3-P04" "CA3-P05" "CA3-P06" "CA4-P04" "CA4-P05" "CA4-P06" "CA5-P04" "CA5-P05" "CA5-P06"
## [34] "CA6-P04" "CA6-P05" "CA6-P06" "EN08" "EN09" "EN10" "EN11" "EN12" "EN13" "EN15" "EN16"
## [45] "EN14" "EN17" "RE19" "RE18" "RE07"rarefa <- t(m_trab_r[c("BB1-P01", "BB1-P02", "BB1-P03"),]) #curva de rarefação para os sítios especificados
class(m_trab)## [1] "data.frame"#rarefa <- t(m_trab_r) #curva de rarefação para todos os sítiosNo código abaixo o valor de q pode ser mudado para q=1 para comparar a diversidade de Shannon e para q=2 para Simpson.
library(iNEXT)
out <- iNEXT(rarefa, q = 0,
datatype = "abundance",
size = NULL,
endpoint = 1500, #define o comprimento de eixo x
knots = 40,
se = TRUE,
conf = 0.95,
nboot = 50)
grafico3 <- ggiNEXT(out, type = 1, facet.var="None") +
theme_bw() +
labs(fill = "Áreas") +
xlab("Número de indivíduos") +
ylab("Riqueza de espécies") +
theme(legend.title=element_blank()) #ver como fica com facet.var="Assemblage"
grafico3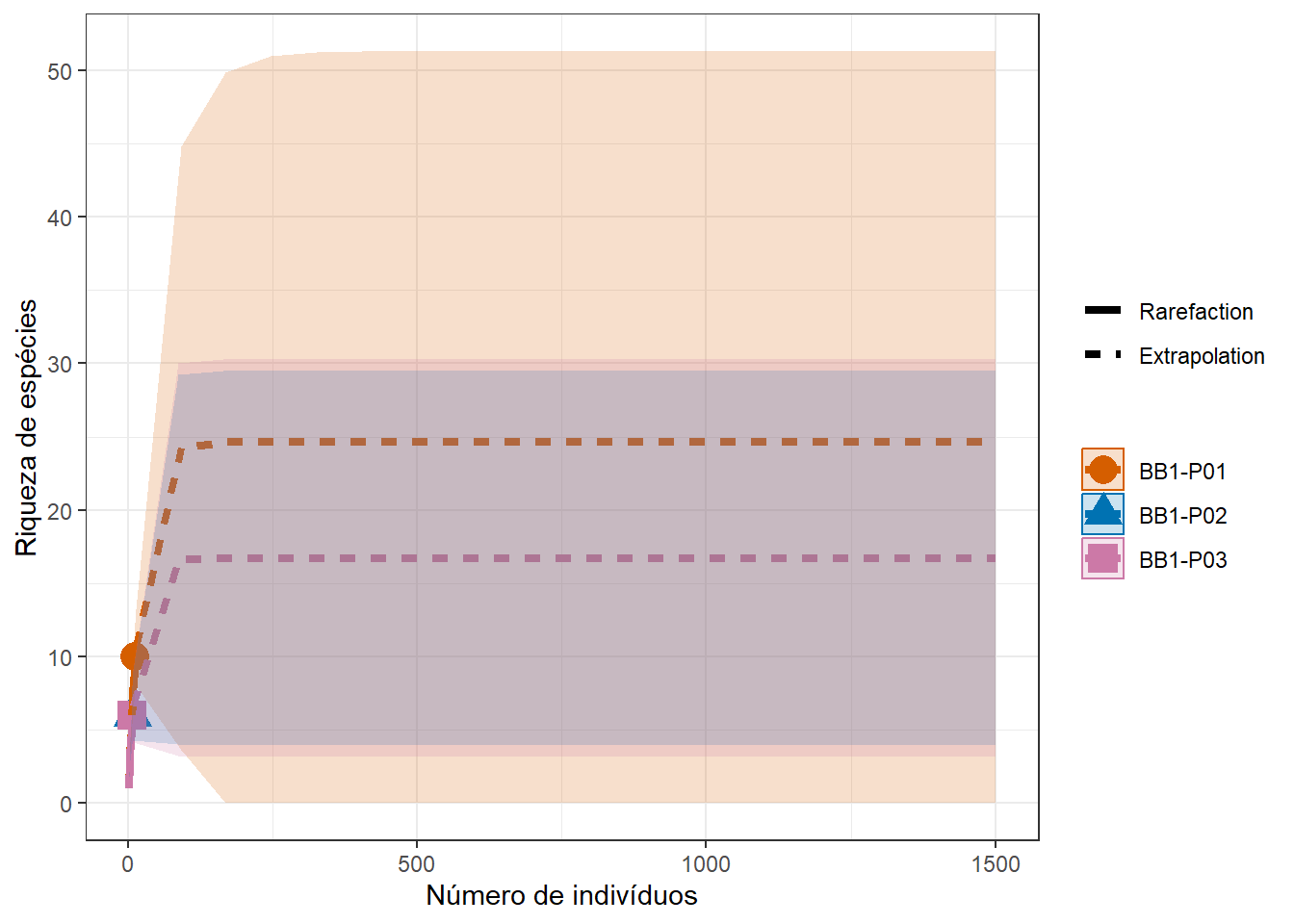
Figure 3.3: Curva de rafefação para sítios específicos.
ggsave(grafico3, dpi = 300, filename = "fig-rare1.png")## Saving 7 x 5 in image## Análise de similaridades
system(paste("open", shQuote("D:/Elvio/OneDrive/MSS/_Zoo-Rebio/R_ZooRebio/mdists.qmd")))