2 Espécies compartilhadas
2.0.1 Organização básica
dev.off()
rm(list=ls(all=TRUE))
cat("\014")
#limpa a memoria, o console e apaga os graficos, se houver algumInstalando os pacotes necessários
install.packages("openxlsx") #importa arquivos do excel
install.packages("gt") #ferramenta para criação de tabelas bonitas e personalizáveis
install.packages("dplyr")
install.packages("eulerr")
install.packages("VennDiagram")
install.packages("ggVennDiagram")
install.packages("ggvenn")
install.packages("gplots") Definindo o diretório de trabalho
getwd()
setwd("C:/Seu/Diretório/De/Trabalho")
dir <- (getwd())Carregado bases de dados dos pontos fixos de amostragem (fixos.xlsx) e dos pontos ao entorno da ReBio (entorno.xlsx)
library(openxlsx)
fixos <- read.xlsx("D:/Elvio/OneDrive/MSS/_Zoo-Rebio/fixos.xlsx",
rowNames = T,
colNames = T,
sheet = "ponto_l")
entorno <- read.xlsx("D:/Elvio/OneDrive/MSS/_Zoo-Rebio/entorno.xlsx",
rowNames = T,
colNames = T,
sheet = "ponto_l")
teste <- read.xlsx("D:/Elvio/OneDrive/MSS/_Zoo-Rebio/teste.xlsx",
rowNames = T,
colNames = T)Aqui são duas bases de dados (pontos fixos 3 e pontos ao entorno 4 da ReBio) que precisam ser importadas e mescladas (Tabela 2.1). Os dados são densidade em no. de indivídos por litro (no.ind/L) (ver Gouveia et al. (2017), Gouveia (2014) e Lima (2022)).
2.1 Particionando a matriz entorno
Deletando colunas e linhas, e excluindo all-zeros e valores N/A e o ponto 20. *Lecane mono* e o Site 20 foram deixados errados de propósito para usar os códigos de deletar uma coluna ou linha, por nome.
entorno <- subset(entorno, select = -Lec.mono) #escolhendo uma coluna pelo nome
#Deletando linhas específicas
del_rows <- c("ST20") #site fake
del_rows
m_part <- entorno[!(row.names(entorno) %in% c(del_rows)),]
m_part
sum <- colSums(m_part)
sum
zero_sum_cols <- names(which(colSums(m_part) == 0))
zero_sum_cols #nomes das espécies zeradas
m_part2 <- m_part[(colSums(m_part) != 0)] #em != a exclamação inverte o sentido
zero_sum_cols2 <- names(which(colSums(m_part2) == 0))
zero_sum_cols2 #nomes das espécies zeradas
sum<-colSums(m_part2)
sum
entorno <- m_part2## [1] "ST20"
## Rotaria.sp Rot.neptunia Dis.aculeata Lep.ovalis Lec.quadridentata Lec.cornuta Lec.luna Lec.lunaris Lec.bulla
## EN08 1.0666667 1.0666667 0.5333333 0.5333333 0.0000000 0.5333333 0.000000 0.5333333 0.0000000
## EN09 0.5333333 0.0000000 1.6000000 0.5333333 0.0000000 0.0000000 0.000000 0.0000000 2.1333333
## EN10 1.0666667 0.0000000 2.6666667 0.0000000 0.0000000 0.0000000 0.000000 0.5333333 1.0666667
## RE19 2.1333333 0.0000000 1.0666667 0.0000000 0.0000000 0.0000000 0.000000 0.0000000 1.0666667
## EN11 0.5333333 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000 72.0000000
## EN12 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.000000 0.0000000 0.5333333
## EN13 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.0000000 0.000000 0.0000000 1.0666667
## EN15 2.6666667 0.0000000 0.0000000 0.0000000 0.0000000 2.6666667 0.000000 0.0000000 12.8000000
## EN16 10.1333333 0.0000000 0.0000000 0.0000000 0.0000000 0.5333333 0.000000 1.6000000 0.0000000
## EN14 9.0666667 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 2.666667 0.5333333 0.0000000
## EN17 10.1333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000 0.0000000
## RE18 3.7333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.000000 0.0000000 2.1333333
## RE07 24.5333333 0.0000000 0.0000000 0.0000000 0.5333333 0.0000000 0.000000 2.6666667 0.0000000
## Euc.dilatata Nauplii Copepodite Lec.curvicornis Bea.eudactylota Lec.leontina Lec.monostyla Chy.sphaericus
## EN08 0.0000000 0.0000000 0.0000000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN09 0.5333333 0.5333333 0.5333333 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN10 0.5333333 0.0000000 0.0000000 1.6 0.0000000 1.066667 0.5333333 0.5333333
## RE19 0.0000000 0.5333333 0.5333333 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN11 0.0000000 0.0000000 0.0000000 1.6 0.0000000 0.000000 0.0000000 0.0000000
## EN12 0.0000000 38.9333333 3.7333333 0.0 0.0000000 0.000000 0.0000000 1.0666667
## EN13 0.0000000 0.5333333 0.0000000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN15 0.0000000 70.9333333 2.1333333 0.0 0.5333333 1.600000 0.5333333 2.1333333
## EN16 0.0000000 49.6000000 2.6666667 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN14 0.0000000 8.0000000 1.6000000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN17 0.0000000 1.6000000 1.0666667 0.0 0.0000000 0.000000 0.0000000 0.0000000
## RE18 0.0000000 0.0000000 1.6000000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## RE07 0.0000000 5.8666667 0.0000000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## Pla.quadricornis Bra.falcatus Lec.furcata Lep.patella Lec.hornemanni Dia.birgei Lec.hastata Mac.subquadratus
## EN08 0.0000000 0.0 0.0000000 0.0000000 0.000000 0.0000000 0.0 0.0000000
## EN09 0.0000000 0.0 0.0000000 0.0000000 0.000000 0.0000000 0.0 0.0000000
## EN10 0.5333333 0.0 0.0000000 0.0000000 0.000000 0.0000000 0.0 0.0000000
## RE19 0.0000000 1.6 0.5333333 0.5333333 0.000000 0.0000000 0.0 0.0000000
## EN11 0.0000000 0.0 10.6666667 53.8666667 3.733333 1.6000000 1.6 0.5333333
## EN12 0.0000000 24.0 0.0000000 0.0000000 0.000000 0.0000000 0.0 0.0000000
## EN13 0.0000000 0.0 0.0000000 1.0666667 0.000000 0.0000000 0.0 0.0000000
## EN15 0.0000000 0.0 1.6000000 0.5333333 0.000000 0.0000000 0.0 0.0000000
## EN16 0.0000000 0.0 0.0000000 0.0000000 0.000000 0.0000000 0.0 0.0000000
## EN14 0.0000000 0.0 0.0000000 0.0000000 0.000000 0.5333333 0.0 0.0000000
## EN17 0.0000000 0.0 0.0000000 2.6666667 0.000000 0.0000000 0.0 0.0000000
## RE18 0.0000000 0.0 0.0000000 1.6000000 0.000000 0.0000000 0.0 0.0000000
## RE07 0.0000000 0.0 0.0000000 3.7333333 0.000000 0.0000000 0.0 0.0000000
## Alonella.sp Bra.quadridentatus Pol.vulgaris Pol.bicerca Dicranop.sp Cyclopoida Asc.saltans Moi.minuta Alo.hamulata
## EN08 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN09 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN10 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## RE19 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN11 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN12 1.6 10.13333 19.2 31.46667 68.8 1.0666667 1.066667 0.5333333 0.5333333
## EN13 1.6 0.00000 0.0 0.00000 0.0 0.5333333 0.000000 0.0000000 0.0000000
## EN15 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## EN16 0.0 0.00000 0.0 26.66667 0.0 1.6000000 0.000000 2.1333333 0.0000000
## EN14 0.0 0.00000 0.0 0.00000 0.0 1.0666667 0.000000 0.0000000 0.5333333
## EN17 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## RE18 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## RE07 0.0 0.00000 0.0 0.00000 0.0 0.0000000 0.000000 0.0000000 0.0000000
## Mac.laticornis Lec.crepida Mac.mira Pro.similis Tri.tetractis Alo.pulchella Dia.spinulosum Macrothrix.sp
## EN08 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN09 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN10 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## RE19 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN11 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN12 1.066667 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN13 0.000000 0.5333333 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN15 1.600000 0.0000000 2.133333 0.5333333 1.6 3.733333 1.6 1.6
## EN16 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN14 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## EN17 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## RE18 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## RE07 0.000000 0.0000000 0.000000 0.0000000 0.0 0.000000 0.0 0.0
## Chydorus.sp Pla.patulus Ily.spinifer Dun.odontoplax Trichocerca.sp
## EN08 0.000000 0.000000 0.000000 0.0000000 0.0
## EN09 0.000000 0.000000 0.000000 0.0000000 0.0
## EN10 0.000000 0.000000 0.000000 0.0000000 0.0
## RE19 0.000000 0.000000 0.000000 0.0000000 0.0
## EN11 0.000000 0.000000 0.000000 0.0000000 0.0
## EN12 0.000000 0.000000 0.000000 0.0000000 0.0
## EN13 0.000000 0.000000 0.000000 0.0000000 0.0
## EN15 1.066667 0.000000 0.000000 0.0000000 0.0
## EN16 0.000000 4.266667 0.000000 0.0000000 0.0
## EN14 0.000000 0.000000 3.733333 0.0000000 0.0
## EN17 0.000000 0.000000 0.000000 0.5333333 0.0
## RE18 0.000000 0.000000 0.000000 0.0000000 0.0
## RE07 0.000000 0.000000 0.000000 0.0000000 1.6
## Rotaria.sp Rot.neptunia Dis.aculeata Lep.ovalis Lec.quadridentata Lec.cornuta
## 65.6000000 1.6000000 5.8666667 1.6000000 0.5333333 4.2666667
## Lec.luna Lec.lunaris Lec.bulla Euc.dilatata Nauplii Copepodite
## 2.6666667 5.8666667 92.8000000 1.0666667 176.5333333 13.8666667
## Lec.curvicornis Bea.eudactylota Lec.leontina Lec.monostyla Chy.sphaericus Pla.quadricornis
## 3.2000000 0.5333333 2.6666667 1.0666667 3.7333333 0.5333333
## Bra.falcatus Lec.furcata Lep.patella Lec.hornemanni Dia.birgei Lec.hastata
## 25.6000000 12.8000000 64.0000000 3.7333333 2.1333333 1.6000000
## Mac.subquadratus Alonella.sp Bra.quadridentatus Pol.vulgaris Pol.bicerca Dicranop.sp
## 0.5333333 3.2000000 10.1333333 19.2000000 58.1333333 68.8000000
## Cyclopoida Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis Lec.crepida
## 4.2666667 1.0666667 2.6666667 1.0666667 2.6666667 0.5333333
## Mac.mira Pro.similis Tri.tetractis Alo.pulchella Dia.spinulosum Macrothrix.sp
## 2.1333333 0.5333333 1.6000000 3.7333333 1.6000000 1.6000000
## Chydorus.sp Pla.patulus Ily.spinifer Dun.odontoplax Trichocerca.sp
## 1.0666667 4.2666667 3.7333333 0.5333333 1.6000000
## character(0)
## character(0)
## Rotaria.sp Rot.neptunia Dis.aculeata Lep.ovalis Lec.quadridentata Lec.cornuta
## 65.6000000 1.6000000 5.8666667 1.6000000 0.5333333 4.2666667
## Lec.luna Lec.lunaris Lec.bulla Euc.dilatata Nauplii Copepodite
## 2.6666667 5.8666667 92.8000000 1.0666667 176.5333333 13.8666667
## Lec.curvicornis Bea.eudactylota Lec.leontina Lec.monostyla Chy.sphaericus Pla.quadricornis
## 3.2000000 0.5333333 2.6666667 1.0666667 3.7333333 0.5333333
## Bra.falcatus Lec.furcata Lep.patella Lec.hornemanni Dia.birgei Lec.hastata
## 25.6000000 12.8000000 64.0000000 3.7333333 2.1333333 1.6000000
## Mac.subquadratus Alonella.sp Bra.quadridentatus Pol.vulgaris Pol.bicerca Dicranop.sp
## 0.5333333 3.2000000 10.1333333 19.2000000 58.1333333 68.8000000
## Cyclopoida Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis Lec.crepida
## 4.2666667 1.0666667 2.6666667 1.0666667 2.6666667 0.5333333
## Mac.mira Pro.similis Tri.tetractis Alo.pulchella Dia.spinulosum Macrothrix.sp
## 2.1333333 0.5333333 1.6000000 3.7333333 1.6000000 1.6000000
## Chydorus.sp Pla.patulus Ily.spinifer Dun.odontoplax Trichocerca.sp
## 1.0666667 4.2666667 3.7333333 0.5333333 1.60000002.1.1 Separando a matriz fixos em suas partes (CA e BB)
Separa-se fixos_bb e fixos_ca e remove-se as colunas zeradas.
fixos_ca <- fixos[1:18,]
names(which(colSums(fixos_ca) == 0)) #all-zero columns## [1] "Lec.curvicornis" "Notholca.sp" "Ascomorpha.sp" "Pom.sulcata" "Harpacticoida" "Epi.senta"fixos_ca <- fixos_ca[, colSums(fixos_ca != 0) > 0]
fixos_bb <- fixos[19:36,]
names(which(colSums(fixos_bb) == 0)) #all-zero columns## [1] "Trichocerca.sp" "Lec.furcata" "Col.geophila" "Lep.dactyliseta" "Bra.calyciflorus" "Bra.caudatus"
## [7] "Pla.patulus" "Lec.elasma" "Pla.quadricornis" "Ker.tropica" "Ker.lenzi" "Alo.dadayi"
## [13] "Mac.collinsi" "Paracyclops.sp" "Asc.ecaudis" "Lep.ovalis" "Polyarthra.sp" "Bra.havanaensis"
## [19] "Hexarthra.sp" "Lec.hastata" "Tri.tetractis" "Ker.serrulata" "Notodiaptomus.sp" "Bra.falcatus"
## [25] "Fil.longiseta" "Myt.crassipes" "Lep.patella" "Euchlanis.sp" "Lec.monostyla" "Lec.ligona"fixos_bb <- fixos_bb[, colSums(fixos_bb != 0) > 0]2.1.1.1 Mesmo procedimento usando o comando grepl() do pacote dplyr
library(dplyr)
entorno_re <- filter(entorno, grepl("RE", row.names(entorno)))
names(which(colSums(entorno_re) == 0)) #all-zero columns## [1] "Rot.neptunia" "Lep.ovalis" "Lec.cornuta" "Lec.luna" "Euc.dilatata"
## [6] "Lec.curvicornis" "Bea.eudactylota" "Lec.leontina" "Lec.monostyla" "Chy.sphaericus"
## [11] "Pla.quadricornis" "Lec.hornemanni" "Dia.birgei" "Lec.hastata" "Mac.subquadratus"
## [16] "Alonella.sp" "Bra.quadridentatus" "Pol.vulgaris" "Pol.bicerca" "Dicranop.sp"
## [21] "Cyclopoida" "Asc.saltans" "Moi.minuta" "Alo.hamulata" "Mac.laticornis"
## [26] "Lec.crepida" "Mac.mira" "Pro.similis" "Tri.tetractis" "Alo.pulchella"
## [31] "Dia.spinulosum" "Macrothrix.sp" "Chydorus.sp" "Pla.patulus" "Ily.spinifer"
## [36] "Dun.odontoplax"entorno_re <- entorno_re[, colSums(entorno_re != 0) > 0]
entorno_en <- filter(entorno, grepl("EN", row.names(entorno)))
names(which(colSums(entorno_en) == 0)) #all-zero columns## [1] "Lec.quadridentata" "Trichocerca.sp"entorno_en <- entorno_en[, colSums(entorno_en != 0) > 0]| Rio | n=315 | ||||
|---|---|---|---|---|---|
| CA | Coleta | Datas | Ponto | Pote | Lâmina |
| BB | 1 | 26-02-2011 | -P1 | . | . |
| . | 2 | 30-04-2011 | -P2 | . | . |
| . | 3 | 30-06-2011 | -P3 | .1 | . |
| . | 4 | 01-09-2011 | . | .2 | . |
| . | 5 | 03-11-2011 | . | .3 | -I |
| . | 6 | 18-01-2012 | . | . | -II |
| . | . | . | . | . | -III |
| 2 | x 12 | . | x 36 | x 108 | = 324 |
2.2 Merging bases de dados fixos e entorno
2.2.1 Taxa compartilhados pelas duas bases de dados
2.2.1.1 Bases de dados em arquivos diferentes fixos e entorno
Nos códigos abaixo, a função intersect() é usada para se obter os nomes de colunas em comum entre fixo e entorno, que são guardados em um vetor. Esses vetores são repassados nos outros argumentos subsequentes. O propósito dos argumentos setdiff() e \<- 0 não é preencher células vazias com zeros, mas adicionar novas colunas em cada data frame com nomes de colunas que estão presentes em um data frame mas não em outro. Se uma coluna está faltando em um data frame, ela será adicionada aquele data frame com o termo NA ou NULL. Para substituir os valores rotulados de NA ou NULL usa-se a função merged[is.na(merged)] <- 0. Ver Apêndices.
shared_spp <- intersect(names(fixos), names(entorno)) #get the common column names
shared_spp## [1] "Lec.leontina" "Lec.bulla" "Lec.cornuta" "Lec.curvicornis" "Trichocerca.sp"
## [6] "Lec.quadridentata" "Lec.lunaris" "Rotaria.sp" "Lec.furcata" "Lec.crepida"
## [11] "Pla.patulus" "Pla.quadricornis" "Macrothrix.sp" "Alonella.sp" "Nauplii"
## [16] "Cyclopoida" "Copepodite" "Lep.ovalis" "Lec.hastata" "Tri.tetractis"
## [21] "Bra.falcatus" "Lep.patella" "Lec.monostyla"fixos_only <- setdiff(names(fixos), names(entorno))
fixos_only## [1] "Bra.angularis" "Lepadella.sp" "Lecane.sp" "Notholca.sp" "Bra.urceolaris" "Lec.kluchor"
## [7] "Aspelta.sp" "Col.geophila" "Lep.dactyliseta" "Bra.calyciflorus" "Bra.caudatus" "Lec.ovalis"
## [13] "Lec.elasma" "Lec.aculeata" "Ker.tropica" "Bdelloidea" "Ker.lenzi" "Con.unicornis"
## [19] "Alo.dadayi" "Chy.eurynotus" "Mac.collinsi" "Paracyclops.sp" "Asc.ecaudis" "Polyarthra.sp"
## [25] "Bra.havanaensis" "Hexarthra.sp" "Ker.serrulata" "Notodiaptomus.sp" "Fil.longiseta" "Myt.crassipes"
## [31] "Euchlanis.sp" "Colurella.sp" "Mytilina.sp" "Lec.ligona" "Ascomorpha.sp" "Pom.sulcata"
## [37] "Harpacticoida" "Epi.senta"entorno_only <- setdiff(names(entorno), names(fixos))
entorno_only## [1] "Rot.neptunia" "Dis.aculeata" "Lec.luna" "Euc.dilatata" "Bea.eudactylota"
## [6] "Chy.sphaericus" "Lec.hornemanni" "Dia.birgei" "Mac.subquadratus" "Bra.quadridentatus"
## [11] "Pol.vulgaris" "Pol.bicerca" "Dicranop.sp" "Asc.saltans" "Moi.minuta"
## [16] "Alo.hamulata" "Mac.laticornis" "Mac.mira" "Pro.similis" "Alo.pulchella"
## [21] "Dia.spinulosum" "Chydorus.sp" "Ily.spinifer" "Dun.odontoplax"Riqueza <- length(shared_spp) + length(fixos_only) + length(entorno_only)
Riqueza## [1] 852.2.1.2 Bases de dados em arquivos diferentes fixos_bb e fixos_ca
shared_fixos <- intersect(names(fixos_bb), names(fixos_ca))
shared_fixos
bb_only <- setdiff(names(fixos_bb), names(fixos_ca))
bb_only
ca_only <- setdiff(names(fixos_ca), names(fixos_bb))
ca_only
Riqueza <- length(shared_fixos) + length(bb_only) + length(ca_only)
Riqueza## [1] "Bra.angularis" "Lepadella.sp" "Lecane.sp" "Lec.leontina" "Lec.bulla"
## [6] "Lec.cornuta" "Bra.urceolaris" "Lec.quadridentata" "Lec.kluchor" "Lec.lunaris"
## [11] "Rotaria.sp" "Aspelta.sp" "Lec.crepida" "Lec.ovalis" "Lec.aculeata"
## [16] "Bdelloidea" "Con.unicornis" "Macrothrix.sp" "Alonella.sp" "Nauplii"
## [21] "Chy.eurynotus" "Cyclopoida" "Copepodite" "Colurella.sp" "Mytilina.sp"
## [1] "Lec.curvicornis" "Notholca.sp" "Ascomorpha.sp" "Pom.sulcata" "Harpacticoida" "Epi.senta"
## [1] "Trichocerca.sp" "Lec.furcata" "Col.geophila" "Lep.dactyliseta" "Bra.calyciflorus" "Bra.caudatus"
## [7] "Pla.patulus" "Lec.elasma" "Pla.quadricornis" "Ker.tropica" "Ker.lenzi" "Alo.dadayi"
## [13] "Mac.collinsi" "Paracyclops.sp" "Asc.ecaudis" "Lep.ovalis" "Polyarthra.sp" "Bra.havanaensis"
## [19] "Hexarthra.sp" "Lec.hastata" "Tri.tetractis" "Ker.serrulata" "Notodiaptomus.sp" "Bra.falcatus"
## [25] "Fil.longiseta" "Myt.crassipes" "Lep.patella" "Euchlanis.sp" "Lec.monostyla" "Lec.ligona"
## [1] 61Criando um data.frame com todas as espécies compartilhadas e exclusivas.
library(tidyverse)
# Create a data frame with all species
all_species <- data.frame(
type = c(rep("Shared", length(shared_spp)),
rep("Fixos_only", length(fixos_only)),
rep("Entorno_only", length(entorno_only))),
species = c(shared_spp, fixos_only, entorno_only)
)
# Create a table using the `table()` function
species_table <- table(all_species$type, all_species$species)
# Convert the table to a data frame and format it
species_table_df <- as.data.frame.matrix(species_table)
rownames(species_table_df) <- c("Entorno_only", "Fixos_only", "Shared")
species_table_df <- t(species_table_df[, order(colnames(species_table_df))])
species_table_df <- as.data.frame(species_table_df)
species_table_df
# Ordenando pelo nome da coluna
species_table_df <- rownames_to_column(species_table_df, var = "Espécies")
species_table_df <- species_table_df[order(species_table_df$Entorno_only, species_table_df$Fixos_only, species_table_df$Shared, decreasing = TRUE),]
library(gt)
gt(species_table_df)Na sequência dos dois próximos tópicos, estou mesclando tudo na base de dados zoorebio (que será a matriz de bruta, subsequentemente) e criando uma copia em .xlsx, .csv e .txt na pasta de trabalho do R.
2.2.2 Definindo a base de dados
library(dplyr)
# Add missing columns with zeros to both data frames
fixos[setdiff(shared_spp, names(fixos))] <- 0
entorno[setdiff(shared_spp, names(entorno))] <- 0
# Combine the two data frames
merged <- bind_rows(fixos, entorno) #merging the two datasets
merged[is.na(merged)] <- 0
# Order the rows by their original data frame names
merged <- merged[order(gsub("[^[:alpha:]]", "", rownames(merged))), ]
# Print the merged data frame
zoorebio <- merged
as.matrix(zoorebio)[1:6, 1:6] ## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## BB1-P01 0.0000000 0.0000000 0 0 1.0666667 0
## BB1-P02 0.5333333 0.1777778 0 0 0.0000000 0
## BB1-P03 0.5333333 0.1777778 0 0 0.3555556 0
## BB2-P01 0.0000000 0.5333333 0 0 0.8888889 0
## BB2-P02 0.0000000 0.1777778 0 0 0.1777778 0
## BB2-P03 0.0000000 0.1777778 0 0 0.0000000 02.2.3 Fazendo uma copia da base de dados zoorebio
write.table(zoorebio, "zoorebio.csv", append = F, quote = TRUE, sep = ";", row.names = T, dec = ",")
#dir <- getwd()
#shell.exec(dir) #abre o diretorio de trabalho no Windows Explorer
zoorebio_csv <- read.csv("zoorebio.csv",
sep = ";", dec = ",",
header = T,
row.names = 1,
na.strings = NA)2.3 Reset point
m_bruta <- zoorebio_csvAqui cria-se o vertor da matriz bruta a partir da base de dados depois de feitos os ajustes necessários.
2.4 Criando uma matriz de médias
#Inserindo coluna para agrupamentos
ncol(m_bruta); nrow(m_bruta) #no. de N colunas x M linhas
m_bruta_g <- cbind(Grupos = rownames(m_bruta), m_bruta)
agrup <- substr(m_bruta_g[, 1], 1,2)
agrup
m_bruta_g <- m_bruta_g %>% mutate(Grupos=c(agrup))
m_avg_part <- aggregate(m_bruta_g[, 2:3], list(m_bruta_g$Grupos), mean)
m_avg <- m_bruta_g %>%
group_by(Grupos) %>%
summarise(across(.cols = everything(), ~ mean(.x, na.rm = TRUE)))
#m_avg <- m_bruta_g %>%
# group_by(Grupos) %>%
# summarise(across(.cols = everything(), list(mean = mean, sd = sd)))
#?across
#Primeira coluna para nomes das linhas
m_avg <- as.data.frame(m_avg)
class(m_avg)
rownames(m_avg) <- m_avg[,1]
m_avg[,1] <- NULL
#Salvando a matriz
write.table(m_avg,
"m_avgcsv.csv",
append = F,
quote = TRUE,
sep = ";", dec = ",",
row.names = T)
m_avg_csv <- read.csv("m_avgcsv.csv",
sep = ";", dec = ",",
header = T,
row.names = 1,
na.strings = NA)2.5 Análise de Espécies Compartilhadas em arquivos diferentes
#Espécies compartilhadas
shared_spp <- intersect(names(fixos), names(entorno)) #get the common column names
shared_spp## [1] "Lec.leontina" "Lec.bulla" "Lec.cornuta" "Lec.curvicornis" "Trichocerca.sp"
## [6] "Lec.quadridentata" "Lec.lunaris" "Rotaria.sp" "Lec.furcata" "Lec.crepida"
## [11] "Pla.patulus" "Pla.quadricornis" "Macrothrix.sp" "Alonella.sp" "Nauplii"
## [16] "Cyclopoida" "Copepodite" "Lep.ovalis" "Lec.hastata" "Tri.tetractis"
## [21] "Bra.falcatus" "Lep.patella" "Lec.monostyla"fixos_only <- setdiff(names(fixos), names(entorno))
fixos_only## [1] "Bra.angularis" "Lepadella.sp" "Lecane.sp" "Notholca.sp" "Bra.urceolaris" "Lec.kluchor"
## [7] "Aspelta.sp" "Col.geophila" "Lep.dactyliseta" "Bra.calyciflorus" "Bra.caudatus" "Lec.ovalis"
## [13] "Lec.elasma" "Lec.aculeata" "Ker.tropica" "Bdelloidea" "Ker.lenzi" "Con.unicornis"
## [19] "Alo.dadayi" "Chy.eurynotus" "Mac.collinsi" "Paracyclops.sp" "Asc.ecaudis" "Polyarthra.sp"
## [25] "Bra.havanaensis" "Hexarthra.sp" "Ker.serrulata" "Notodiaptomus.sp" "Fil.longiseta" "Myt.crassipes"
## [31] "Euchlanis.sp" "Colurella.sp" "Mytilina.sp" "Lec.ligona" "Ascomorpha.sp" "Pom.sulcata"
## [37] "Harpacticoida" "Epi.senta"entorno_only <- setdiff(names(entorno), names(fixos))
entorno_only## [1] "Rot.neptunia" "Dis.aculeata" "Lec.luna" "Euc.dilatata" "Bea.eudactylota"
## [6] "Chy.sphaericus" "Lec.hornemanni" "Dia.birgei" "Mac.subquadratus" "Bra.quadridentatus"
## [11] "Pol.vulgaris" "Pol.bicerca" "Dicranop.sp" "Asc.saltans" "Moi.minuta"
## [16] "Alo.hamulata" "Mac.laticornis" "Mac.mira" "Pro.similis" "Alo.pulchella"
## [21] "Dia.spinulosum" "Chydorus.sp" "Ily.spinifer" "Dun.odontoplax"Riqueza <- length(shared_spp) + length(fixos_only) + length(entorno_only)
Riqueza## [1] 85shared_fixos <- intersect(names(fixos_bb), names(fixos_ca))
shared_fixos## [1] "Bra.angularis" "Lepadella.sp" "Lecane.sp" "Lec.leontina" "Lec.bulla"
## [6] "Lec.cornuta" "Bra.urceolaris" "Lec.quadridentata" "Lec.kluchor" "Lec.lunaris"
## [11] "Rotaria.sp" "Aspelta.sp" "Lec.crepida" "Lec.ovalis" "Lec.aculeata"
## [16] "Bdelloidea" "Con.unicornis" "Macrothrix.sp" "Alonella.sp" "Nauplii"
## [21] "Chy.eurynotus" "Cyclopoida" "Copepodite" "Colurella.sp" "Mytilina.sp"bb_only <- setdiff(names(fixos_bb), names(fixos_ca))
bb_only## [1] "Lec.curvicornis" "Notholca.sp" "Ascomorpha.sp" "Pom.sulcata" "Harpacticoida" "Epi.senta"ca_only <- setdiff(names(fixos_ca), names(fixos_bb))
ca_only## [1] "Trichocerca.sp" "Lec.furcata" "Col.geophila" "Lep.dactyliseta" "Bra.calyciflorus" "Bra.caudatus"
## [7] "Pla.patulus" "Lec.elasma" "Pla.quadricornis" "Ker.tropica" "Ker.lenzi" "Alo.dadayi"
## [13] "Mac.collinsi" "Paracyclops.sp" "Asc.ecaudis" "Lep.ovalis" "Polyarthra.sp" "Bra.havanaensis"
## [19] "Hexarthra.sp" "Lec.hastata" "Tri.tetractis" "Ker.serrulata" "Notodiaptomus.sp" "Bra.falcatus"
## [25] "Fil.longiseta" "Myt.crassipes" "Lep.patella" "Euchlanis.sp" "Lec.monostyla" "Lec.ligona"Riqueza <- length(shared_fixos) + length(bb_only) + length(ca_only)
Riqueza## [1] 612.6 Bases de dados no mesmo arquivo zoorebio
Pode-se usar a matriz teste.xlsx para testar e verificar os resultados dos comandos em comparação com as matrizes reais. O comando fix() permite editar uma matriz. A matriz de teste tem 9 linhas e 20 colunas, com os nomes A1, A2, A3, B1, B2, B3, C1, C2, C3 nas linhas e SP1, SP2, SP3, SP4, SP5, SP6, SP7, SP8, SP9, SP10, SP11, SP12, SP13, SP14, SP15, SP16, SP17, SP18, SP19, SP20 como os nomes das colunas. Códigos baseados nas letras A, B e C, portanto, se referem a matriz de teste.
A seguir separamos a matriz em suas partes.
data <- (teste)
data## SP1 SP2 SP3 SP4 SP5 SP6 SP7 SP8 SP9 SP10 SP11 SP12 SP13 SP14 SP15 SP16 SP17 SP18 SP19 SP20
## A1 1 0 0 1 0 1 1 1 0 0 1 0 0 1 0 1 1 1 0 0
## A2 1 0 0 1 0 1 1 0 0 0 1 0 0 1 0 1 1 0 0 0
## A3 1 0 0 1 0 1 1 0 0 0 1 0 0 1 0 1 1 0 0 0
## B1 0 1 0 1 1 0 1 0 1 0 0 1 0 1 1 0 1 0 1 0
## B2 0 1 0 1 1 0 1 0 0 0 0 1 0 1 1 0 1 0 0 0
## B3 0 1 0 1 1 0 1 0 0 0 0 1 0 1 1 0 1 0 0 0
## C1 0 0 1 0 1 1 1 0 0 1 0 0 1 0 1 1 1 0 0 1
## C2 0 0 1 0 1 1 1 0 0 0 0 0 1 0 1 1 1 0 0 0
## C3 0 0 1 0 1 1 1 0 0 0 0 0 1 0 1 1 1 0 0 0library(dplyr)
A <- filter(data, grepl("A", row.names(data)))
names(which(colSums(A) == 0)) #all-zero columns## [1] "SP2" "SP3" "SP5" "SP9" "SP10" "SP12" "SP13" "SP15" "SP19" "SP20"A <- A[, colSums(A != 0) > 0]
B <- filter(data, grepl("B", row.names(data)))
names(which(colSums(B) == 0)) #all-zero columns## [1] "SP1" "SP3" "SP6" "SP8" "SP10" "SP11" "SP13" "SP16" "SP18" "SP20"B <- B[, colSums(B != 0) > 0]
C <- filter(data, grepl("C", row.names(data)))
names(which(colSums(C) == 0)) #all-zero columns## [1] "SP1" "SP2" "SP4" "SP8" "SP9" "SP11" "SP12" "SP14" "SP18" "SP19"C <- C[, colSums(C != 0) > 0]2.8 Encontrando espécies exclusivas
2.8.1 Escolhendo as LINHAS OU GRUPOS para comparar
# Get the row indices where 'A' occurs
rownames(data)## [1] "BB1-P01" "BB1-P02" "BB1-P03" "BB2-P01" "BB2-P02" "BB2-P03" "BB3-P01" "BB3-P02" "BB3-P03" "BB4-P01" "BB4-P02"
## [12] "BB4-P03" "BB5-P01" "BB5-P02" "BB5-P03" "BB6-P01" "BB6-P02" "BB6-P03" "CA1-P04" "CA1-P05" "CA1-P06" "CA2-P04"
## [23] "CA2-P05" "CA2-P06" "CA3-P04" "CA3-P05" "CA3-P06" "CA4-P04" "CA4-P05" "CA4-P06" "CA5-P04" "CA5-P05" "CA5-P06"
## [34] "CA6-P04" "CA6-P05" "CA6-P06" "EN08" "EN09" "EN10" "EN11" "EN12" "EN13" "EN15" "EN16"
## [45] "EN14" "EN17" "RE19" "RE18" "RE07"rows <- grep("A1", rownames(data))
rows <- grep("E", rownames(data))
rows <- c(1:36)
rows## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 362.8.1.1 Espécies exclusivas que ocorrem em 1 LINHA E OUTRA, OU EM 1 GRUPO E OUTRO (#EM TESTE)
# Initialize an empty vector to store species exclusive to 'A' rows
species_only_in <- character(0)
# Iterate over each column
for (col in colnames(data)) {
# Check if the species occurs only in 'A' rows
if (all(data[rows, col] != 0) && !any(data[-rows, col] != 0)) {
species_only_in <- c(species_only_in, col)
}
}
# Print species that only occur in 'A' rows
print(species_only_in)## character(0)data## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta Lec.curvicornis Notholca.sp
## BB1-P01 0.0000000 0.0000000 0.0000000 0 1.0666667 0.0000000 0.0000000 0.0000000
## BB1-P02 0.5333333 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB1-P03 0.5333333 0.1777778 0.0000000 0 0.3555556 0.0000000 0.0000000 0.0000000
## BB2-P01 0.0000000 0.5333333 0.0000000 0 0.8888889 0.0000000 0.0000000 0.0000000
## BB2-P02 0.0000000 0.1777778 0.0000000 0 0.1777778 0.0000000 0.0000000 0.0000000
## BB2-P03 0.0000000 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB3-P01 0.0000000 0.0000000 0.1777778 0 0.5333333 0.1777778 0.0000000 0.0000000
## BB3-P02 0.0000000 0.0000000 0.0000000 0 1.6000000 0.0000000 0.0000000 0.0000000
## BB3-P03 0.0000000 0.0000000 0.0000000 0 1.4222222 0.1777778 0.0000000 0.0000000
## BB4-P01 0.0000000 0.0000000 0.0000000 0 1.4222222 0.0000000 0.1777778 0.0000000
## BB4-P02 0.0000000 0.0000000 0.0000000 0 0.3555556 0.0000000 0.0000000 0.1777778
## Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata
## BB1-P01 0.1777778 0 0 0.1777778 0.3555556 0.0000000 0.1777778 0
## BB1-P02 0.0000000 0 0 0.0000000 0.7111111 1.0666667 0.0000000 0
## BB1-P03 0.0000000 0 0 0.0000000 1.0666667 0.0000000 0.0000000 0
## BB2-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB2-P02 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB2-P03 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB3-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P02 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P03 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P02 0.0000000 0 0 0.0000000 0.0000000 0.3555556 0.0000000 0
## Col.geophila Lep.dactyliseta Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.0000000 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.1777778 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis Alo.dadayi Macrothrix.sp
## BB1-P01 0 1.6000000 0 0.0000000 0 0.0000000 0 0.1777778
## BB1-P02 0 0.8888889 0 0.0000000 0 0.0000000 0 0.0000000
## BB1-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P01 0 0.1777778 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P02 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB3-P01 0 0.0000000 0 0.1777778 0 0.0000000 0 0.0000000
## BB3-P02 0 0.0000000 0 0.3555556 0 0.5333333 0 0.0000000
## BB3-P03 0 0.0000000 0 0.7111111 0 0.3555556 0 0.0000000
## BB4-P01 0 0.0000000 0 3.5555556 0 0.0000000 0 0.0000000
## BB4-P02 0 0.0000000 0 1.2444444 0 0.0000000 0 0.0000000
## Alonella.sp Nauplii Chy.eurynotus Cyclopoida Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis
## BB1-P01 0 0.7111111 0.3555556 0.0000000 0 0.0000000 0 0 0
## BB1-P02 0 0.8888889 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB1-P03 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P01 0 0.8888889 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB2-P02 0 0.7111111 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P01 0 0.0000000 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P02 0 0.3555556 0.0000000 0.1777778 0 0.1777778 0 0 0
## BB3-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB4-P01 0 1.4222222 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB4-P02 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## Polyarthra.sp Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp Bra.falcatus
## BB1-P01 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0
## Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp Mytilina.sp Lec.monostyla Lec.ligona
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.1777778 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.0000000 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Ascomorpha.sp Pom.sulcata Harpacticoida Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata
## BB1-P01 0.1777778 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P02 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB2-P01 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P02 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P02 0.0000000 0.0000000 0.1777778 0.0000000 0 0 0 0
## BB3-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P02 0.0000000 0.0000000 0.0000000 0.1777778 0 0 0 0
## Bea.eudactylota Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## BB1-P01 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis Mac.mira Pro.similis Alo.pulchella
## BB1-P01 0 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0 0
## Dia.spinulosum Chydorus.sp Ily.spinifer Dun.odontoplax
## BB1-P01 0 0 0 0
## BB1-P02 0 0 0 0
## BB1-P03 0 0 0 0
## BB2-P01 0 0 0 0
## BB2-P02 0 0 0 0
## BB2-P03 0 0 0 0
## BB3-P01 0 0 0 0
## BB3-P02 0 0 0 0
## BB3-P03 0 0 0 0
## BB4-P01 0 0 0 0
## BB4-P02 0 0 0 0
## [ reached 'max' / getOption("max.print") -- omitted 38 rows ]2.9 Espécies exclusivas para 1 LINHA OU 1 GRUPO - Baseado na soma dos grupos (#FUNCIONA)
# Get the row indices where 'A' occurs
rows## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36# Calculate the sum of occurrences of each species in 'A' rows
sum_of <- colSums(data[rows, ])
sum_of## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 1.7777778 11.7333333 0.7111111 1.2444444 32.0000000 1.4222222
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 0.1777778 0.1777778 0.5333333 3.3777778 1.0666667 1.2444444
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 4.6222222 8.7111111 1.9555556 1.2444444 0.3555556 6.5777778
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.1777778 0.3555556 1.0666667 0.5333333 0.5333333 0.1777778
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.1777778 3.9111111 0.1777778 60.5333333 0.7111111 1.0666667
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 1.6000000 0.3555556 1.6000000 57.5111111 1.4222222 3.2000000
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.3555556 9.4222222 0.3555556 0.1777778 0.7111111 0.1777778
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.1777778 0.1777778 0.1777778 0.7111111 0.1777778 0.3555556
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 0.1777778 0.1777778 0.7111111 1.6000000 2.6666667 7.8222222
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 1.9555556 0.7111111 1.6000000 0.1777778 0.7111111 0.1777778
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.1777778 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Dun.odontoplax
## 0.0000000# Initialize an empty vector to store species exclusive to 'A' rows
species_only_in <- character(0)
# Iterate over each column
for (col in colnames(data)) {
# Check if the species occurs only in 'A' rows
if (sum(data[rows, col]) == sum_of[col] && !any(data[-rows, col] != 0)) {
species_only_in <- c(species_only_in, col)
}
}
# Print species that only occur in 'A' rows
print(species_only_in)## [1] "Bra.angularis" "Lepadella.sp" "Lecane.sp" "Notholca.sp" "Bra.urceolaris" "Lec.kluchor"
## [7] "Aspelta.sp" "Col.geophila" "Lep.dactyliseta" "Bra.calyciflorus" "Bra.caudatus" "Lec.ovalis"
## [13] "Lec.elasma" "Lec.aculeata" "Ker.tropica" "Bdelloidea" "Ker.lenzi" "Con.unicornis"
## [19] "Alo.dadayi" "Chy.eurynotus" "Mac.collinsi" "Paracyclops.sp" "Asc.ecaudis" "Polyarthra.sp"
## [25] "Bra.havanaensis" "Hexarthra.sp" "Ker.serrulata" "Notodiaptomus.sp" "Fil.longiseta" "Myt.crassipes"
## [31] "Euchlanis.sp" "Colurella.sp" "Mytilina.sp" "Lec.ligona" "Ascomorpha.sp" "Pom.sulcata"
## [37] "Harpacticoida" "Epi.senta"entorno_only## [1] "Rot.neptunia" "Dis.aculeata" "Lec.luna" "Euc.dilatata" "Bea.eudactylota"
## [6] "Chy.sphaericus" "Lec.hornemanni" "Dia.birgei" "Mac.subquadratus" "Bra.quadridentatus"
## [11] "Pol.vulgaris" "Pol.bicerca" "Dicranop.sp" "Asc.saltans" "Moi.minuta"
## [16] "Alo.hamulata" "Mac.laticornis" "Mac.mira" "Pro.similis" "Alo.pulchella"
## [21] "Dia.spinulosum" "Chydorus.sp" "Ily.spinifer" "Dun.odontoplax"data## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta Lec.curvicornis Notholca.sp
## BB1-P01 0.0000000 0.0000000 0.0000000 0 1.0666667 0.0000000 0.0000000 0.0000000
## BB1-P02 0.5333333 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB1-P03 0.5333333 0.1777778 0.0000000 0 0.3555556 0.0000000 0.0000000 0.0000000
## BB2-P01 0.0000000 0.5333333 0.0000000 0 0.8888889 0.0000000 0.0000000 0.0000000
## BB2-P02 0.0000000 0.1777778 0.0000000 0 0.1777778 0.0000000 0.0000000 0.0000000
## BB2-P03 0.0000000 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB3-P01 0.0000000 0.0000000 0.1777778 0 0.5333333 0.1777778 0.0000000 0.0000000
## BB3-P02 0.0000000 0.0000000 0.0000000 0 1.6000000 0.0000000 0.0000000 0.0000000
## BB3-P03 0.0000000 0.0000000 0.0000000 0 1.4222222 0.1777778 0.0000000 0.0000000
## BB4-P01 0.0000000 0.0000000 0.0000000 0 1.4222222 0.0000000 0.1777778 0.0000000
## BB4-P02 0.0000000 0.0000000 0.0000000 0 0.3555556 0.0000000 0.0000000 0.1777778
## Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata
## BB1-P01 0.1777778 0 0 0.1777778 0.3555556 0.0000000 0.1777778 0
## BB1-P02 0.0000000 0 0 0.0000000 0.7111111 1.0666667 0.0000000 0
## BB1-P03 0.0000000 0 0 0.0000000 1.0666667 0.0000000 0.0000000 0
## BB2-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB2-P02 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB2-P03 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB3-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P02 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P03 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P02 0.0000000 0 0 0.0000000 0.0000000 0.3555556 0.0000000 0
## Col.geophila Lep.dactyliseta Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.0000000 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.1777778 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis Alo.dadayi Macrothrix.sp
## BB1-P01 0 1.6000000 0 0.0000000 0 0.0000000 0 0.1777778
## BB1-P02 0 0.8888889 0 0.0000000 0 0.0000000 0 0.0000000
## BB1-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P01 0 0.1777778 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P02 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB3-P01 0 0.0000000 0 0.1777778 0 0.0000000 0 0.0000000
## BB3-P02 0 0.0000000 0 0.3555556 0 0.5333333 0 0.0000000
## BB3-P03 0 0.0000000 0 0.7111111 0 0.3555556 0 0.0000000
## BB4-P01 0 0.0000000 0 3.5555556 0 0.0000000 0 0.0000000
## BB4-P02 0 0.0000000 0 1.2444444 0 0.0000000 0 0.0000000
## Alonella.sp Nauplii Chy.eurynotus Cyclopoida Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis
## BB1-P01 0 0.7111111 0.3555556 0.0000000 0 0.0000000 0 0 0
## BB1-P02 0 0.8888889 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB1-P03 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P01 0 0.8888889 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB2-P02 0 0.7111111 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P01 0 0.0000000 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P02 0 0.3555556 0.0000000 0.1777778 0 0.1777778 0 0 0
## BB3-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB4-P01 0 1.4222222 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB4-P02 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## Polyarthra.sp Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp Bra.falcatus
## BB1-P01 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0
## Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp Mytilina.sp Lec.monostyla Lec.ligona
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.1777778 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.0000000 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Ascomorpha.sp Pom.sulcata Harpacticoida Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata
## BB1-P01 0.1777778 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P02 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB2-P01 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P02 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P02 0.0000000 0.0000000 0.1777778 0.0000000 0 0 0 0
## BB3-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P02 0.0000000 0.0000000 0.0000000 0.1777778 0 0 0 0
## Bea.eudactylota Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## BB1-P01 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis Mac.mira Pro.similis Alo.pulchella
## BB1-P01 0 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0 0
## Dia.spinulosum Chydorus.sp Ily.spinifer Dun.odontoplax
## BB1-P01 0 0 0 0
## BB1-P02 0 0 0 0
## BB1-P03 0 0 0 0
## BB2-P01 0 0 0 0
## BB2-P02 0 0 0 0
## BB2-P03 0 0 0 0
## BB3-P01 0 0 0 0
## BB3-P02 0 0 0 0
## BB3-P03 0 0 0 0
## BB4-P01 0 0 0 0
## BB4-P02 0 0 0 0
## [ reached 'max' / getOption("max.print") -- omitted 38 rows ]rows## [1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 362.10 Espécies exclusivas para 2 LINHAS OU 2 GRUPOs - Baseado na soma dos grupos (#FUNCIONA)
# Get the row indices where 'A' occurs
rows <- grep("EN", rownames(data))
rows2 <- grep("RE", rownames(data))
rows <- as.vector(rbind(c(rows, rows2)))
rows## [1] 37 38 39 40 41 42 43 44 45 46 47 48 49# Calculate the sum of occurrences of each species in 'A' rows
sum_of <- colSums(data[rows, ])
sum_of## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta
## 0.0000000 0.0000000 0.0000000 2.6666667 92.8000000 4.2666667
## Lec.curvicornis Notholca.sp Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor
## 3.2000000 0.0000000 0.0000000 1.6000000 0.5333333 0.0000000
## Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata Col.geophila Lep.dactyliseta
## 5.8666667 65.6000000 0.0000000 12.8000000 0.0000000 0.0000000
## Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## 0.0000000 0.0000000 0.5333333 4.2666667 0.0000000 0.0000000
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis
## 0.5333333 0.0000000 0.0000000 0.0000000 0.0000000 0.0000000
## Alo.dadayi Macrothrix.sp Alonella.sp Nauplii Chy.eurynotus Cyclopoida
## 0.0000000 1.6000000 3.2000000 176.5333333 0.0000000 4.2666667
## Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis Polyarthra.sp
## 0.0000000 13.8666667 0.0000000 0.0000000 1.6000000 0.0000000
## Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp
## 0.0000000 0.0000000 1.6000000 1.6000000 0.0000000 0.0000000
## Bra.falcatus Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp
## 25.6000000 0.0000000 0.0000000 64.0000000 0.0000000 0.0000000
## Mytilina.sp Lec.monostyla Lec.ligona Ascomorpha.sp Pom.sulcata Harpacticoida
## 0.0000000 1.0666667 0.0000000 0.0000000 0.0000000 0.0000000
## Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata Bea.eudactylota
## 0.0000000 1.6000000 5.8666667 2.6666667 1.0666667 0.5333333
## Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## 3.7333333 3.7333333 2.1333333 0.5333333 10.1333333 19.2000000
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis
## 58.1333333 68.8000000 1.0666667 2.6666667 1.0666667 2.6666667
## Mac.mira Pro.similis Alo.pulchella Dia.spinulosum Chydorus.sp Ily.spinifer
## 2.1333333 0.5333333 3.7333333 1.6000000 1.0666667 3.7333333
## Dun.odontoplax
## 0.5333333# Initialize an empty vector to store species exclusive to 'A' rows
species_only_in <- character(0)
# Iterate over each column
for (col in colnames(data)) {
# Check if the species occurs only in 'A' rows
if (sum(data[rows, col]) == sum_of[col] && !any(data[-rows, col] != 0)) {
species_only_in <- c(species_only_in, col)
}
}
# Print species that only occur in 'A' rows
print(species_only_in)## [1] "Rot.neptunia" "Dis.aculeata" "Lec.luna" "Euc.dilatata" "Bea.eudactylota"
## [6] "Chy.sphaericus" "Lec.hornemanni" "Dia.birgei" "Mac.subquadratus" "Bra.quadridentatus"
## [11] "Pol.vulgaris" "Pol.bicerca" "Dicranop.sp" "Asc.saltans" "Moi.minuta"
## [16] "Alo.hamulata" "Mac.laticornis" "Mac.mira" "Pro.similis" "Alo.pulchella"
## [21] "Dia.spinulosum" "Chydorus.sp" "Ily.spinifer" "Dun.odontoplax"entorno_only## [1] "Rot.neptunia" "Dis.aculeata" "Lec.luna" "Euc.dilatata" "Bea.eudactylota"
## [6] "Chy.sphaericus" "Lec.hornemanni" "Dia.birgei" "Mac.subquadratus" "Bra.quadridentatus"
## [11] "Pol.vulgaris" "Pol.bicerca" "Dicranop.sp" "Asc.saltans" "Moi.minuta"
## [16] "Alo.hamulata" "Mac.laticornis" "Mac.mira" "Pro.similis" "Alo.pulchella"
## [21] "Dia.spinulosum" "Chydorus.sp" "Ily.spinifer" "Dun.odontoplax"data## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta Lec.curvicornis Notholca.sp
## BB1-P01 0.0000000 0.0000000 0.0000000 0 1.0666667 0.0000000 0.0000000 0.0000000
## BB1-P02 0.5333333 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB1-P03 0.5333333 0.1777778 0.0000000 0 0.3555556 0.0000000 0.0000000 0.0000000
## BB2-P01 0.0000000 0.5333333 0.0000000 0 0.8888889 0.0000000 0.0000000 0.0000000
## BB2-P02 0.0000000 0.1777778 0.0000000 0 0.1777778 0.0000000 0.0000000 0.0000000
## BB2-P03 0.0000000 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB3-P01 0.0000000 0.0000000 0.1777778 0 0.5333333 0.1777778 0.0000000 0.0000000
## BB3-P02 0.0000000 0.0000000 0.0000000 0 1.6000000 0.0000000 0.0000000 0.0000000
## BB3-P03 0.0000000 0.0000000 0.0000000 0 1.4222222 0.1777778 0.0000000 0.0000000
## BB4-P01 0.0000000 0.0000000 0.0000000 0 1.4222222 0.0000000 0.1777778 0.0000000
## BB4-P02 0.0000000 0.0000000 0.0000000 0 0.3555556 0.0000000 0.0000000 0.1777778
## Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata
## BB1-P01 0.1777778 0 0 0.1777778 0.3555556 0.0000000 0.1777778 0
## BB1-P02 0.0000000 0 0 0.0000000 0.7111111 1.0666667 0.0000000 0
## BB1-P03 0.0000000 0 0 0.0000000 1.0666667 0.0000000 0.0000000 0
## BB2-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB2-P02 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB2-P03 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB3-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P02 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P03 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P02 0.0000000 0 0 0.0000000 0.0000000 0.3555556 0.0000000 0
## Col.geophila Lep.dactyliseta Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.0000000 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.1777778 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis Alo.dadayi Macrothrix.sp
## BB1-P01 0 1.6000000 0 0.0000000 0 0.0000000 0 0.1777778
## BB1-P02 0 0.8888889 0 0.0000000 0 0.0000000 0 0.0000000
## BB1-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P01 0 0.1777778 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P02 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB3-P01 0 0.0000000 0 0.1777778 0 0.0000000 0 0.0000000
## BB3-P02 0 0.0000000 0 0.3555556 0 0.5333333 0 0.0000000
## BB3-P03 0 0.0000000 0 0.7111111 0 0.3555556 0 0.0000000
## BB4-P01 0 0.0000000 0 3.5555556 0 0.0000000 0 0.0000000
## BB4-P02 0 0.0000000 0 1.2444444 0 0.0000000 0 0.0000000
## Alonella.sp Nauplii Chy.eurynotus Cyclopoida Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis
## BB1-P01 0 0.7111111 0.3555556 0.0000000 0 0.0000000 0 0 0
## BB1-P02 0 0.8888889 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB1-P03 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P01 0 0.8888889 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB2-P02 0 0.7111111 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P01 0 0.0000000 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P02 0 0.3555556 0.0000000 0.1777778 0 0.1777778 0 0 0
## BB3-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB4-P01 0 1.4222222 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB4-P02 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## Polyarthra.sp Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp Bra.falcatus
## BB1-P01 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0
## Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp Mytilina.sp Lec.monostyla Lec.ligona
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.1777778 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.0000000 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Ascomorpha.sp Pom.sulcata Harpacticoida Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata
## BB1-P01 0.1777778 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P02 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB2-P01 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P02 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P02 0.0000000 0.0000000 0.1777778 0.0000000 0 0 0 0
## BB3-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P02 0.0000000 0.0000000 0.0000000 0.1777778 0 0 0 0
## Bea.eudactylota Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## BB1-P01 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis Mac.mira Pro.similis Alo.pulchella
## BB1-P01 0 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0 0
## Dia.spinulosum Chydorus.sp Ily.spinifer Dun.odontoplax
## BB1-P01 0 0 0 0
## BB1-P02 0 0 0 0
## BB1-P03 0 0 0 0
## BB2-P01 0 0 0 0
## BB2-P02 0 0 0 0
## BB2-P03 0 0 0 0
## BB3-P01 0 0 0 0
## BB3-P02 0 0 0 0
## BB3-P03 0 0 0 0
## BB4-P01 0 0 0 0
## BB4-P02 0 0 0 0
## [ reached 'max' / getOption("max.print") -- omitted 38 rows ]rows## [1] 37 38 39 40 41 42 43 44 45 46 47 48 49identical(entorno_only, species_only_in)## [1] TRUEintersect(entorno_only, species_only_in) #shared column names## [1] "Rot.neptunia" "Dis.aculeata" "Lec.luna" "Euc.dilatata" "Bea.eudactylota"
## [6] "Chy.sphaericus" "Lec.hornemanni" "Dia.birgei" "Mac.subquadratus" "Bra.quadridentatus"
## [11] "Pol.vulgaris" "Pol.bicerca" "Dicranop.sp" "Asc.saltans" "Moi.minuta"
## [16] "Alo.hamulata" "Mac.laticornis" "Mac.mira" "Pro.similis" "Alo.pulchella"
## [21] "Dia.spinulosum" "Chydorus.sp" "Ily.spinifer" "Dun.odontoplax"setdiff(entorno_only, species_only_in) #only in 1st vector## character(0)length(intersect(entorno_only, species_only_in)) #how many## [1] 242.11 Encontrando espécies compartilhadas ENTRE DOIS GRUPOS
Encontra espécies que são compartilhadas (ocorrem em TODAS AS LINHAS) dentro do grupo analisado. Mas, não significa que elas estejam apenas nestas linhas. A função any() indica que QUALQUER linha em comum entre duas colunas faz com que elas tenha esses colunas compartilhadas. Já a função all() indica que, para duas colunas terem linhas compartilhadas os grupos das colunas tem que compartilhar TODAS as suas linhas. Uma análise sobre esse loop
# Get the rows corresponding to A and B (and C)
rows_A <- grep("A", rownames(data))
rows_B <- grep("B", rownames(data))
rows_C <- grep("C", rownames(data))
rows <- as.vector(cbind(c(rows_A, rows_B, rows_C)))
# Initialize an empty vector to store species that A and B have in common
species_in <- character(0)
# Iterate over each column
for (col in colnames(data)) {
# Check if the species has at least one non-zero value in both A and B rows
if (any(data[rows_A, col] != 0) && any(data[rows_B, col] != 0) && any(data[rows_C, col] != 0)) {
species_in <- c(species_in, col)
}
}
# Print species that A and B have in common
print(species_in)## [1] "Bra.angularis" "Lepadella.sp" "Lecane.sp" "Lec.leontina" "Lec.bulla"
## [6] "Lec.cornuta" "Bra.urceolaris" "Lec.quadridentata" "Lec.kluchor" "Lec.lunaris"
## [11] "Rotaria.sp" "Aspelta.sp" "Lec.crepida" "Lec.ovalis" "Lec.aculeata"
## [16] "Bdelloidea" "Con.unicornis" "Macrothrix.sp" "Alonella.sp" "Nauplii"
## [21] "Chy.eurynotus" "Cyclopoida" "Copepodite" "Colurella.sp" "Mytilina.sp"data## Bra.angularis Lepadella.sp Lecane.sp Lec.leontina Lec.bulla Lec.cornuta Lec.curvicornis Notholca.sp
## BB1-P01 0.0000000 0.0000000 0.0000000 0 1.0666667 0.0000000 0.0000000 0.0000000
## BB1-P02 0.5333333 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB1-P03 0.5333333 0.1777778 0.0000000 0 0.3555556 0.0000000 0.0000000 0.0000000
## BB2-P01 0.0000000 0.5333333 0.0000000 0 0.8888889 0.0000000 0.0000000 0.0000000
## BB2-P02 0.0000000 0.1777778 0.0000000 0 0.1777778 0.0000000 0.0000000 0.0000000
## BB2-P03 0.0000000 0.1777778 0.0000000 0 0.0000000 0.0000000 0.0000000 0.0000000
## BB3-P01 0.0000000 0.0000000 0.1777778 0 0.5333333 0.1777778 0.0000000 0.0000000
## BB3-P02 0.0000000 0.0000000 0.0000000 0 1.6000000 0.0000000 0.0000000 0.0000000
## BB3-P03 0.0000000 0.0000000 0.0000000 0 1.4222222 0.1777778 0.0000000 0.0000000
## BB4-P01 0.0000000 0.0000000 0.0000000 0 1.4222222 0.0000000 0.1777778 0.0000000
## BB4-P02 0.0000000 0.0000000 0.0000000 0 0.3555556 0.0000000 0.0000000 0.1777778
## Bra.urceolaris Trichocerca.sp Lec.quadridentata Lec.kluchor Lec.lunaris Rotaria.sp Aspelta.sp Lec.furcata
## BB1-P01 0.1777778 0 0 0.1777778 0.3555556 0.0000000 0.1777778 0
## BB1-P02 0.0000000 0 0 0.0000000 0.7111111 1.0666667 0.0000000 0
## BB1-P03 0.0000000 0 0 0.0000000 1.0666667 0.0000000 0.0000000 0
## BB2-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB2-P02 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB2-P03 0.0000000 0 0 0.0000000 0.1777778 0.0000000 0.0000000 0
## BB3-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P02 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB3-P03 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P01 0.0000000 0 0 0.0000000 0.0000000 0.0000000 0.0000000 0
## BB4-P02 0.0000000 0 0 0.0000000 0.0000000 0.3555556 0.0000000 0
## Col.geophila Lep.dactyliseta Bra.calyciflorus Bra.caudatus Lec.crepida Pla.patulus Lec.ovalis Lec.elasma
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.0000000 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.1777778 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Pla.quadricornis Lec.aculeata Ker.tropica Bdelloidea Ker.lenzi Con.unicornis Alo.dadayi Macrothrix.sp
## BB1-P01 0 1.6000000 0 0.0000000 0 0.0000000 0 0.1777778
## BB1-P02 0 0.8888889 0 0.0000000 0 0.0000000 0 0.0000000
## BB1-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P01 0 0.1777778 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P02 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB2-P03 0 0.0000000 0 0.0000000 0 0.0000000 0 0.0000000
## BB3-P01 0 0.0000000 0 0.1777778 0 0.0000000 0 0.0000000
## BB3-P02 0 0.0000000 0 0.3555556 0 0.5333333 0 0.0000000
## BB3-P03 0 0.0000000 0 0.7111111 0 0.3555556 0 0.0000000
## BB4-P01 0 0.0000000 0 3.5555556 0 0.0000000 0 0.0000000
## BB4-P02 0 0.0000000 0 1.2444444 0 0.0000000 0 0.0000000
## Alonella.sp Nauplii Chy.eurynotus Cyclopoida Mac.collinsi Copepodite Paracyclops.sp Asc.ecaudis Lep.ovalis
## BB1-P01 0 0.7111111 0.3555556 0.0000000 0 0.0000000 0 0 0
## BB1-P02 0 0.8888889 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB1-P03 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P01 0 0.8888889 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB2-P02 0 0.7111111 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB2-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P01 0 0.0000000 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB3-P02 0 0.3555556 0.0000000 0.1777778 0 0.1777778 0 0 0
## BB3-P03 0 0.3555556 0.0000000 0.0000000 0 0.0000000 0 0 0
## BB4-P01 0 1.4222222 0.0000000 0.0000000 0 0.1777778 0 0 0
## BB4-P02 0 0.1777778 0.0000000 0.0000000 0 0.0000000 0 0 0
## Polyarthra.sp Bra.havanaensis Hexarthra.sp Lec.hastata Tri.tetractis Ker.serrulata Notodiaptomus.sp Bra.falcatus
## BB1-P01 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0
## Fil.longiseta Myt.crassipes Lep.patella Euchlanis.sp Colurella.sp Mytilina.sp Lec.monostyla Lec.ligona
## BB1-P01 0 0 0 0 0.0000000 0 0 0
## BB1-P02 0 0 0 0 0.0000000 0 0 0
## BB1-P03 0 0 0 0 0.1777778 0 0 0
## BB2-P01 0 0 0 0 0.0000000 0 0 0
## BB2-P02 0 0 0 0 0.0000000 0 0 0
## BB2-P03 0 0 0 0 0.0000000 0 0 0
## BB3-P01 0 0 0 0 0.0000000 0 0 0
## BB3-P02 0 0 0 0 0.0000000 0 0 0
## BB3-P03 0 0 0 0 0.0000000 0 0 0
## BB4-P01 0 0 0 0 0.0000000 0 0 0
## BB4-P02 0 0 0 0 0.0000000 0 0 0
## Ascomorpha.sp Pom.sulcata Harpacticoida Epi.senta Rot.neptunia Dis.aculeata Lec.luna Euc.dilatata
## BB1-P01 0.1777778 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P02 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB1-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB2-P01 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P02 0.0000000 0.3555556 0.0000000 0.0000000 0 0 0 0
## BB2-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB3-P02 0.0000000 0.0000000 0.1777778 0.0000000 0 0 0 0
## BB3-P03 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P01 0.0000000 0.0000000 0.0000000 0.0000000 0 0 0 0
## BB4-P02 0.0000000 0.0000000 0.0000000 0.1777778 0 0 0 0
## Bea.eudactylota Chy.sphaericus Lec.hornemanni Dia.birgei Mac.subquadratus Bra.quadridentatus Pol.vulgaris
## BB1-P01 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0
## Pol.bicerca Dicranop.sp Asc.saltans Moi.minuta Alo.hamulata Mac.laticornis Mac.mira Pro.similis Alo.pulchella
## BB1-P01 0 0 0 0 0 0 0 0 0
## BB1-P02 0 0 0 0 0 0 0 0 0
## BB1-P03 0 0 0 0 0 0 0 0 0
## BB2-P01 0 0 0 0 0 0 0 0 0
## BB2-P02 0 0 0 0 0 0 0 0 0
## BB2-P03 0 0 0 0 0 0 0 0 0
## BB3-P01 0 0 0 0 0 0 0 0 0
## BB3-P02 0 0 0 0 0 0 0 0 0
## BB3-P03 0 0 0 0 0 0 0 0 0
## BB4-P01 0 0 0 0 0 0 0 0 0
## BB4-P02 0 0 0 0 0 0 0 0 0
## Dia.spinulosum Chydorus.sp Ily.spinifer Dun.odontoplax
## BB1-P01 0 0 0 0
## BB1-P02 0 0 0 0
## BB1-P03 0 0 0 0
## BB2-P01 0 0 0 0
## BB2-P02 0 0 0 0
## BB2-P03 0 0 0 0
## BB3-P01 0 0 0 0
## BB3-P02 0 0 0 0
## BB3-P03 0 0 0 0
## BB4-P01 0 0 0 0
## BB4-P02 0 0 0 0
## [ reached 'max' / getOption("max.print") -- omitted 38 rows ]Uma análise feita pelo ChatGPT sobre o loop “for…{…if{…}}” acima e os outros anteriores é apresentada aqui
2.12 Diagrama de Venn
Primeiro é necessário criar uma matriz binária para os valores das colunas nas linhas. Isso é feito a seguir.
m_venn <- as.data.frame(t(m_avg_csv))
m_venn## BB CA EN RE
## Bra.angularis 0.059259259 0.039506173 0.00000000 0.0000000
## Lepadella.sp 0.108641975 0.543209877 0.00000000 0.0000000
## Lecane.sp 0.009876543 0.029629630 0.00000000 0.0000000
## Lec.leontina 0.019753086 0.049382716 0.26666667 0.0000000
## Lec.bulla 0.780246914 0.997530864 8.96000000 1.0666667
## Lec.cornuta 0.029629630 0.049382716 0.42666667 0.0000000
## Lec.curvicornis 0.009876543 0.000000000 0.32000000 0.0000000
## Notholca.sp 0.009876543 0.000000000 0.00000000 0.0000000
## Bra.urceolaris 0.009876543 0.019753086 0.00000000 0.0000000
## Trichocerca.sp 0.000000000 0.187654321 0.00000000 0.5333333
## Lec.quadridentata 0.009876543 0.049382716 0.00000000 0.1777778
## Lec.kluchor 0.009876543 0.059259259 0.00000000 0.0000000
## Lec.lunaris 0.148148148 0.108641975 0.32000000 0.8888889
## Rotaria.sp 0.167901235 0.316049383 3.52000000 10.1333333
## Aspelta.sp 0.009876543 0.098765432 0.00000000 0.0000000
## Lec.furcata 0.000000000 0.069135802 1.22666667 0.1777778
## Col.geophila 0.000000000 0.019753086 0.00000000 0.0000000
## Lep.dactyliseta 0.000000000 0.365432099 0.00000000 0.0000000
## Bra.calyciflorus 0.000000000 0.009876543 0.00000000 0.0000000
## Bra.caudatus 0.000000000 0.019753086 0.00000000 0.0000000
## Lec.crepida 0.009876543 0.049382716 0.05333333 0.0000000
## Pla.patulus 0.000000000 0.029629630 0.42666667 0.0000000
## Lec.ovalis 0.019753086 0.009876543 0.00000000 0.0000000
## Lec.elasma 0.000000000 0.009876543 0.00000000 0.0000000
## Pla.quadricornis 0.000000000 0.009876543 0.05333333 0.0000000
## Lec.aculeata 0.148148148 0.069135802 0.00000000 0.0000000
## Ker.tropica 0.000000000 0.009876543 0.00000000 0.0000000
## Bdelloidea 1.743209877 1.619753086 0.00000000 0.0000000
## Ker.lenzi 0.000000000 0.039506173 0.00000000 0.0000000
## Con.unicornis 0.049382716 0.009876543 0.00000000 0.0000000
## Alo.dadayi 0.000000000 0.088888889 0.00000000 0.0000000
## Macrothrix.sp 0.009876543 0.009876543 0.16000000 0.0000000
## Alonella.sp 0.049382716 0.039506173 0.32000000 0.0000000
## Nauplii 0.597530864 2.597530864 17.01333333 2.1333333
## Chy.eurynotus 0.039506173 0.039506173 0.00000000 0.0000000
## Cyclopoida 0.019753086 0.158024691 0.42666667 0.0000000
## Mac.collinsi 0.000000000 0.019753086 0.00000000 0.0000000
## Copepodite 0.088888889 0.434567901 1.17333333 0.7111111
## Paracyclops.sp 0.000000000 0.019753086 0.00000000 0.0000000
## Asc.ecaudis 0.000000000 0.009876543 0.00000000 0.0000000
## Lep.ovalis 0.000000000 0.039506173 0.16000000 0.0000000
## Polyarthra.sp 0.000000000 0.009876543 0.00000000 0.0000000
## Bra.havanaensis 0.000000000 0.009876543 0.00000000 0.0000000
## Hexarthra.sp 0.000000000 0.009876543 0.00000000 0.0000000
## Lec.hastata 0.000000000 0.009876543 0.16000000 0.0000000
## Tri.tetractis 0.000000000 0.039506173 0.16000000 0.0000000
## Ker.serrulata 0.000000000 0.009876543 0.00000000 0.0000000
## Notodiaptomus.sp 0.000000000 0.019753086 0.00000000 0.0000000
## Bra.falcatus 0.000000000 0.009876543 2.40000000 0.5333333
## Fil.longiseta 0.000000000 0.009876543 0.00000000 0.0000000
## Myt.crassipes 0.000000000 0.039506173 0.00000000 0.0000000
## Lep.patella 0.000000000 0.088888889 5.81333333 1.9555556
## Euchlanis.sp 0.000000000 0.148148148 0.00000000 0.0000000
## Colurella.sp 0.009876543 0.424691358 0.00000000 0.0000000
## Mytilina.sp 0.019753086 0.088888889 0.00000000 0.0000000
## Lec.monostyla 0.000000000 0.039506173 0.10666667 0.0000000
## Lec.ligona 0.000000000 0.088888889 0.00000000 0.0000000
## Ascomorpha.sp 0.009876543 0.000000000 0.00000000 0.0000000
## Pom.sulcata 0.039506173 0.000000000 0.00000000 0.0000000
## Harpacticoida 0.009876543 0.000000000 0.00000000 0.0000000
## Epi.senta 0.009876543 0.000000000 0.00000000 0.0000000
## Rot.neptunia 0.000000000 0.000000000 0.16000000 0.0000000
## Dis.aculeata 0.000000000 0.000000000 0.48000000 0.3555556
## Lec.luna 0.000000000 0.000000000 0.26666667 0.0000000
## Euc.dilatata 0.000000000 0.000000000 0.10666667 0.0000000
## Bea.eudactylota 0.000000000 0.000000000 0.05333333 0.0000000
## Chy.sphaericus 0.000000000 0.000000000 0.37333333 0.0000000
## Lec.hornemanni 0.000000000 0.000000000 0.37333333 0.0000000
## Dia.birgei 0.000000000 0.000000000 0.21333333 0.0000000
## Mac.subquadratus 0.000000000 0.000000000 0.05333333 0.0000000
## Bra.quadridentatus 0.000000000 0.000000000 1.01333333 0.0000000
## Pol.vulgaris 0.000000000 0.000000000 1.92000000 0.0000000
## Pol.bicerca 0.000000000 0.000000000 5.81333333 0.0000000
## Dicranop.sp 0.000000000 0.000000000 6.88000000 0.0000000
## Asc.saltans 0.000000000 0.000000000 0.10666667 0.0000000
## Moi.minuta 0.000000000 0.000000000 0.26666667 0.0000000
## Alo.hamulata 0.000000000 0.000000000 0.10666667 0.0000000
## Mac.laticornis 0.000000000 0.000000000 0.26666667 0.0000000
## Mac.mira 0.000000000 0.000000000 0.21333333 0.0000000
## Pro.similis 0.000000000 0.000000000 0.05333333 0.0000000
## Alo.pulchella 0.000000000 0.000000000 0.37333333 0.0000000
## Dia.spinulosum 0.000000000 0.000000000 0.16000000 0.0000000
## Chydorus.sp 0.000000000 0.000000000 0.10666667 0.0000000
## Ily.spinifer 0.000000000 0.000000000 0.37333333 0.0000000
## Dun.odontoplax 0.000000000 0.000000000 0.05333333 0.0000000m_venn[m_venn !=0] <- 1 #matriz binária 2.13 Usando o pacote eulerr
library("eulerr")
#set.seed() #this seed changes the orientation of the sets
plot(euler(m_venn), counts = TRUE, fontface = 1) 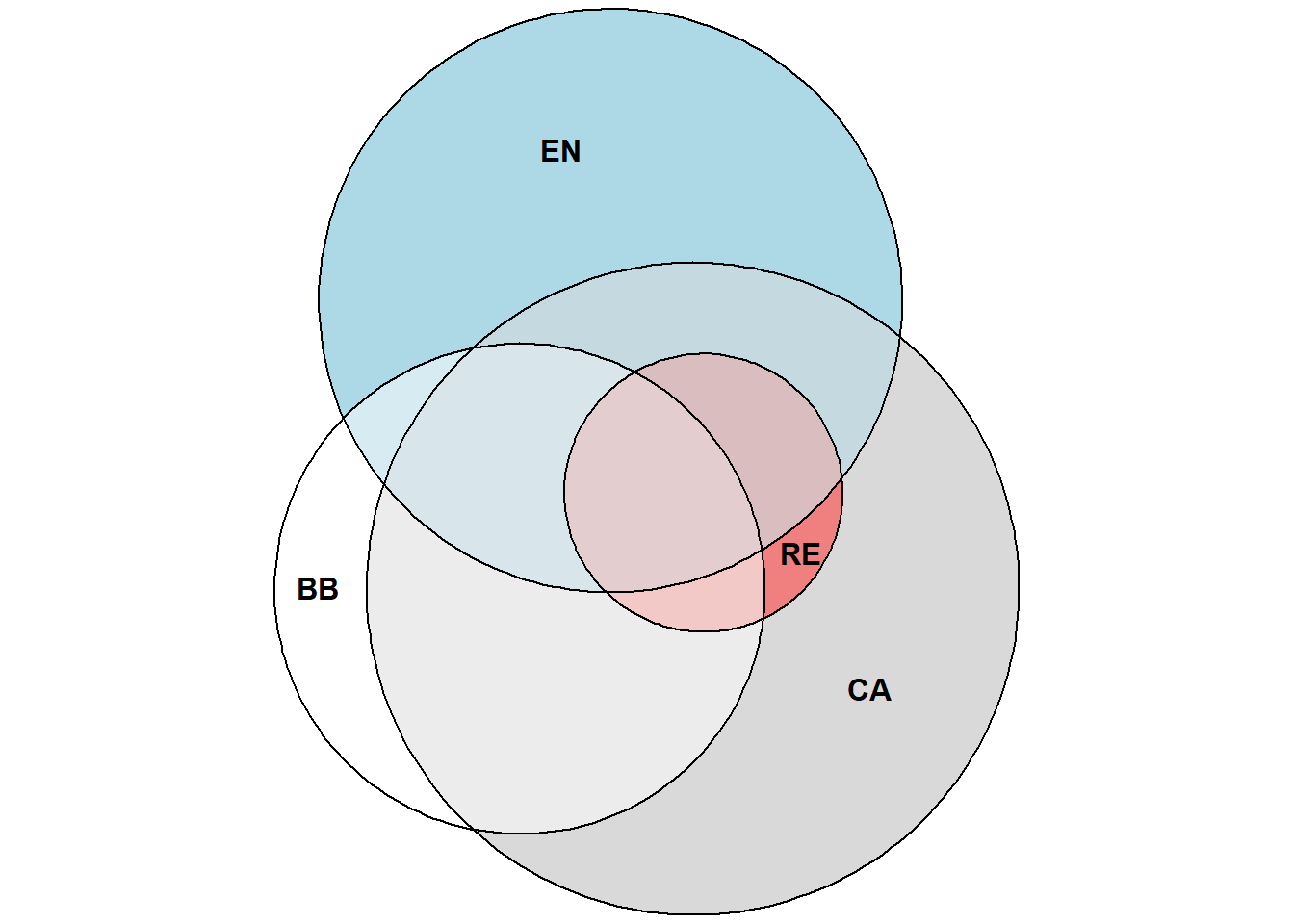
2.14 Usando o pacote VennDiagram5
# Load required libraries
library(VennDiagram)
library(ggvenn)
BB <- nrow(subset(m_venn, BB == 1))
CA <- nrow(subset(m_venn, CA == 1))
EN <- nrow(subset(m_venn, EN == 1))
RE <- nrow(subset(m_venn, RE == 1))
BB_CA <- nrow(subset(m_venn, BB == 1 & CA == 1))
BB_EN <- nrow(subset(m_venn, BB == 1 & EN == 1))
BB_RE <- nrow(subset(m_venn, BB == 1 & RE == 1))
CA_EN <- nrow(subset(m_venn, CA == 1 & EN == 1))
CA_RE <- nrow(subset(m_venn, CA == 1 & RE == 1))
EN_RE <- nrow(subset(m_venn, EN == 1 & RE == 1))
BB_CA_EN <- nrow(subset(m_venn, BB == 1 & CA == 1 & EN == 1))
BB_CA_RE <- nrow(subset(m_venn, BB == 1 & CA == 1 & RE == 1))
BB_EN_RE <- nrow(subset(m_venn, BB == 1 & EN == 1 & RE == 1))
CA_EN_RE <- nrow(subset(m_venn, CA == 1 & EN == 1 & RE == 1))
BB_CA_EN_RE <- nrow(subset(m_venn, BB == 1 & CA == 1 & EN == 1 & RE == 1))
grid.newpage()
draw.single.venn(area = BB, category = "BB")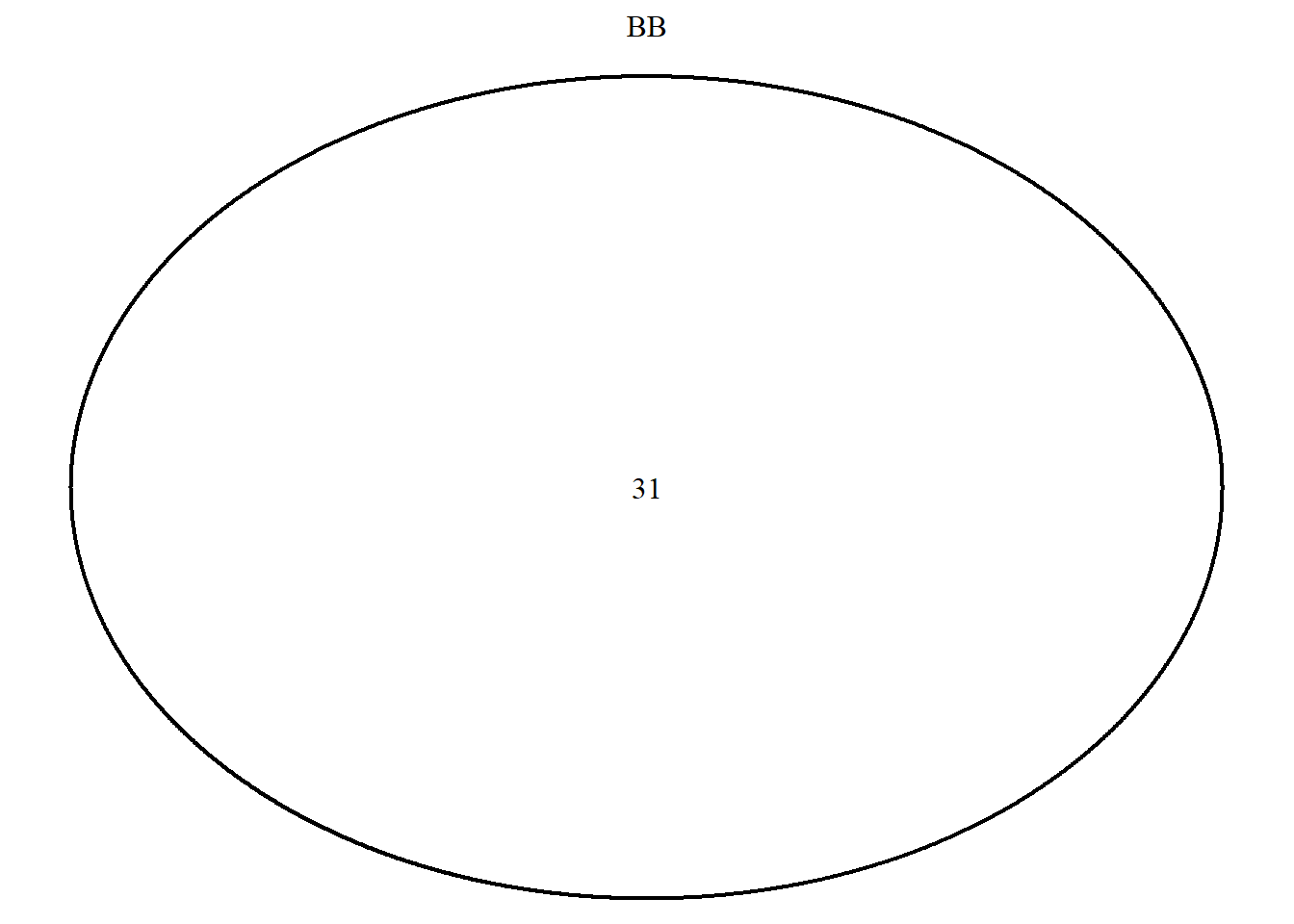
## (polygon[GRID.polygon.9838], polygon[GRID.polygon.9839], text[GRID.text.9840], text[GRID.text.9841])grid.newpage()
draw.single.venn(BB, category = "BB",
lty = "blank",
fill = "cornflower blue",
alpha = 0.5)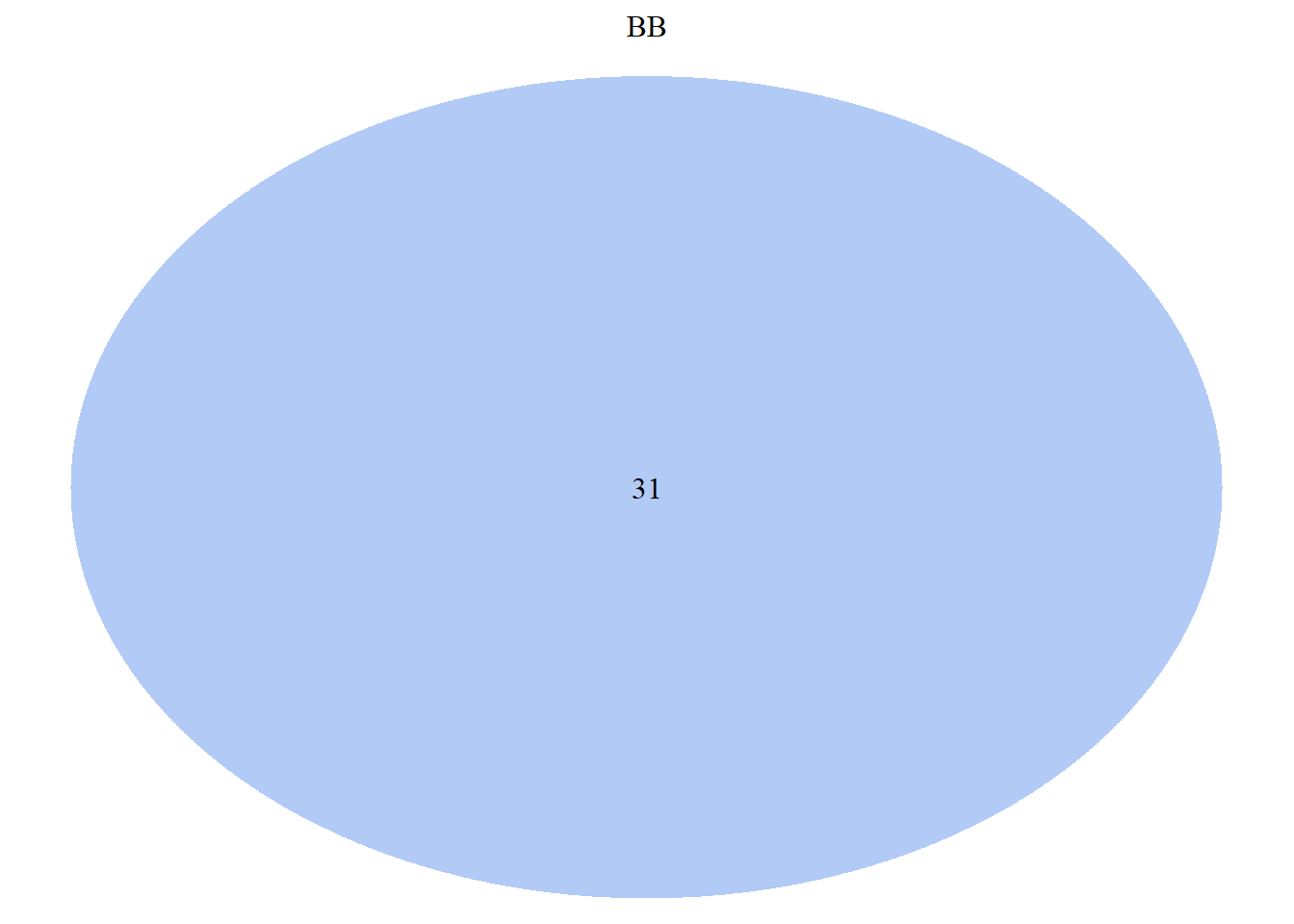
## (polygon[GRID.polygon.9842], polygon[GRID.polygon.9843], text[GRID.text.9844], text[GRID.text.9845])##lty - outline of cirlces, ## fill - colour, ## alpha - colour transparency
grid.newpage()
draw.pairwise.venn(BB, CA, BB_CA,
category = c("BB","CA"))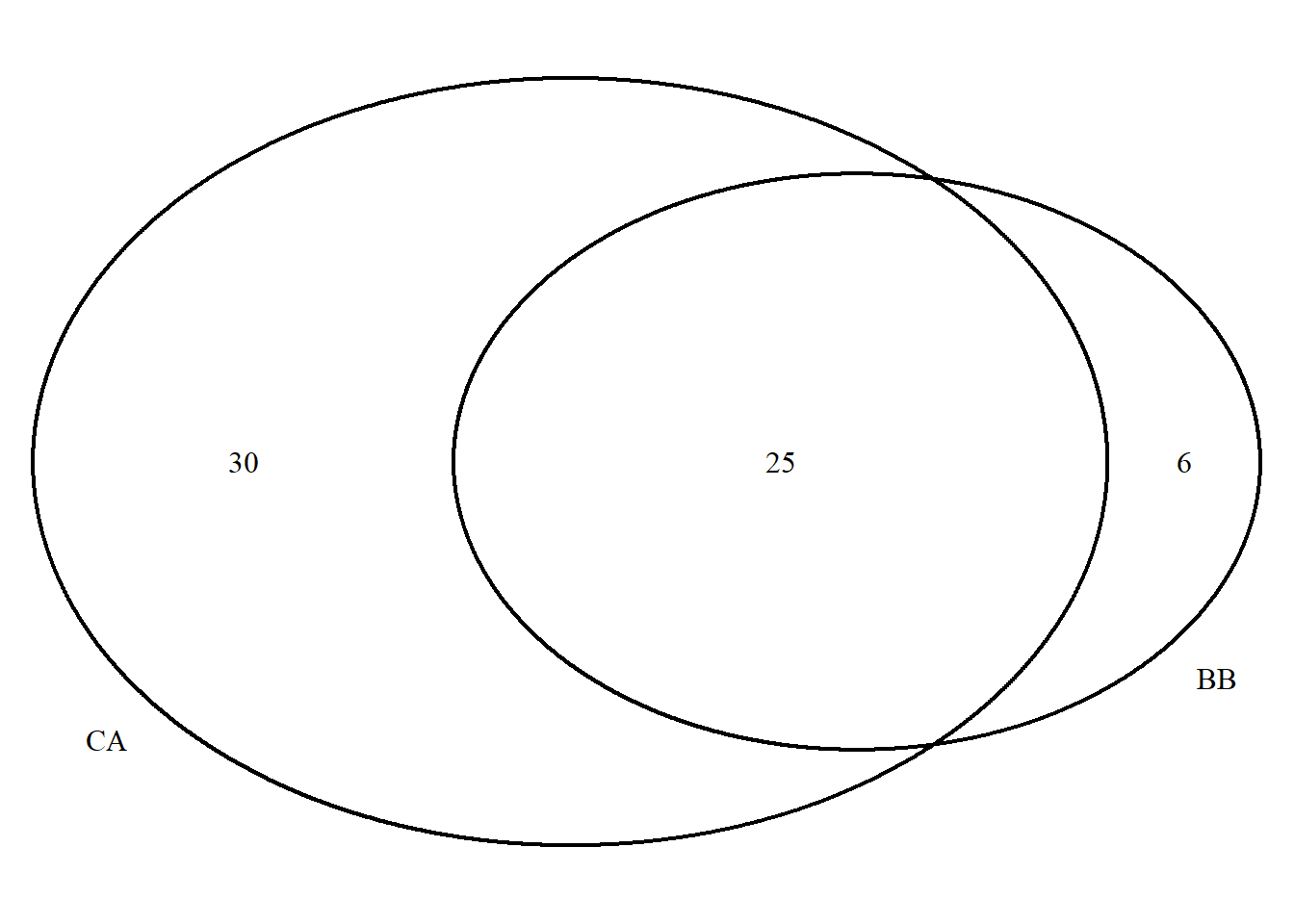
## (polygon[GRID.polygon.9846], polygon[GRID.polygon.9847], polygon[GRID.polygon.9848], polygon[GRID.polygon.9849], text[GRID.text.9850], text[GRID.text.9851], text[GRID.text.9852], text[GRID.text.9853], text[GRID.text.9854])grid.newpage()
draw.pairwise.venn(BB, CA, BB_CA,
category = c("BB","CA"),
lty = rep("blank",2),
fill = c("light blue", "pink"),
alpha = rep(0.5,2),
cat.pos = c(0,0),
cat.dist = rep(0.025,2))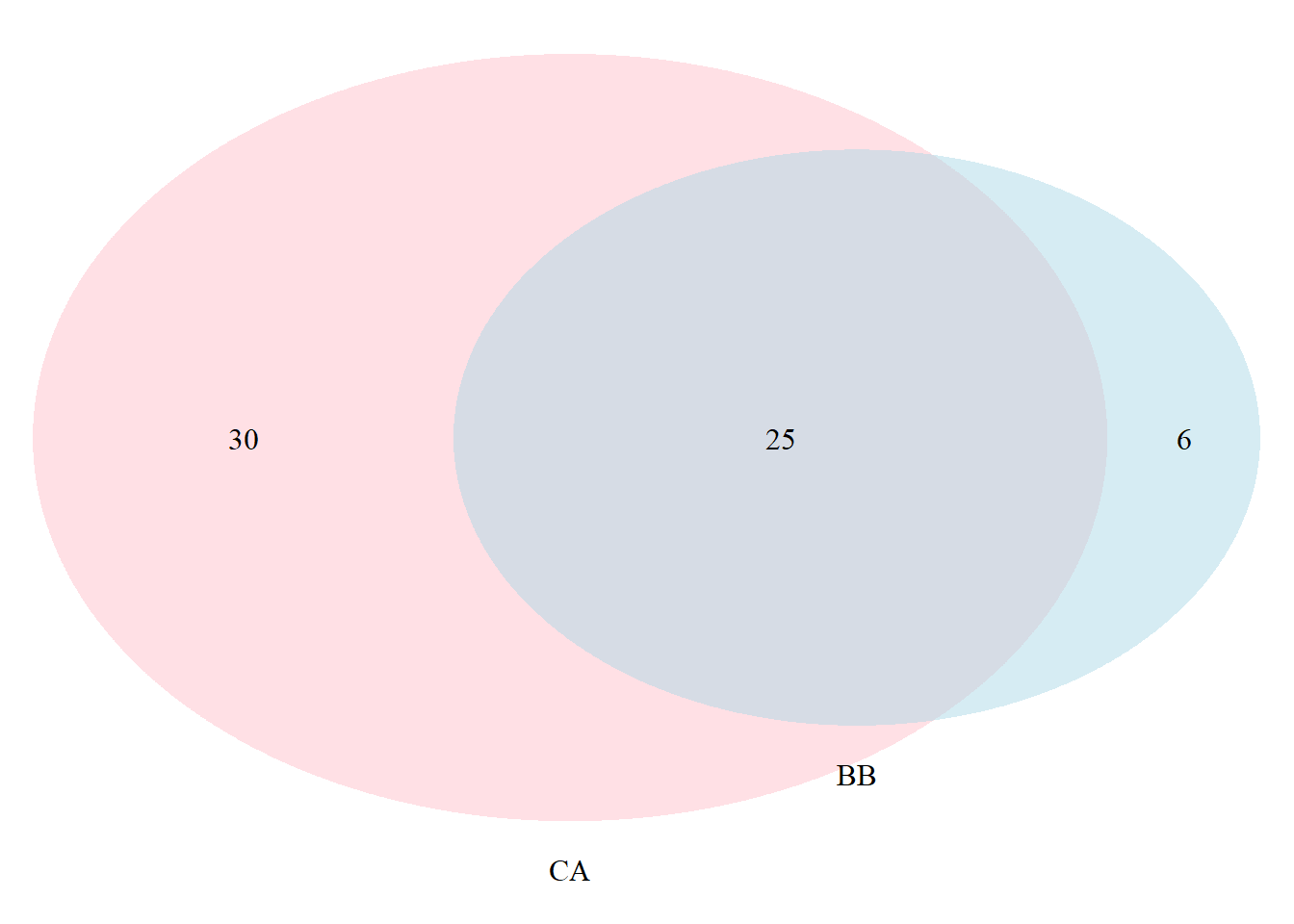
## (polygon[GRID.polygon.9855], polygon[GRID.polygon.9856], polygon[GRID.polygon.9857], polygon[GRID.polygon.9858], text[GRID.text.9859], text[GRID.text.9860], text[GRID.text.9861], text[GRID.text.9862], text[GRID.text.9863])## cat.pos - position of category titles, represented by degree from the
## middle of the circle
## cat.dist - distance of the category titles from the edge of the circle
grid.newpage()
draw.pairwise.venn(BB, CA, BB_CA,
category = c("BB", "CA"),
lty = rep("blank",2),
fill = c("light blue", "pink"),
alpha = rep(0.5, 2),
cat.pos = c(0,0),
cat.dist = rep(0.025, 2),
scaled = FALSE)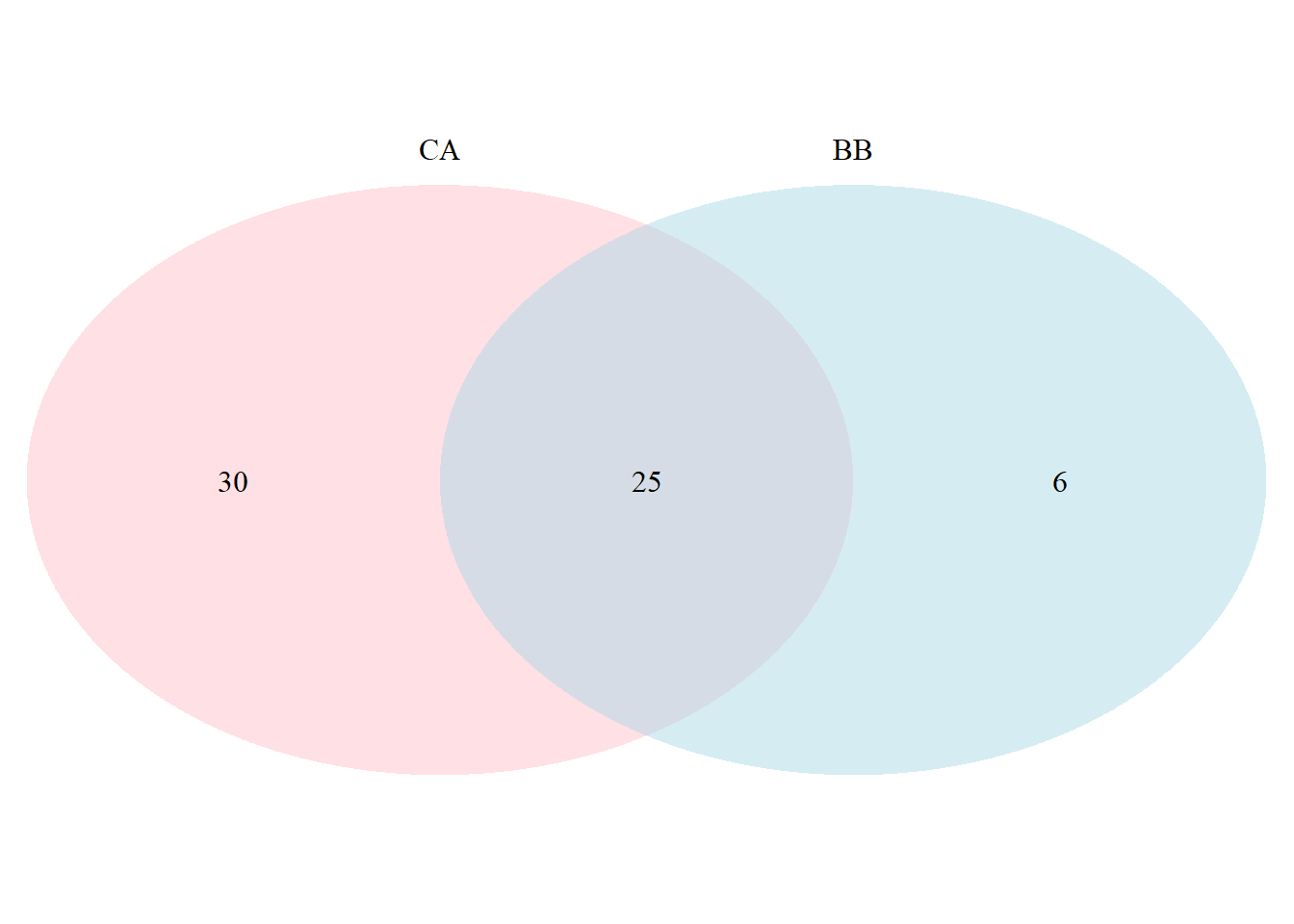
## (polygon[GRID.polygon.9864], polygon[GRID.polygon.9865], polygon[GRID.polygon.9866], polygon[GRID.polygon.9867], text[GRID.text.9868], text[GRID.text.9869], text[GRID.text.9870], text[GRID.text.9871], text[GRID.text.9872])## scaled - TRUE for scaled or FALSE for unscaled cirlces
grid.newpage()
draw.pairwise.venn(area1 = BB, area2 = CA, cross.area = 0,
category = c("BB","CA"),
lty = rep("blank",2),
fill = c("light blue", "green"),
alpha = rep(0.5, 2),
cat.pos = c(0, 180),
euler.d = TRUE, sep.dist = 0.03,
rotation.degree = 45)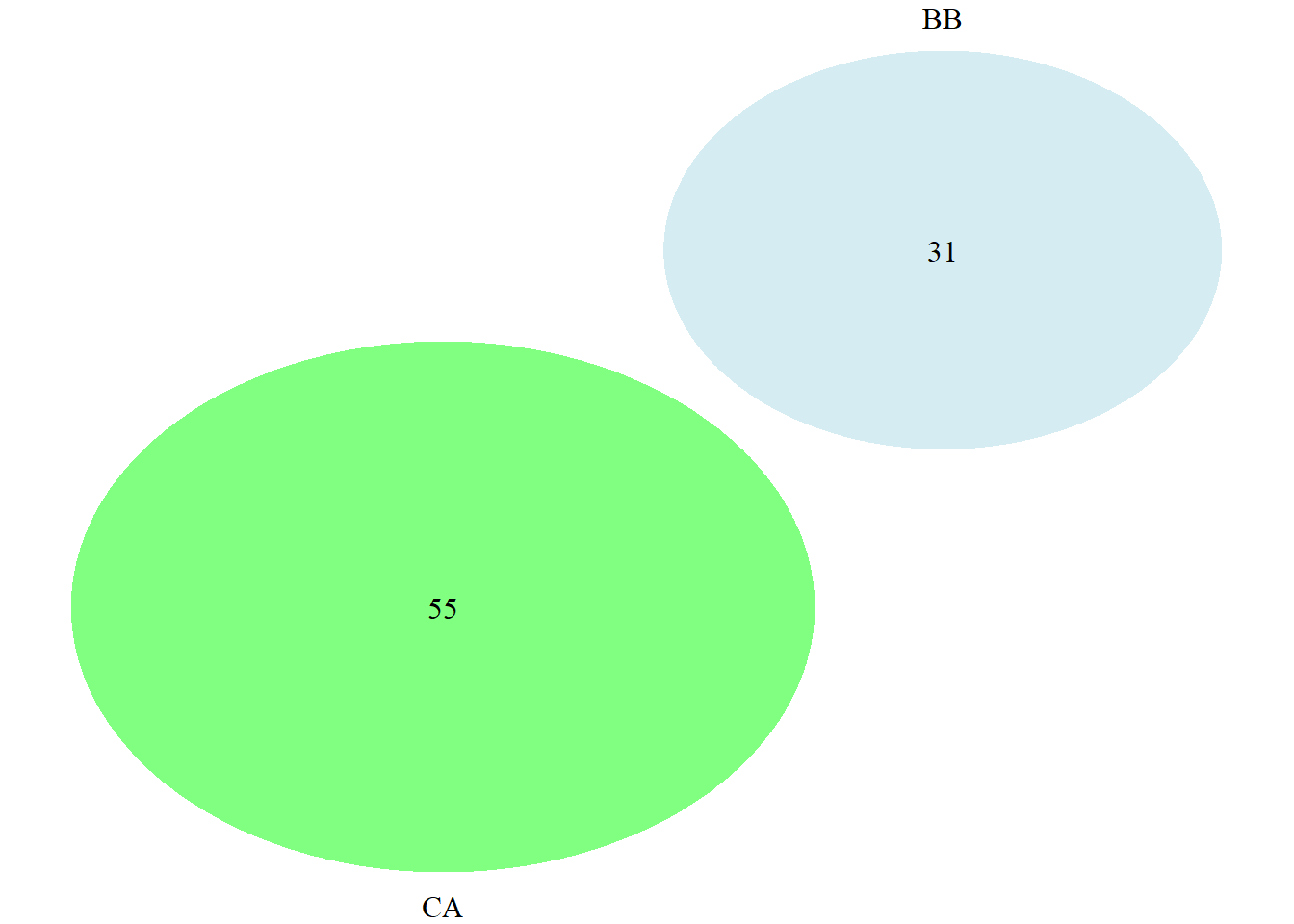
## (polygon[GRID.polygon.9873], polygon[GRID.polygon.9874], polygon[GRID.polygon.9875], polygon[GRID.polygon.9876], text[GRID.text.9877], text[GRID.text.9878], text[GRID.text.9879], text[GRID.text.9880])## euler.d - TRUE for movable circles; FALSE for unmovable circles. Must be
## TRUE to have space between non-overlapping circles.
## sep.dist - distance between circles
## rotation.degree - degrees the diagram is rotated
grid.newpage()
draw.triple.venn(area1 = BB, area2 = CA, area3 = EN,
n12 = BB_CA, n23 = CA_EN, n13 = BB_EN,
n123 = BB_CA_EN,
category = c("Rio Barro Branco", "Rio Caiana", "Entorno da REBio"),
lty = "blank",
fill = c("skyblue", "pink1", "mediumorchid"),
scaled = TRUE)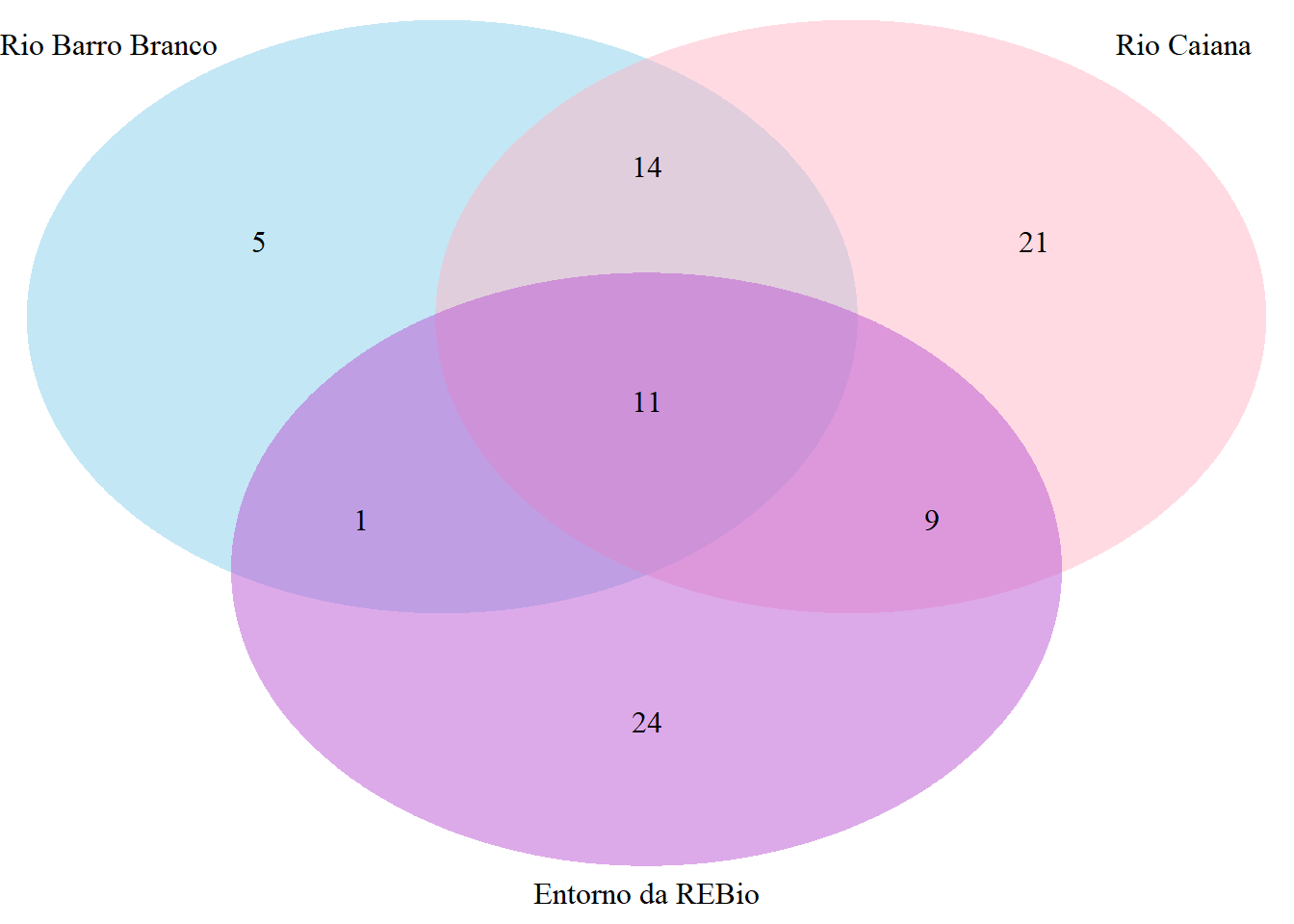
## (polygon[GRID.polygon.9881], polygon[GRID.polygon.9882], polygon[GRID.polygon.9883], polygon[GRID.polygon.9884], polygon[GRID.polygon.9885], polygon[GRID.polygon.9886], text[GRID.text.9887], text[GRID.text.9888], text[GRID.text.9889], text[GRID.text.9890], text[GRID.text.9891], text[GRID.text.9892], text[GRID.text.9893], text[GRID.text.9894], text[GRID.text.9895], text[GRID.text.9896])grid.newpage()
draw.quad.venn(area1 = BB, area2 = CA, area3 = EN, area4 = RE,
n12 = BB_CA, n23 = CA_EN, n13 = BB_EN,
n14 = BB_RE, n24 = CA_RE, n34 = EN_RE,
n123 = BB_CA_EN, n1234 = BB_CA_EN_RE,
n124 = BB_CA_RE, n134 = BB_EN_RE,
n234 = CA_EN_RE,
category = c("BB", "CA", "EN", "RE"),
lty = "blank",
fill = c("skyblue", "pink1", "mediumorchid", "orange"),
scaled = TRUE)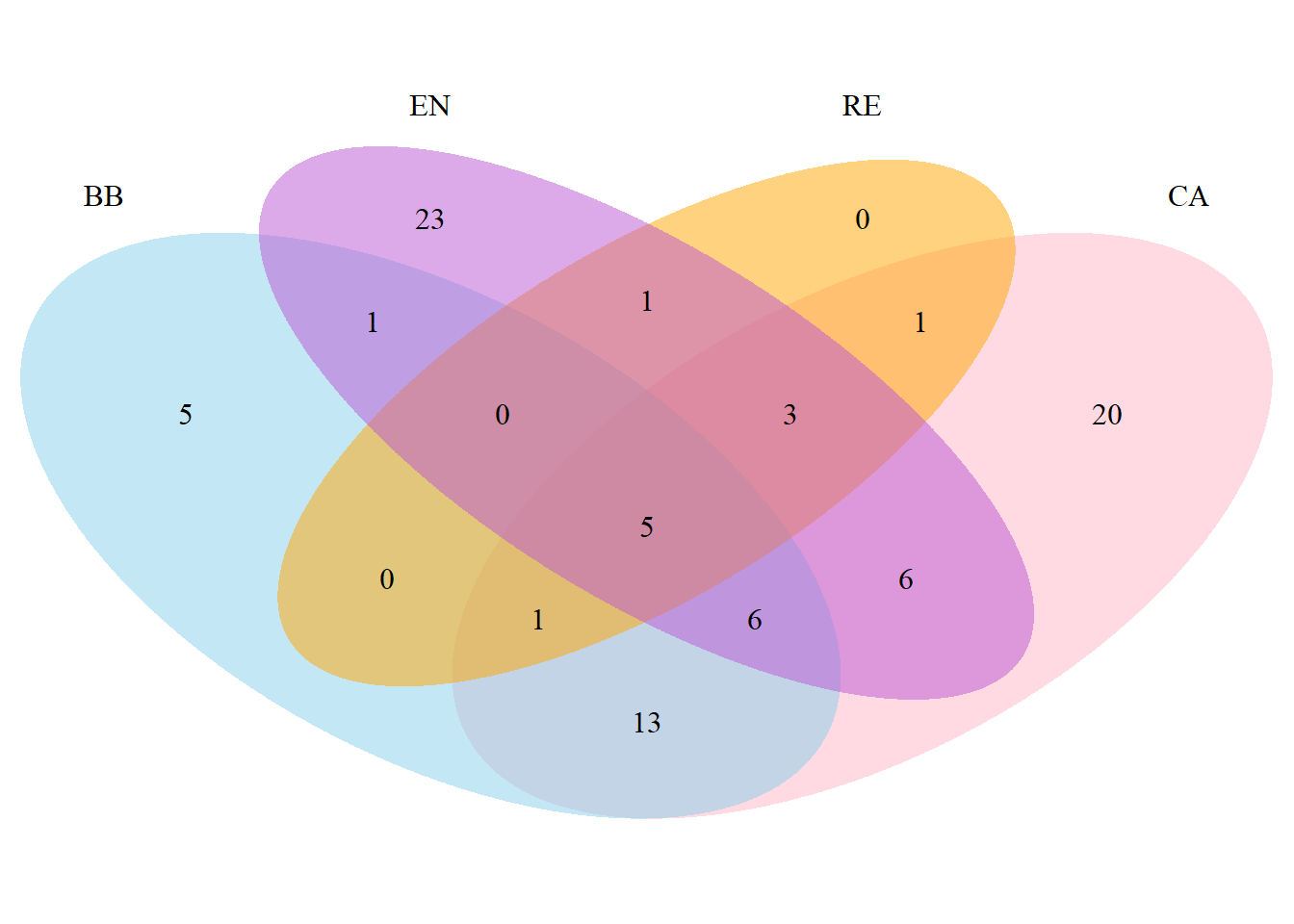
## (polygon[GRID.polygon.9897], polygon[GRID.polygon.9898], polygon[GRID.polygon.9899], polygon[GRID.polygon.9900], polygon[GRID.polygon.9901], polygon[GRID.polygon.9902], polygon[GRID.polygon.9903], polygon[GRID.polygon.9904], text[GRID.text.9905], text[GRID.text.9906], text[GRID.text.9907], text[GRID.text.9908], text[GRID.text.9909], text[GRID.text.9910], text[GRID.text.9911], text[GRID.text.9912], text[GRID.text.9913], text[GRID.text.9914], text[GRID.text.9915], text[GRID.text.9916], text[GRID.text.9917], text[GRID.text.9918], text[GRID.text.9919], text[GRID.text.9920], text[GRID.text.9921], text[GRID.text.9922], text[GRID.text.9923])2.15 Diagrama de Venn - BASEADO EM PALAVRAS
lista <- m_venn
lista## BB CA EN RE
## Bra.angularis 1 1 0 0
## Lepadella.sp 1 1 0 0
## Lecane.sp 1 1 0 0
## Lec.leontina 1 1 1 0
## Lec.bulla 1 1 1 1
## Lec.cornuta 1 1 1 0
## Lec.curvicornis 1 0 1 0
## Notholca.sp 1 0 0 0
## Bra.urceolaris 1 1 0 0
## Trichocerca.sp 0 1 0 1
## Lec.quadridentata 1 1 0 1
## Lec.kluchor 1 1 0 0
## Lec.lunaris 1 1 1 1
## Rotaria.sp 1 1 1 1
## Aspelta.sp 1 1 0 0
## Lec.furcata 0 1 1 1
## Col.geophila 0 1 0 0
## Lep.dactyliseta 0 1 0 0
## Bra.calyciflorus 0 1 0 0
## Bra.caudatus 0 1 0 0
## Lec.crepida 1 1 1 0
## Pla.patulus 0 1 1 0
## Lec.ovalis 1 1 0 0
## Lec.elasma 0 1 0 0
## Pla.quadricornis 0 1 1 0
## Lec.aculeata 1 1 0 0
## Ker.tropica 0 1 0 0
## Bdelloidea 1 1 0 0
## Ker.lenzi 0 1 0 0
## Con.unicornis 1 1 0 0
## Alo.dadayi 0 1 0 0
## Macrothrix.sp 1 1 1 0
## Alonella.sp 1 1 1 0
## Nauplii 1 1 1 1
## Chy.eurynotus 1 1 0 0
## Cyclopoida 1 1 1 0
## Mac.collinsi 0 1 0 0
## Copepodite 1 1 1 1
## Paracyclops.sp 0 1 0 0
## Asc.ecaudis 0 1 0 0
## Lep.ovalis 0 1 1 0
## Polyarthra.sp 0 1 0 0
## Bra.havanaensis 0 1 0 0
## Hexarthra.sp 0 1 0 0
## Lec.hastata 0 1 1 0
## Tri.tetractis 0 1 1 0
## Ker.serrulata 0 1 0 0
## Notodiaptomus.sp 0 1 0 0
## Bra.falcatus 0 1 1 1
## Fil.longiseta 0 1 0 0
## Myt.crassipes 0 1 0 0
## Lep.patella 0 1 1 1
## Euchlanis.sp 0 1 0 0
## Colurella.sp 1 1 0 0
## Mytilina.sp 1 1 0 0
## Lec.monostyla 0 1 1 0
## Lec.ligona 0 1 0 0
## Ascomorpha.sp 1 0 0 0
## Pom.sulcata 1 0 0 0
## Harpacticoida 1 0 0 0
## Epi.senta 1 0 0 0
## Rot.neptunia 0 0 1 0
## Dis.aculeata 0 0 1 1
## Lec.luna 0 0 1 0
## Euc.dilatata 0 0 1 0
## Bea.eudactylota 0 0 1 0
## Chy.sphaericus 0 0 1 0
## Lec.hornemanni 0 0 1 0
## Dia.birgei 0 0 1 0
## Mac.subquadratus 0 0 1 0
## Bra.quadridentatus 0 0 1 0
## Pol.vulgaris 0 0 1 0
## Pol.bicerca 0 0 1 0
## Dicranop.sp 0 0 1 0
## Asc.saltans 0 0 1 0
## Moi.minuta 0 0 1 0
## Alo.hamulata 0 0 1 0
## Mac.laticornis 0 0 1 0
## Mac.mira 0 0 1 0
## Pro.similis 0 0 1 0
## Alo.pulchella 0 0 1 0
## Dia.spinulosum 0 0 1 0
## Chydorus.sp 0 0 1 0
## Ily.spinifer 0 0 1 0
## Dun.odontoplax 0 0 1 0bb <- rownames(lista)[which(lista$BB !=0)]
ca <- rownames(lista)[which(lista$CA !=0)]
en <- rownames(lista)[which(lista$EN !=0)]
re <- rownames(lista)[which(lista$RE !=0)]
venn.diagram(#salva o diagrama em um arquivo
x = list(bb, ca, en, re),
category.names = c("BB" , "CA" , "EN", "RE"),
filename = 'fig-venn_diagramm.png',
height = 3000, width = 3000, resolution = 500,
disable.logging = T, scaled = T,
output=F,
lty = "blank",
fill = c("skyblue", "pink1", "mediumorchid", "orange"
))## INFO [2024-03-15 12:15:21] $x
## INFO [2024-03-15 12:15:21] list(bb, ca, en, re)
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $category.names
## INFO [2024-03-15 12:15:21] c("BB", "CA", "EN", "RE")
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $filename
## INFO [2024-03-15 12:15:21] [1] "fig-venn_diagramm.png"
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $height
## INFO [2024-03-15 12:15:21] [1] 3000
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $width
## INFO [2024-03-15 12:15:21] [1] 3000
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $resolution
## INFO [2024-03-15 12:15:21] [1] 500
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $disable.logging
## INFO [2024-03-15 12:15:21] T
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $scaled
## INFO [2024-03-15 12:15:21] T
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $output
## INFO [2024-03-15 12:15:21] F
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $lty
## INFO [2024-03-15 12:15:21] [1] "blank"
## INFO [2024-03-15 12:15:21]
## INFO [2024-03-15 12:15:21] $fill
## INFO [2024-03-15 12:15:21] c("skyblue", "pink1", "mediumorchid", "orange")
## INFO [2024-03-15 12:15:21]## [1] 1library("ggVennDiagram")
x <- list(
bb = rownames(lista)[which(lista$BB !=0)],
ca = rownames(lista)[which(lista$CA !=0)],
en = rownames(lista)[which(lista$EN !=0)],
re = rownames(lista)[which(lista$RE !=0)])
ggvenn(
x,
fill_color = c("#0073C2FF", "#EFC000FF", "#868686FF", "#CD534CFF"),
stroke_size = 0.5, set_name_size = 4
)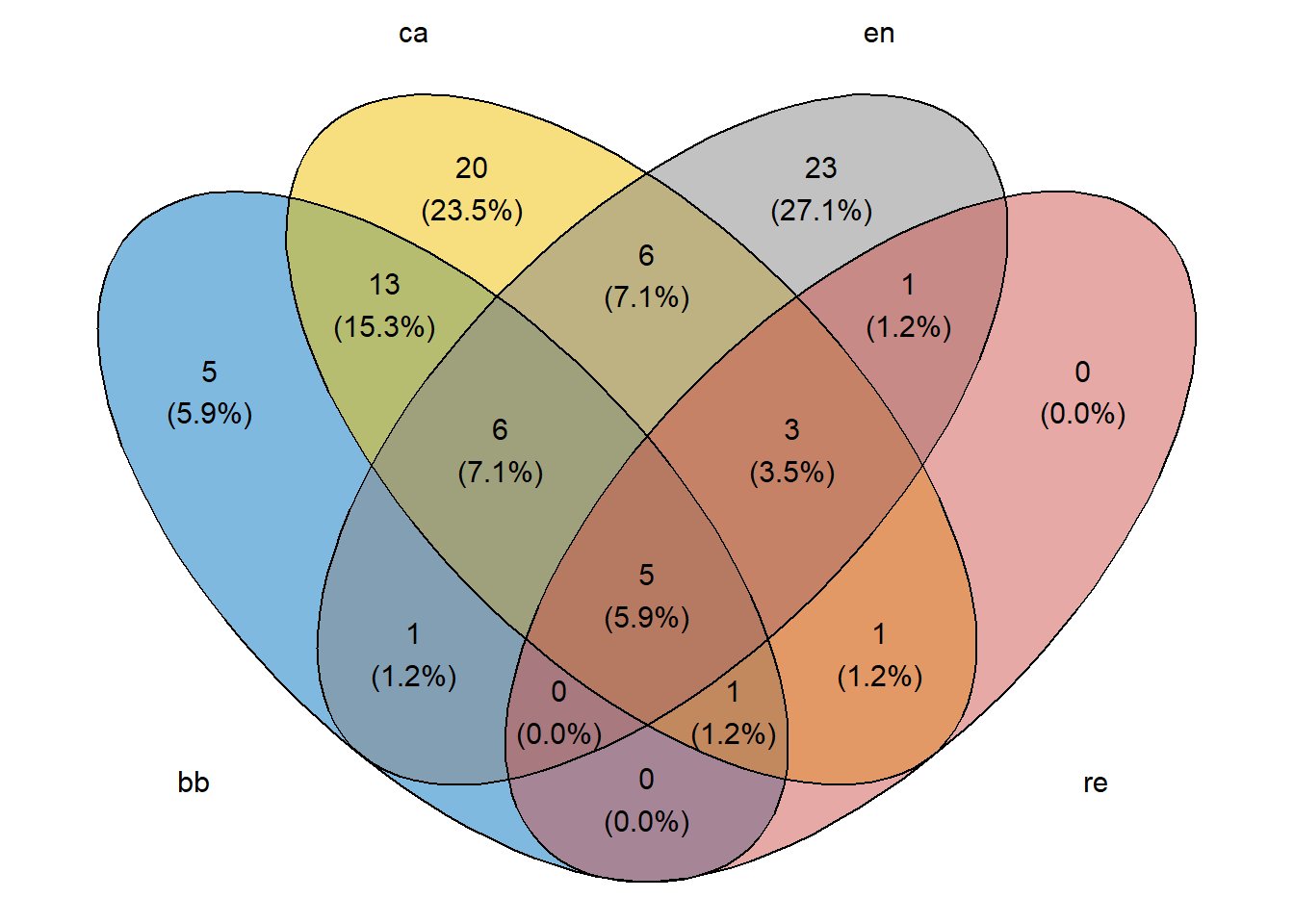
# Default plot
ggVennDiagram(
x, label_alpha = 0,
category.names = c("bb","ca","en", "re")
) +
ggplot2::scale_fill_gradient(low="yellow",high = "green")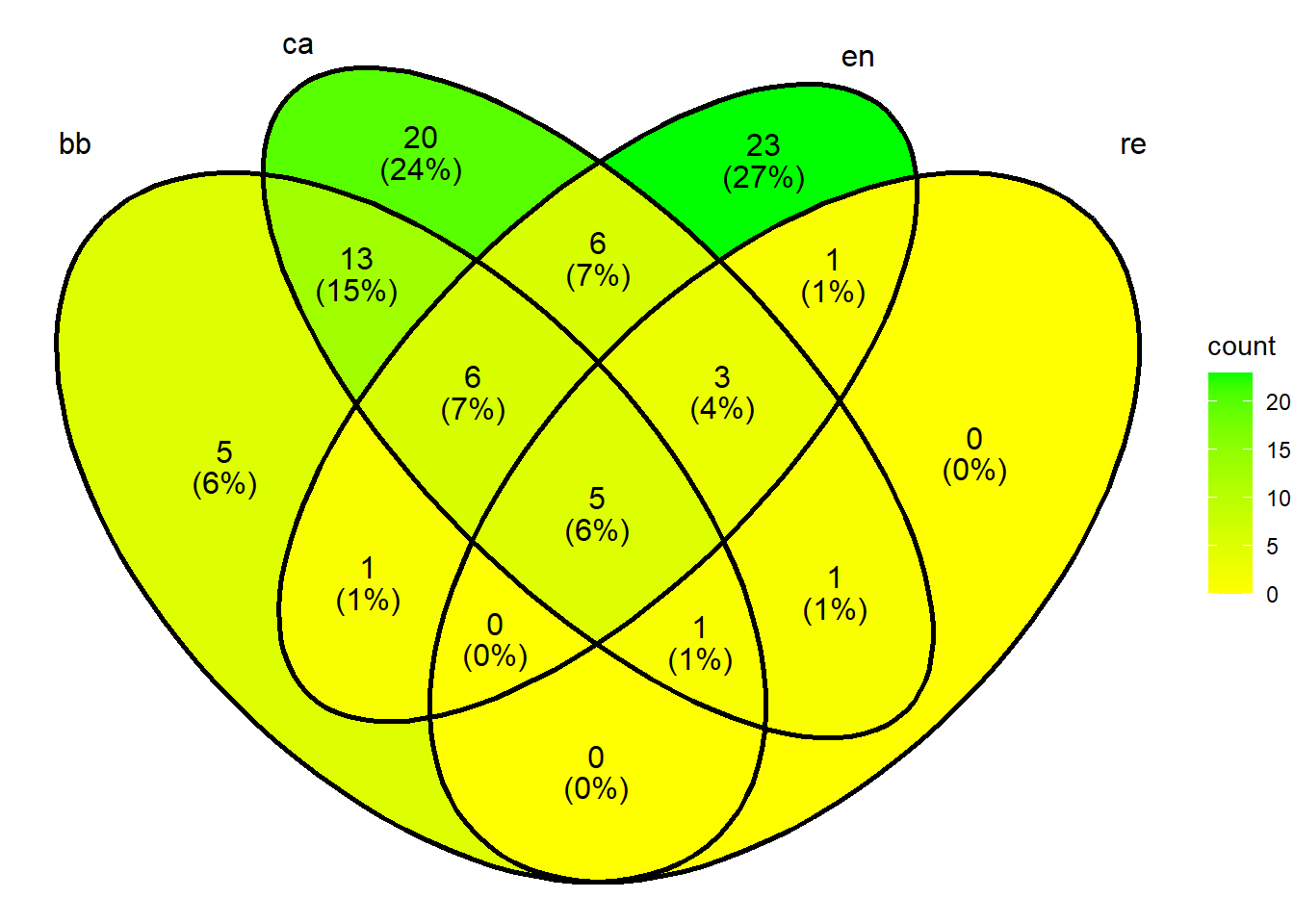
grid.newpage()
ggVennDiagram(x[1:2], label_alpha = 0)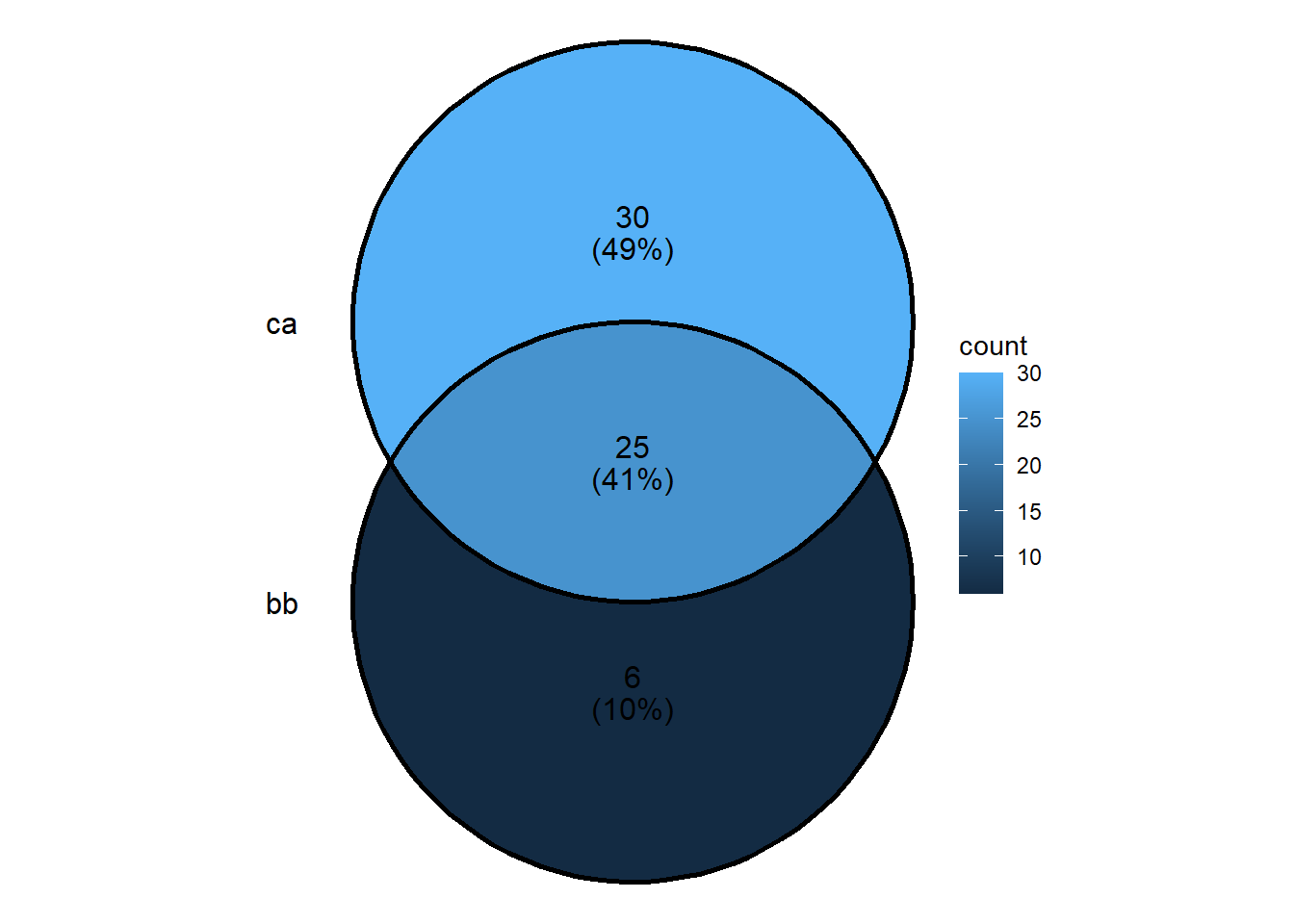
library("ggvenn")
grid.newpage()
ggvenn(x)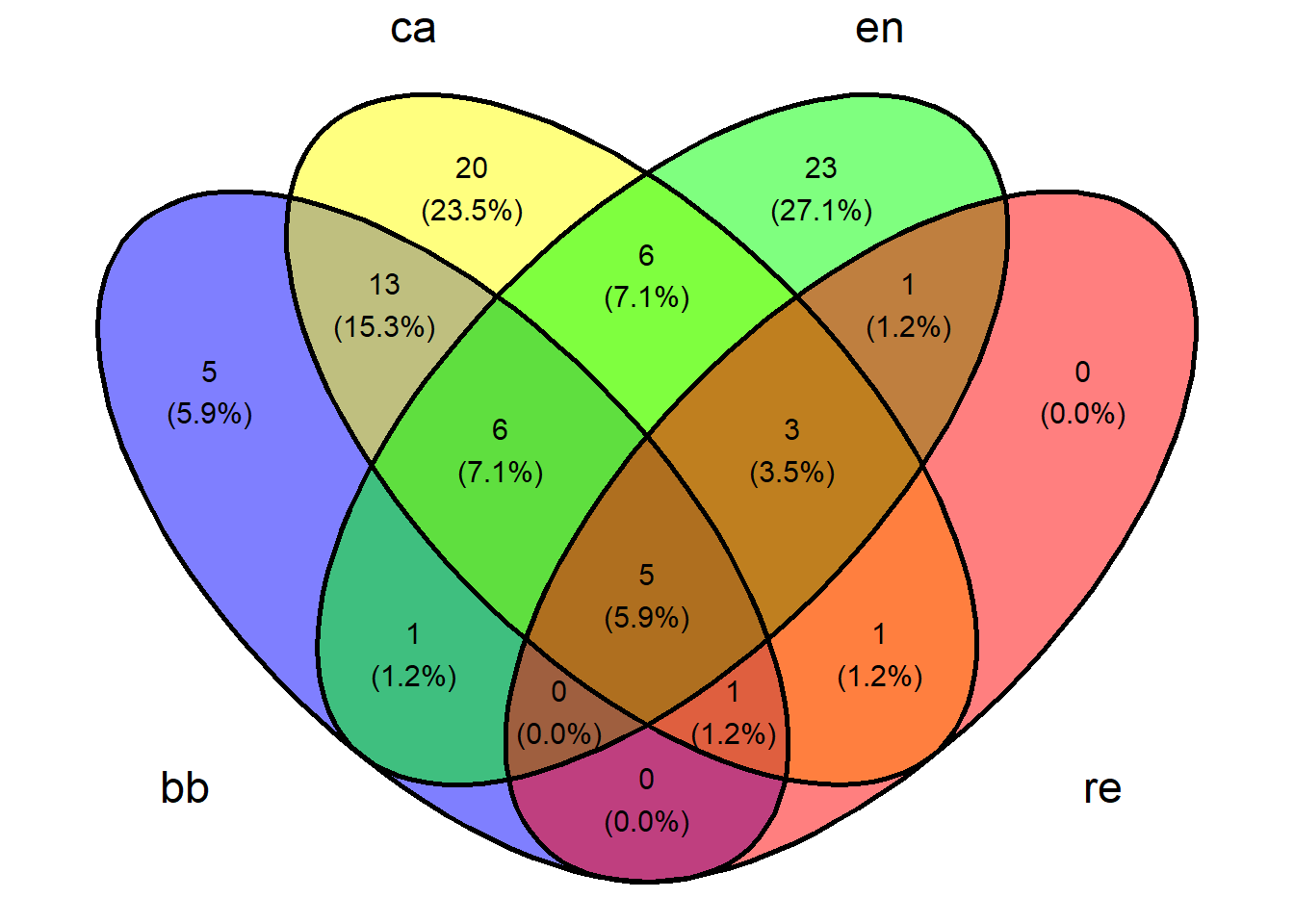
library("gplots")
grid.newpage()
v.table <- venn(x)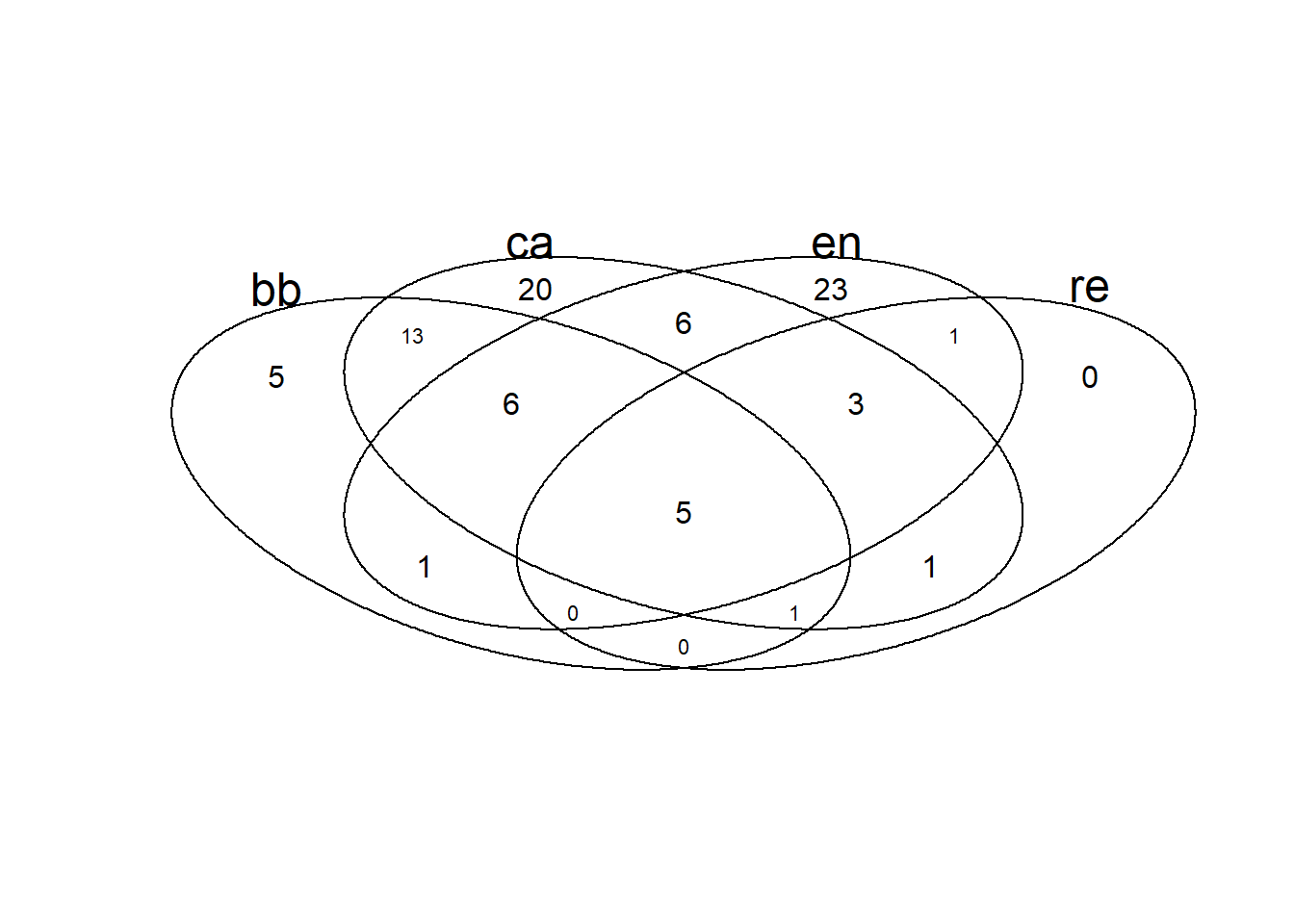
v.table <- as.data.frame(v.table)
print(v.table)## Warning in format.data.frame(if (omit) x[seq_len(n0), , drop = FALSE] else x, : data frame corrompido: colunas serão
## truncadas ou completadas com NAs## x.num x.bb x.ca x.en x.re
## 1 0 0 0 0 0
## 2 0 0 0 0 1
## 3 23 0 0 1 0
## 4 1 0 0 1 1
## 5 20 0 1 0 0
## 6 1 0 1 0 1
## 7 6 0 1 1 0
## 8 3 0 1 1 1
## 9 5 1 0 0 0
## 10 0 1 0 0 1
## 11 1 1 0 1 0
## 12 0 1 0 1 1
## 13 13 1 1 0 0
## 14 1 1 1 0 1
## 15 6 1 1 1 0
## 16 5 1 1 1 1
## 17 <NA> <NA> <NA> <NA> <NA>
## 18 <NA> <NA> <NA> <NA> <NA>
## 19 <NA> <NA> <NA> <NA> <NA>
## 20 <NA> <NA> <NA> <NA> <NA>
## 21 <NA> <NA> <NA> <NA> <NA>
## 22 <NA> <NA> <NA> <NA> <NA>
## 23 <NA> <NA> <NA> <NA> <NA>
## 24 <NA> <NA> <NA> <NA> <NA>
## 25 <NA> <NA> <NA> <NA> <NA>
## 26 <NA> <NA> <NA> <NA> <NA>
## 27 <NA> <NA> <NA> <NA> <NA>
## 28 <NA> <NA> <NA> <NA> <NA>
## 29 <NA> <NA> <NA> <NA> <NA>
## 30 <NA> <NA> <NA> <NA> <NA>
## 31 <NA> <NA> <NA> <NA> <NA>
## 32 <NA> <NA> <NA> <NA> <NA>
## 33 <NA> <NA> <NA> <NA> <NA>
## 34 <NA> <NA> <NA> <NA> <NA>
## 35 <NA> <NA> <NA> <NA> <NA>
## 36 <NA> <NA> <NA> <NA> <NA>
## 37 <NA> <NA> <NA> <NA> <NA>
## 38 <NA> <NA> <NA> <NA> <NA>
## 39 <NA> <NA> <NA> <NA> <NA>
## 40 <NA> <NA> <NA> <NA> <NA>
## 41 <NA> <NA> <NA> <NA> <NA>
## 42 <NA> <NA> <NA> <NA> <NA>
## 43 <NA> <NA> <NA> <NA> <NA>
## 44 <NA> <NA> <NA> <NA> <NA>
## 45 <NA> <NA> <NA> <NA> <NA>
## 46 <NA> <NA> <NA> <NA> <NA>
## 47 <NA> <NA> <NA> <NA> <NA>
## 48 <NA> <NA> <NA> <NA> <NA>
## 49 <NA> <NA> <NA> <NA> <NA>
## 50 <NA> <NA> <NA> <NA> <NA>
## 51 <NA> <NA> <NA> <NA> <NA>
## 52 <NA> <NA> <NA> <NA> <NA>
## 53 <NA> <NA> <NA> <NA> <NA>
## 54 <NA> <NA> <NA> <NA> <NA>
## 55 <NA> <NA> <NA> <NA> <NA>
## 56 <NA> <NA> <NA> <NA> <NA>
## 57 <NA> <NA> <NA> <NA> <NA>
## 58 <NA> <NA> <NA> <NA> <NA>
## 59 <NA> <NA> <NA> <NA> <NA>
## 60 <NA> <NA> <NA> <NA> <NA>
## 61 <NA> <NA> <NA> <NA> <NA>
## 62 <NA> <NA> <NA> <NA> <NA>
## 63 <NA> <NA> <NA> <NA> <NA>
## 64 <NA> <NA> <NA> <NA> <NA>
## 65 <NA> <NA> <NA> <NA> <NA>
## 66 <NA> <NA> <NA> <NA> <NA>
## 67 <NA> <NA> <NA> <NA> <NA>
## 68 <NA> <NA> <NA> <NA> <NA>
## 69 <NA> <NA> <NA> <NA> <NA>
## 70 <NA> <NA> <NA> <NA> <NA>
## 71 <NA> <NA> <NA> <NA> <NA>
## 72 <NA> <NA> <NA> <NA> <NA>
## 73 <NA> <NA> <NA> <NA> <NA>
## 74 <NA> <NA> <NA> <NA> <NA>
## 75 <NA> <NA> <NA> <NA> <NA>
## 76 <NA> <NA> <NA> <NA> <NA>
## 77 <NA> <NA> <NA> <NA> <NA>
## 78 <NA> <NA> <NA> <NA> <NA>
## 79 <NA> <NA> <NA> <NA> <NA>
## 80 <NA> <NA> <NA> <NA> <NA># Estrutura da comunidade
system(paste("open", shQuote("D:/Elvio/OneDrive/MSS/_Zoo-Rebio/R_ZooRebio/strcom.qmd")))References
São os três pontos com 6 coletas cada em dois rios, o Barro Branco (dentro da reserva) e o Caiana (fora da reserva).↩︎
Esses são os 13 pontos (para zooplâncton) com apenas uma amostra para cada, no entorno da reserva, sendo que os pontos “Inhão” e “Ponto14” estão dentro da reserva.↩︎
https://rstudio-pubs-static.s3.amazonaws.com/13301_6641d73cfac741a59c0a851feb99e98b.html↩︎