7. Propensity Score Matching
Key Concepts
- Propensity scores can be used as an attempt to deal with treatment bias in non-experimental data.
- It is more powerful than simply including many covariates because it attempts to minimize observed differences through predictions from logistic regression.
Methods Matter, Chapter 12
The following example comes from Murnane and Willett (2010), chapter 12.
NELS-88 Public vs. Catholic School Math Achievement
“We investigate the impact on a student’s twelfth-grade mathematics achievement of attending a Catholic (versus a public) high school while attempting to remove bias due to observed differences in base-year family income, parental educational attainment and expectations, and base-year student academic achievement and behavior.” This is based on research by Altonji, Elder, and Taber (2005).
The sample contains the 5,671 students from the NELS-88 dataset who were living in families with annual income of less than $75,000 (in 1988 dollars) in the base year of the survey.
# load packages
library(tidyverse)
library(haven)
library(gt)
library(janitor)
library(psych)
library(apaTables)
library(broom)
library(modelsummary)
source("data/apafunction.R")
source("data/apa_msummary.R")
source("data/corstars.R")labelled::var_label(nels88) %>%
unlist() %>%
matrix(nrow=25, byrow=T) %>%
as.data.frame() %>%
rename("description" = V1) %>%
add_column(variable = names(nels88), .before=1) %>%
apa("Key Variables")| Key Variables | |
|---|---|
| variable | description |
| id | student id code |
| read12 | 12th grade standardized reading score |
| math12 | 12th grade standardized mathematics score |
| hsgrad | graduated hs on time? |
| inpse | entered pse after hs? |
| catholic | attended catholic hs? |
| read8 | 8th grade standardized reading score |
| math8 | 8th grade standardized mathematics score |
| female | student is female? |
| race | r^s race/ethnic background |
| white | student is white? |
| black | student is black? |
| hisp | student is hispanic? |
| api | student is asian/pacific islander? |
| nativam | student is native american? |
| parmar8 | parents marital status in 8th grade |
| faminc8 | total annual family income in 8th grade |
| fathed8 | father^s highest level of education |
| mothed8 | mother^s highest level of education |
| fhowfar | how far in schl r^s father wants r to go |
| mhowfar | how far in schl r^s mother wants r to go |
| fight8 | r got into fight with another student |
| nohw8 | student rarely completes homework |
| disrupt8 | student frequently disruptive |
| riskdrop8 | # of risk factors for later dropout |
Descriptive Statistics
describe(nels88) %>%
as.data.frame() %>%
select(n, mean, sd, min, max, skew, kurtosis) %>%
rownames_to_column("variables") %>%
slice(3, 6, 17) %>%
mutate(across(where(is.numeric), round, 3)) %>%
apa("Descriptive statistics for mathematics score (math12), type of high school (catholic), and family income (faminc8)")| Descriptive statistics for mathematics score (math12), type of high school (catholic), and family income (faminc8) | |||||||
|---|---|---|---|---|---|---|---|
| variables | n | mean | sd | min | max | skew | kurtosis |
| math12 | 5671 | 51.051 | 9.502 | 29.88 | 71.37 | -0.057 | -0.929 |
| catholic | 5671 | 0.104 | 0.306 | 0.00 | 1.00 | 2.587 | 4.693 |
| faminc8 | 5671 | 9.526 | 2.218 | 1.00 | 12.00 | -1.268 | 1.446 |
nels88 %>%
mutate(catholic = as_factor(catholic),
faminc8 = as_factor(faminc8)) %>%
tabyl(faminc8, catholic) %>%
adorn_totals(c("row", "col")) %>%
apa("Catholic school attendance by family income") %>%
tab_spanner("Attended Catholic school?", vars(no, yes))| Catholic school attendance by family income | |||
|---|---|---|---|
| faminc8 | Attended Catholic school? | Total | |
| no | yes | ||
| none | 17 | 1 | 18 |
| <$1000 | 41 | 1 | 42 |
| $1000-$2999 | 84 | 0 | 84 |
| $3000-$4999 | 79 | 6 | 85 |
| $5000-$7499 | 138 | 6 | 144 |
| 7500-$9999 | 169 | 6 | 175 |
| $10000-$14999 | 427 | 20 | 447 |
| $15000-$19999 | 410 | 31 | 441 |
| $20000-$24999 | 608 | 47 | 655 |
| $25000-$34999 | 1137 | 130 | 1267 |
| 35000-$49999 | 1221 | 198 | 1419 |
| 50000-$74999 | 748 | 146 | 894 |
| $75000-$99999 | 0 | 0 | 0 |
| $100000-199999 | 0 | 0 | 0 |
| $200000 or more | 0 | 0 | 0 |
| Total | 5079 | 592 | 5671 |
##
## Pearson's Chi-squared test
##
## data: .
## X-squared = 111.41, df = 11, p-value < 2.2e-16nels88 %>%
mutate(faminc8 = as_factor(faminc8),
catholic = as_factor(catholic)) %>%
ggplot()+
coord_flip()+
geom_point(aes(x=faminc8, y=catholic, color=catholic),
position = position_jitter())+
guides(color = guide_legend(reverse=T))+
labs(title="Distribution of individuals by school type and income")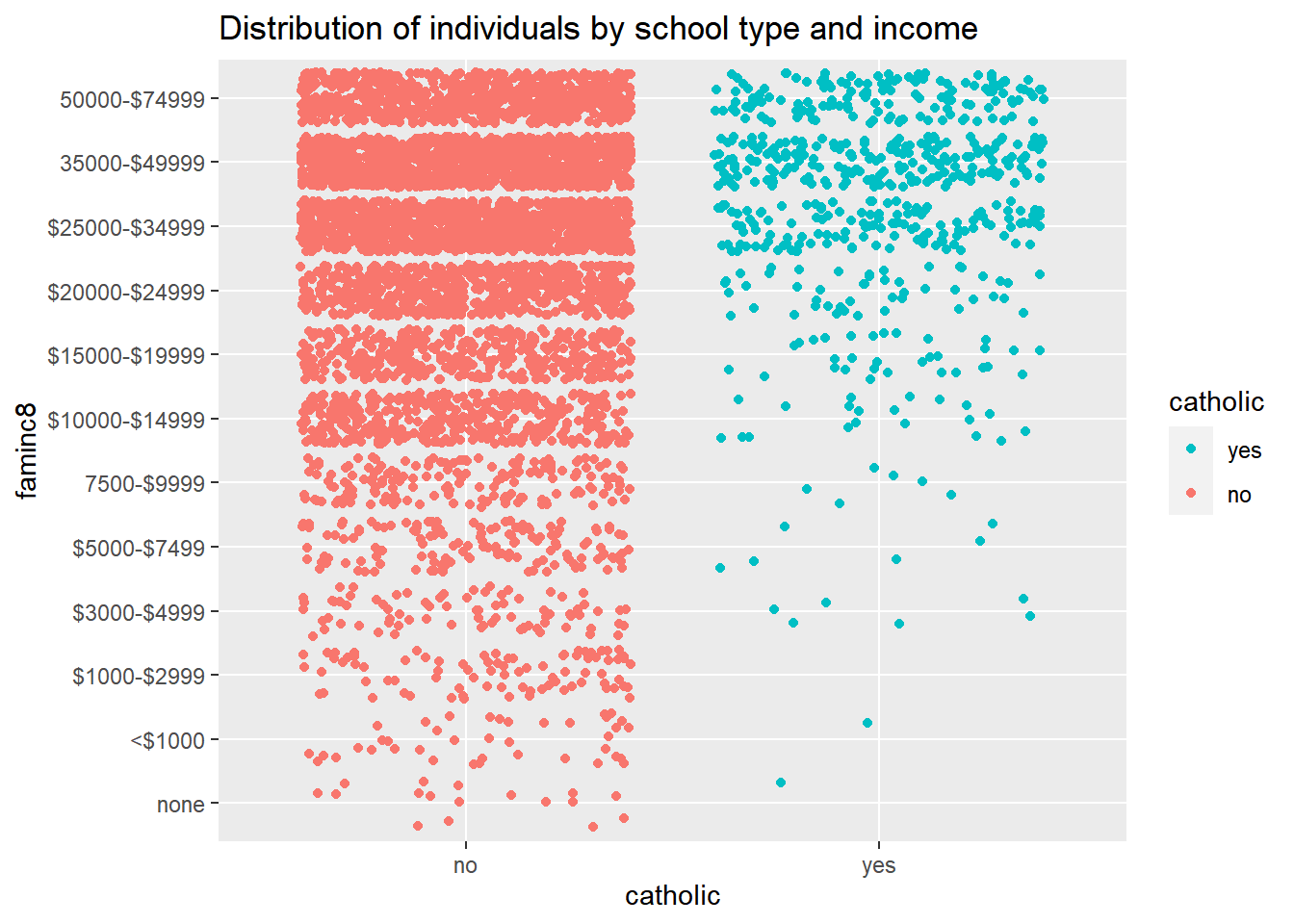
Generating Variable Categories
The book examples uses generated variables faminc8, which uses the actual mid-values of each income range, scaled.
Determining the Best Model
Before generating propensity scores, it is important to determine the best model that predicts group membership through an iterative process.
# this functions adds chi-square to model summary outputs
glance_custom.glm <- function(x) {
lrchi2 <- lmtest::lrtest(x)[2,4]
lrchi2out <- tibble::tibble(`LR Chi-Square:` = lrchi2)
return(lrchi2out)
}
nels88_modela <- glm(catholic ~ (inc8*math8),
data=nels88, family="binomial")
nels88_modelb <- glm(catholic ~ I(inc8^2) + (inc8*math8),
data=nels88, family="binomial")
nels88_modelc <- glm(catholic ~ I(inc8^2) + (inc8*math8) + fhowfar + mhowfar + fight8 + nohw8 + disrupt8 + riskdrop8,
data=nels88, family="binomial")
msummary(list(
"Model A" = nels88_modela,
"Model B" = nels88_modelb,
"Model c" = nels88_modelc)) %>%
apa_msummary("Parameter estimates and approximate p-values for a set of fitted logistic regression models in which attendance at a public or a Catholic high school (CATHOLIC) has been regressed on hypothesized selection predictors (INC8 and MATH8) that describe the base-year annual family income and student mathematics achievement (n = 5,671)")| Parameter estimates and approximate p-values for a set of fitted logistic regression models in which attendance at a public or a Catholic high school (CATHOLIC) has been regressed on hypothesized selection predictors (INC8 and MATH8) that describe the base-year annual family income and student mathematics achievement (n = 5,671) | |||
|---|---|---|---|
| Model A | Model B | Model c | |
| (Intercept) | -5.209 | -5.362 | -4.982 |
| (0.586) | (0.619) | (0.703) | |
| inc8 | 0.062 | 0.087 | 0.054 |
| (0.014) | (0.017) | (0.019) | |
| inc8:math8 | -0.001 | -0.001 | -0.000 |
| (0.000) | (0.000) | (0.000) | |
| math8 | 0.043 | 0.036 | 0.022 |
| (0.011) | (0.012) | (0.012) | |
| I(inc8^2) | -0.000 | -0.000 | |
| (0.000) | (0.000) | ||
| disrupt8 | 0.693 | ||
| (0.386) | |||
| fhowfar | 0.196 | ||
| (0.087) | |||
| fight8 | -0.474 | ||
| (0.325) | |||
| mhowfar | 0.026 | ||
| (0.087) | |||
| nohw8 | -0.688 | ||
| (0.176) | |||
| riskdrop8 | -0.303 | ||
| (0.084) | |||
| Num.Obs. | 5671 | 5671 | 5671 |
| AIC | 3683.2 | 3677.1 | 3630.3 |
| BIC | 3709.8 | 3710.3 | 3703.3 |
| Log.Lik. | -1837.592 | -1833.541 | -1804.125 |
| LR Chi-Square: | 120.129 | 128.231 | 187.063 |
Interpretations
Model C was determined to be the best based on having smaller model fit statistics AIC/BIC. Also, since Model B is nested within Model C, we can perform a Likelihood-Ratio Chi-squared test between the model deviances (i.e., \(-2\times LL\)): \((-2 \times -1833.541) - (-2\times -1804.125) = 58.8\), which is distibuted as a Chi-squared with degrees of freedom equal the difference in the number of paramters between models (10-4 = 6): \(\chi^2\)(6)=58.8, p < .001.
Note: a polynomial term (\(inc8^2\)) to account for a non-linear relationship between catholic and inc8. See Box-Tidwell (i.e., adding interactions between the continuous variables and its natural log, if it’s significant, it suggest non-linearity) and link tests (i.e., this uses the predicted values and square of predicted values to test whether the model is properly specified, the squared term should not be statistically significant) for logistic regression.
Examining the Region of Common Support
These graphics show the overlap of the propensity scores of the two groups, suggesting they share a lot of common support on the covariates in the model. Common support indicates the regions of stratification share enough members of the treatment and control groups.
# Create Propensity Scores
nels88_propscore <- data.frame(pr_score = predict(nels88_modelc,
type="response")) %>%
add_column(nels88_modelc$data)
nels88_propscore %>%
mutate(catholic = ifelse(catholic == 1, "Catholic", "Public")) %>%
ggplot()+
geom_histogram(aes(x=pr_score, y=..density..,), color="white", bins=30)+
geom_density(aes(x=pr_score))+
facet_wrap(~catholic, ncol=1)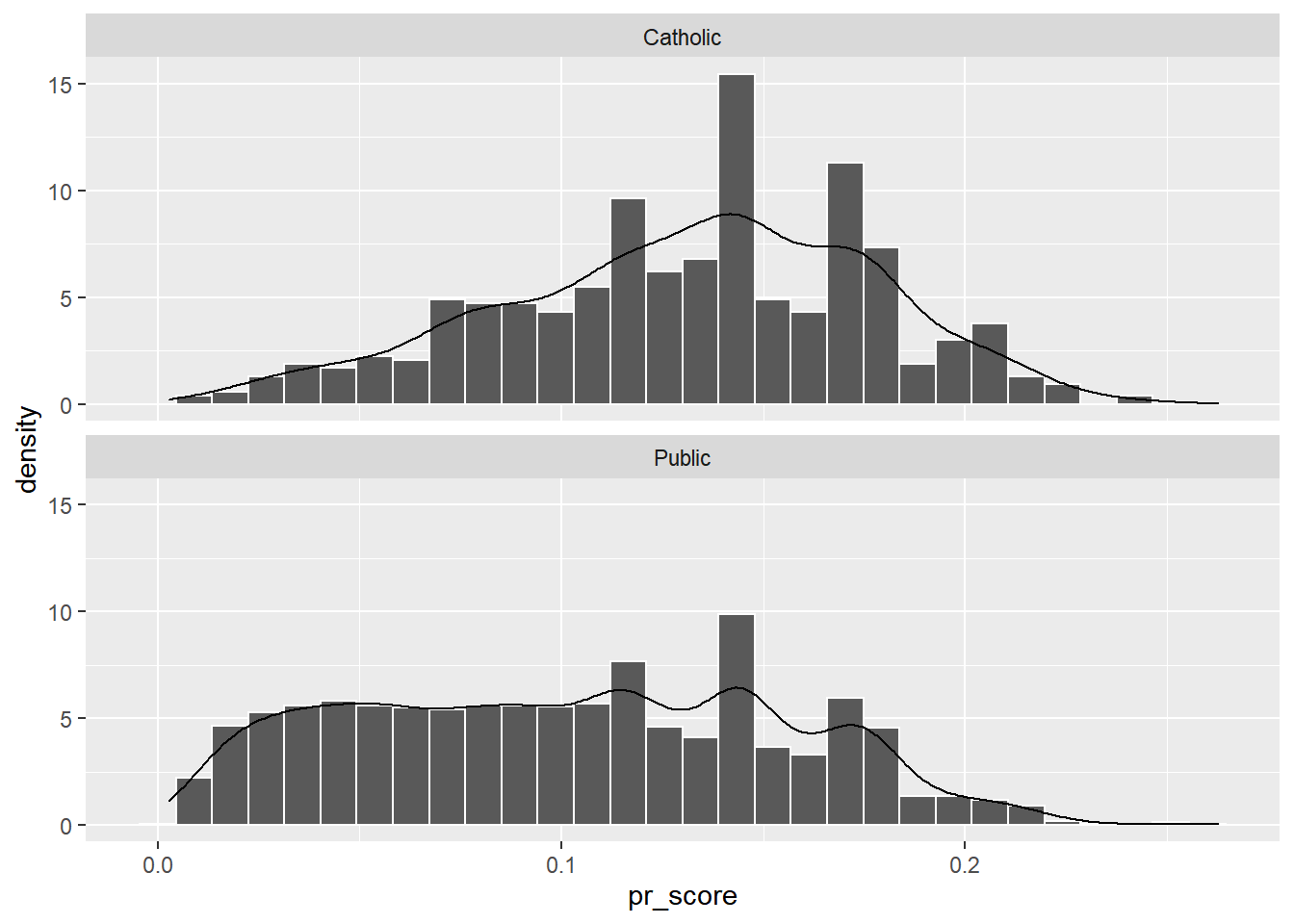
nels88_propscore %>%
mutate(catholic = ifelse(catholic == 1, "Catholic", "Public")) %>%
ggplot()+
geom_histogram(aes(x=pr_score, y=..density.., fill=catholic), color="white", bins=30, position="identity", alpha=.3)+
geom_density(aes(x=pr_score, color=catholic))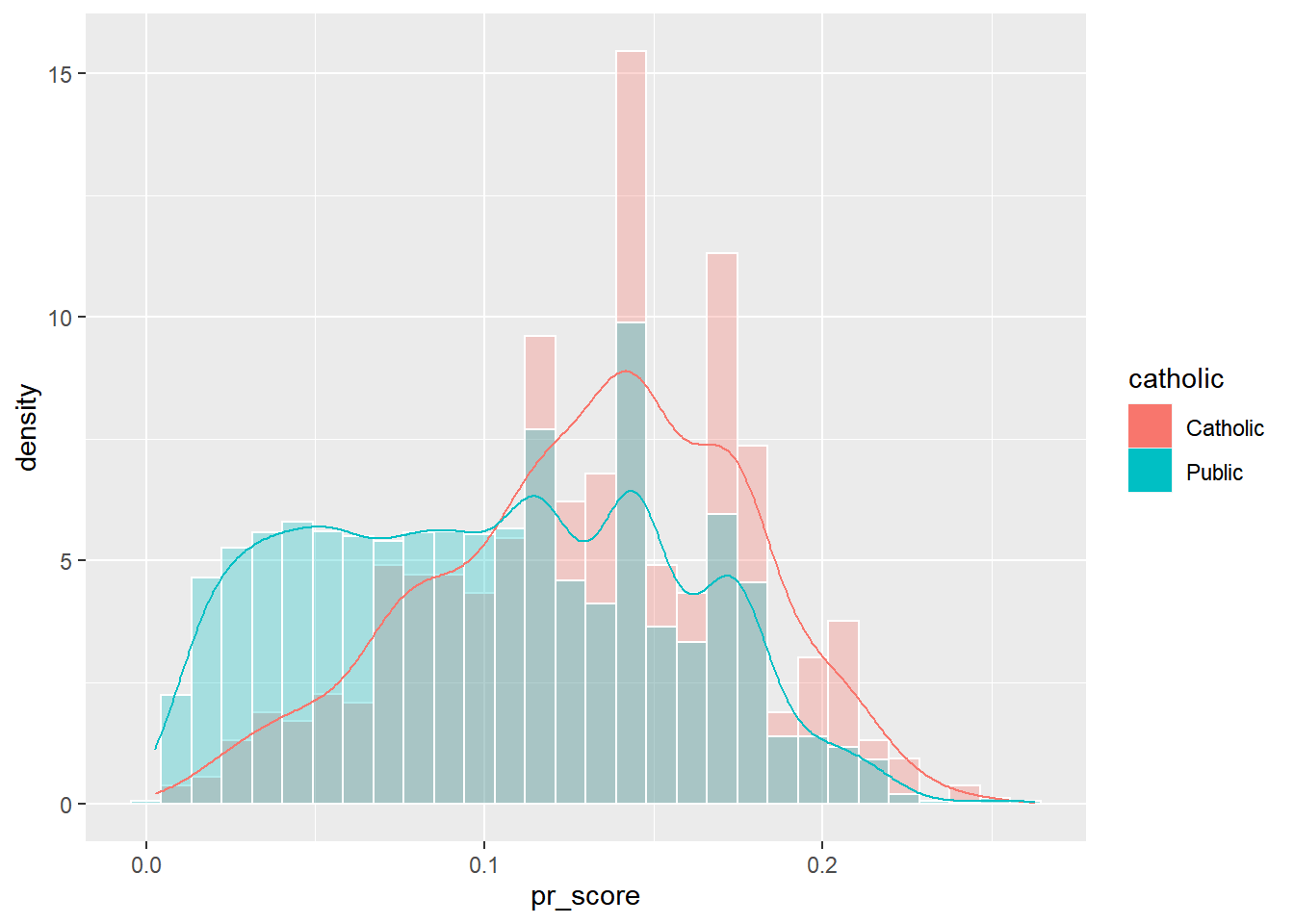
Stratifying on the Propensities
Stratification into 5 blocks based on quintiles
Checking balance on the propensities
do(nels88_5blocks, tidy(t.test(.$pr_score ~ .$catholic,
alternative = "greater",
paired = FALSE,
var.equal = TRUE,
conf.level = 0.95
))) %>%
select(blocks, p.value) %>%
ungroup() %>%
apa("Testing balance of blocks based on quntiles")| Testing balance of blocks based on quntiles | |
|---|---|
| blocks | p.value |
| 1 | 0.9846739 |
| 2 | 0.9996522 |
| 3 | 0.9851109 |
| 4 | 0.7720567 |
| 5 | 0.9508310 |
Interpretation
The table above shows t-tests per block between propensity scores and treatment groups. Using a one-sided test where the expectation is a greater score, none of the differences are significant. This suggests balance.
Note: if you choose “less”, you will get different results becuase it is the lower side of the one-sided t-test. You’d be testing whether the expectation is a lower score. You may also want to check individual covariate balance, as below.
Stratification into 5 blocks based on cut scores
The following stratification comes from p. 320
nels88_5blocks_2 <- nels88_propscore %>%
mutate(blocks = case_when(
pr_score < .05 ~ 1,
pr_score = .05 & pr_score <.1 ~2,
pr_score =.1 & pr_score <.15 ~ 3,
pr_score = .15 & pr_score < .2 ~ 4,
pr_score = .2 & pr_score < .3 ~ 5
),
catholic = factor(catholic, labels = c(
"Public", "Catholic"
))) %>%
group_by(blocks)
do(nels88_5blocks_2, tidy(t.test(.$pr_score ~ .$catholic,
alternative = "greater",
paired = FALSE,
var.equal = TRUE,
conf.level = 0.95
))) %>%
select(blocks, p.value) %>%
ungroup() %>%
apa("Testing balance of blocks based on the book's example")| Testing balance of blocks based on the book's example | |
|---|---|
| blocks | p.value |
| 1 | 0.9788125 |
| 2 | 0.9884155 |
| 3 | 0.9776620 |
| 4 | 0.9145168 |
| 5 | 0.2712632 |
Estimate the ATE and ATT of Stratified Propensity Scores
# Summarize Block
nels88_5blocks_sum <- nels88_5blocks_2 %>%
group_by(blocks, catholic) %>%
summarize(n = n(),
pr_score = mean(pr_score),
income_mean=mean(inc8),
mathmean = mean(math8),
math_achievment = mean(math12)) %>%
pivot_wider(names_from=catholic, values_from = c(n, pr_score, income_mean, mathmean, math_achievment)) %>%
mutate(diff = math_achievment_Catholic - math_achievment_Public,
n = n_Catholic + n_Public,
mutate(across(where(is.numeric), round, 3))) %>%
ungroup()## `summarise()` regrouping output by 'blocks' (override with `.groups` argument)| Five propensity-score blocks, based on predicted values from final Model C (which contains selected covariates in addition to those included in Model B). Within-block sample statistics include: (a) frequencies, (b) average propensity scores, and (c) average twelfth grade mathematics achievement by type of high school, and their difference (n = 5,671) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| blocks | n_Public | n_Catholic | pr_score_Public | pr_score_Catholic | income_mean_Public | income_mean_Catholic | mathmean_Public | mathmean_Catholic | math_achievment_Public | math_achievment_Catholic | diff | n |
| 1 | 1089 | 34 | 0.030 | 0.035 | 13.316 | 13.169 | 44.542 | 46.039 | 43.664 | 46.014 | 2.350 | 1123 |
| 2 | 1431 | 110 | 0.075 | 0.078 | 27.017 | 26.675 | 49.412 | 49.956 | 48.853 | 51.002 | 2.149 | 1541 |
| 3 | 1599 | 253 | 0.127 | 0.129 | 35.930 | 37.362 | 53.891 | 53.971 | 53.623 | 55.383 | 1.760 | 1852 |
| 4 | 829 | 160 | 0.172 | 0.173 | 52.289 | 52.891 | 58.048 | 57.814 | 56.869 | 57.346 | 0.477 | 989 |
| 5 | 131 | 35 | 0.213 | 0.212 | 59.752 | 60.214 | 51.304 | 51.475 | 52.506 | 55.013 | 2.507 | 166 |
#Weighted Average Treatment Effect
nels88_5blocks_sum %>%
mutate(weighted = diff * n) %>%
pull(weighted) %>%
sum()/sum(nels88_5blocks_sum$n)## [1] 1.780655#Weighted Average Treatment on the Treated Effect
nels88_5blocks_sum %>%
mutate(weighted = diff * n_Catholic) %>%
pull(weighted) %>%
sum()/sum(nels88_5blocks_sum$n_Catholic)## [1] 1.563573Using MatchIt
Method from http://www.practicalpropensityscore.com/matching.html
library(MatchIt)
set.seed(2020)
#generate propensity scores using Model C
nels88_ps <- matchit(catholic ~ I(inc8^2) + (inc8*math8) + fhowfar + mhowfar + fight8 + nohw8 + disrupt8 + riskdrop8,
data=nels88,
method="nearest", #nearest neighbors
replace=T, #with replacement
ratio=1) #one to one matching - each treated with one control
#A user may want to consider adding a caliper option to only select cases within a certain range, caliper = .2Diagnose Covariate Balance
This table shows the maximum standardized mean difference in propensity scores ? between groups.
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.000000 0.004342 0.020411 0.021566 0.036718 0.051833The results show Small differences between groups on the covariates. (Max = .05).
##
## FALSE
## 11This table shows that only there are no differences greater than .1 (FALSE 11)
Estimate Treatment Effects
#obtain matched data
matched_data <- match.data(nels88_ps)
# use weights to estimate ATT
library(survey)
design.nels88 <- svydesign(ids=~1, weights=~weights,
data=matched_data)
#estimate the ATT
model.nels88 <- svyglm(math12~catholic + I(inc8^2) + (inc8*math8) + fhowfar + mhowfar + fight8 + nohw8 + disrupt8 + riskdrop8, design.nels88, family=gaussian())
summary(model.nels88)##
## Call:
## svyglm(formula = math12 ~ catholic + I(inc8^2) + (inc8 * math8) +
## fhowfar + mhowfar + fight8 + nohw8 + disrupt8 + riskdrop8,
## design = design.nels88, family = gaussian())
##
## Survey design:
## svydesign(ids = ~1, weights = ~weights, data = matched_data)
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 5.2344350 2.5150613 2.081 0.037641 *
## catholic 1.2250485 0.3128480 3.916 9.56e-05 ***
## I(inc8^2) -0.0013863 0.0006069 -2.284 0.022542 *
## inc8 0.2410835 0.0681540 3.537 0.000421 ***
## math8 0.8123924 0.0426961 19.027 < 2e-16 ***
## fhowfar -0.0421049 0.4213556 -0.100 0.920420
## mhowfar 0.4604611 0.4064090 1.133 0.257459
## fight8 0.3835736 1.5872490 0.242 0.809089
## nohw8 -1.9228815 0.6605490 -2.911 0.003674 **
## disrupt8 -0.3809514 1.7834881 -0.214 0.830899
## riskdrop8 -0.1132044 0.3294263 -0.344 0.731181
## inc8:math8 -0.0020930 0.0010155 -2.061 0.039521 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for gaussian family taken to be 25.95404)
##
## Number of Fisher Scoring iterations: 2Because of unmatched controls (4,546; see
summary(nels88_ps)), we can typically only estimate the ATT since we don’t typically include the entire sample. Weighting by the inverse of the propensity score then running the analysis is one way to get the ATE. You can also get ATE from stratification.
Another MatchIt Method
From Propensity Scores: A Practical Introduction Using R
Visualize Propensity Scores
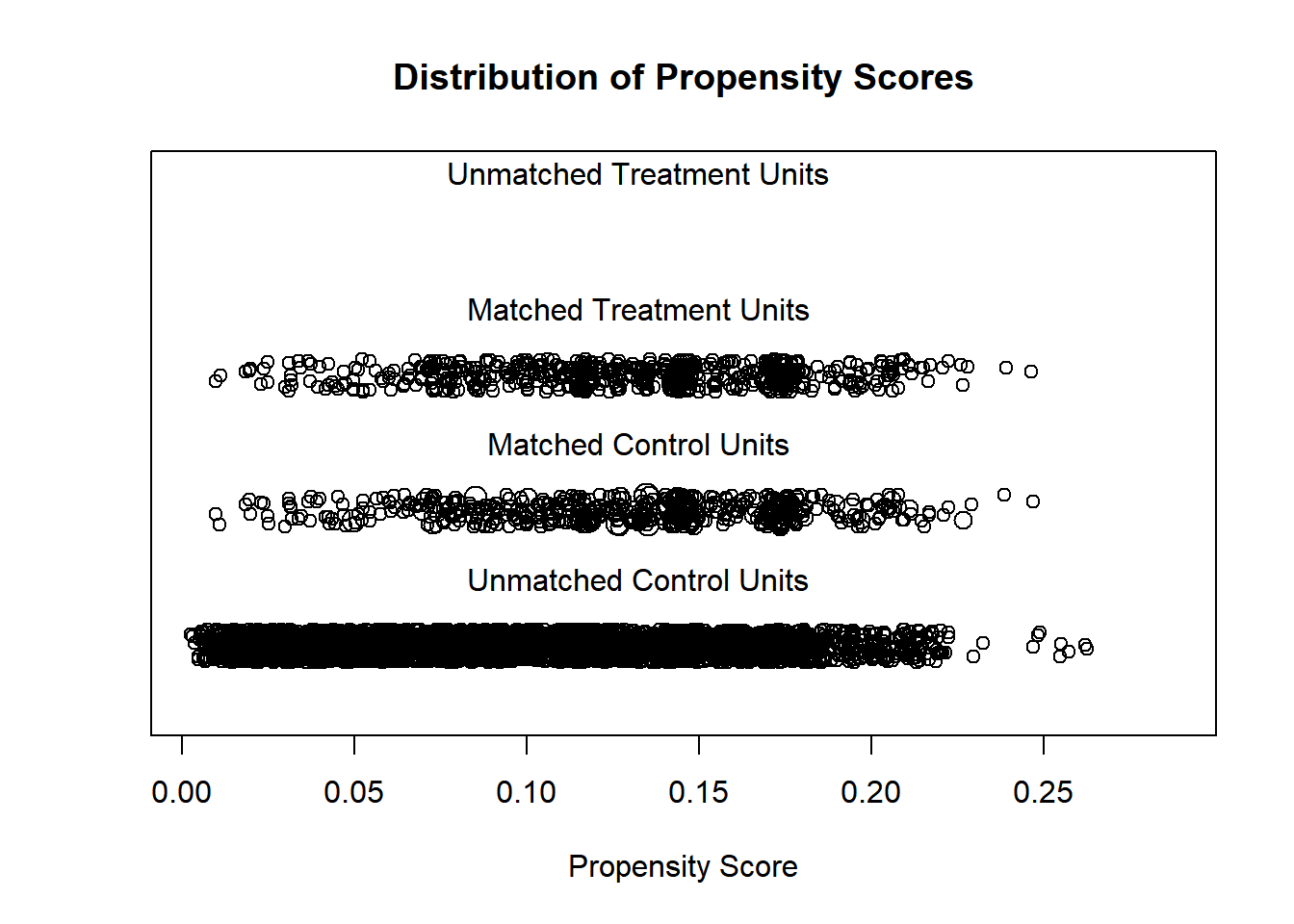
## [1] "To identify the units, use first mouse button; to stop, use second."## integer(0)Prepare Matched Data
# save matched data
matches <- data.frame(nels88_ps$match.matrix)
# find the matches
group1 <- match(row.names(matches), row.names(matched_data))
group2 <- match(matches$X1, row.names(matched_data))
# extract outcome values for matches
treatment <- matched_data$math12[group1]
control <- matched_data$math12[group2]
# create matched data set
outcomes <- cbind(matches, treatment, control)
# t-test
t.test(outcomes$treatment, outcomes$control, paired = T) %>%
tidy() %>%
select(estimate:p.value, method, alternative) %>%
apa("Outcome Analysis")| Outcome Analysis | ||||
|---|---|---|---|---|
| estimate | statistic | p.value | method | alternative |
| 0.939831 | 2.22596 | 0.02639323 | Paired t-test | two.sided |
.93 is the ATT. It is lower than what’s found in the Methods Matter, but it also uses a slightly different method. One thing we’d want to do is assess the robustness of these results by altering parameters of the matching algorithm (e.g., adding a caliper, altering the values of the caliper .2 to .15, matching with and without replacement, ratio matching 1:5) or by choosing a different matching algorithm (e.g., optimal matching, full matching). These type of tests help demonstrate that results do not depend on the particular matching method chosen.
Stratification with MatchIt
Based on Practical Propensity Score Methods Using R (Leite, 2019)
formula <- formula("catholic ~ I(inc8^2) + (inc8*math8) + fhowfar + mhowfar + fight8 + nohw8 + disrupt8 + riskdrop8")
# make the subclasses
nels_stratified <- matchit(formula, #formula
distance=nels88_propscore$pr_score, #propensity scores
data = nels88_propscore, #data
method = "subclass", #statifying method
sub.by = "treat",
subclass=5) # quintiles
# get the matched data
data.stratification <- match.data(nels_stratified) %>%
mutate(catholic = factor(catholic, labels=c("Public", "Catholic")))
#preview
data.stratification %>%
janitor::tabyl(catholic, subclass)## catholic 1 2 3 4 5
## Public 2198 1016 702 616 547
## Catholic 119 118 118 118 119Covariate Balance Evaluation
# obtain standardize mean differences
balance.stratification <- summary(nels_stratified, standardize=T)
# the following extracts only the standardized mean differences for all strata
balance.stratification$q.table[,3,] %>%
as.data.frame() %>%
pivot_longer("Subclass 1":"Subclass 5", names_to = "strata", values_to="mean_diff") %>%
group_by(strata) %>%
summarize(min = min(mean_diff),
mean = mean(mean_diff),
max = max(mean_diff))## # A tibble: 5 x 4
## strata min mean max
## <chr> <dbl> <dbl> <dbl>
## 1 Subclass 1 -0.565 0.00616 0.271
## 2 Subclass 2 -0.0656 0.0478 0.144
## 3 Subclass 3 -0.0764 0.00674 0.0739
## 4 Subclass 4 -0.102 -0.00880 0.0317
## 5 Subclass 5 -0.0108 0.0328 0.0837Two stratas have greater than .1 and 1 has greater than .25. This is OK, but not great.
Obtaining ATE and ATE Estimates
design <- svydesign(ids=~0,
weights=~weights,
data=data.stratification)
# Robust SE through bootstrapping
# Good for when not known or easy formula
set.seed(2020)
surveyDesignBoot <- as.svrepdesign(design, type=c("bootstrap"),replicates=1000)
#obtain means and standard errors by stratum and treatment group combinations
subclassMeans <- svyby(formula=~math12,
by=~catholic+subclass,
design=surveyDesignBoot,
FUN=svymean,
covmat=TRUE)
# get weights for estimation
weights <- data.stratification %>%
janitor::tabyl(catholic, subclass) %>%
pivot_longer(`1`:`5`, names_to = "strat", values_to = "n") %>%
pivot_wider(names_from = catholic, values_from=n) %>%
mutate(strat_sum = Public + Catholic,
total_treated = sum(Catholic),
total_n = sum(strat_sum),
ATE_wk = strat_sum/total_n,
ATT_wk = Catholic/total_treated)
# estimation via linear/nonlinear contrasts
pooledEffects <- svycontrast(subclassMeans, list(
ATE=c(rbind(-weights$ATE_wk, weights$ATE_wk)),
ATT=c(rbind(-weights$ATT_wk, weights$ATT_wk))))
pooledEffects## contrast SE
## ATE 2.1079 0.404
## ATT 1.6107 0.364