Chapter 8 Visualizing and characterizing variability
[This “chapter” is essentially the same as 2, but with some of the notes I wrote in the document as we went.]
This workshop will attempt to teach you some of the basics of the data science workflow, as imagined by Wickham and Grolemund:
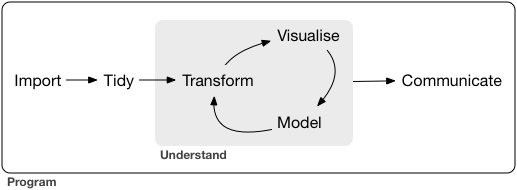
via Wickham and Grolemund (2017)
Notice that this graphic already includes a cycle– this is an important part of the process! It is unlikely (impossible) that you could work linearly through a data science project.
It is tempting to focus on the modeling aspect of the cycle, and miss the other elements. But, recall
“90% of data science is wrangling data, the other 10% is complaining about wrangling data.”@evelgab at #rstatsnyc
— David Robinson (@drob) April 20, 2018
Statistics is the practice of using data from a sample to make inferences about some broader population of interest. Data is a key word here. In the pre-bootcamp prep, you explored the first simple steps of a statistical analysis: familiarizing yourself with the structure of your data and conducting some simple univariate analyses.
We are doing all our work in the programming language R, and I am going to show you one way to do the tasks. However, one of the powers of R is that there are many ways to “say” the same thing in R. In fact, part of my research is about studying the various syntaxes in R (sometimes called Domain Specific Languages). I characterize the three main syntaxes as:
- base
R. If you have usedRbefore, it is highly likely you have seen this syntax before. I don’t use it much (anymore). This syntax is characterized by dollar signs$and square brackets[ , ]. - formula syntax. We will be using this syntax for modeling, but you can also do many other tasks in it, using
mosaicfor statistics, and thenlatticeofggformulagraphics - tidyverse syntax. This is most of what I use for data wrangling and data visualization. Main packages include
ggplot2anddplyr. You can use the convenience packagetidyverseto install and load many of the packages.
If you want to see a comparison between these three syntaxes, see my cheatsheet in the “contributed” section of the RStudio Cheatsheets page.
8.1 Pre-bootcamp review
What did you learn about
hiphop_cand_lyrics?Did exploring the
hiphop_cand_lyricsdataset raise any questions for you?What was the variability like in the
salaryvariable in theSalariesdataset?What do you think might explain the variability in
salary?
We are going to try to use plots and models to explain this variability.
8.1.1 Variable types, redux
It may be useful to think about variables as
- response variable: this is the one we’re the most interested in, which we think might “respond.”
- explanatory variable(s): these are the ones we think might “explain” the variation in the response.
Before building any models, the first crucial step in understanding relationships is to build informative visualizations.
8.1.2 Visualizing variability (in breakout rooms)
Run through the following exercises which introduce different approaches to visualizing relationships. In doing so, for each plot:
- examine what the plot tells us about relationship trends & strength (degree of variability from the trend) as well as outliers or deviations from the trend.
- think: what’s the take-home message?
We’ll do the first exercise together.
You (hopefully!) need to re-load the Salaries dataset. (If you don’t, that probably means you saved your workspace the last time you closed RStudio, talk to me or a TA about how to clear your workspace.)
- Sketches How might we visualize the relationships among the following pairs of variables? Checking out the structure on just a few people might help:
## rank discipline yrs.since.phd yrs.service sex salary
## 1 Prof B 19 18 Male 139750
## 2 Prof B 20 16 Male 173200
## 3 AsstProf B 4 3 Male 79750
## 4 Prof B 45 39 Male 115000
## 5 Prof B 40 41 Male 141500
## 6 AssocProf B 6 6 Male 97000
## 7 Prof B 30 23 Male 175000
## 8 Prof B 45 45 Male 147765
## 9 Prof B 21 20 Male 119250
## 10 Prof B 18 18 Female 129000salaryvs.yrs.since.phd(scatterplot, geom_point)salaryvs.yrs.service(scatterplot, geom_point)salaryvs.discipline(side-by-side boxplots, side-by-histograms)salaryvs.yrs.since.phdanddiscipline(in one plot) (scatterplot with colored dots)salaryvs.yrs.since.phdandsex(in one plot) (scatterplot with colored dots)
- Scatterplots of 2 quantitative variables
By default, the response variable (\(Y\)) goes on the y-axis and the predictor (\(X\)) goes on the x-axis.
Again, you (hopefully!) need to re-load ggplot2 in order to run this code.
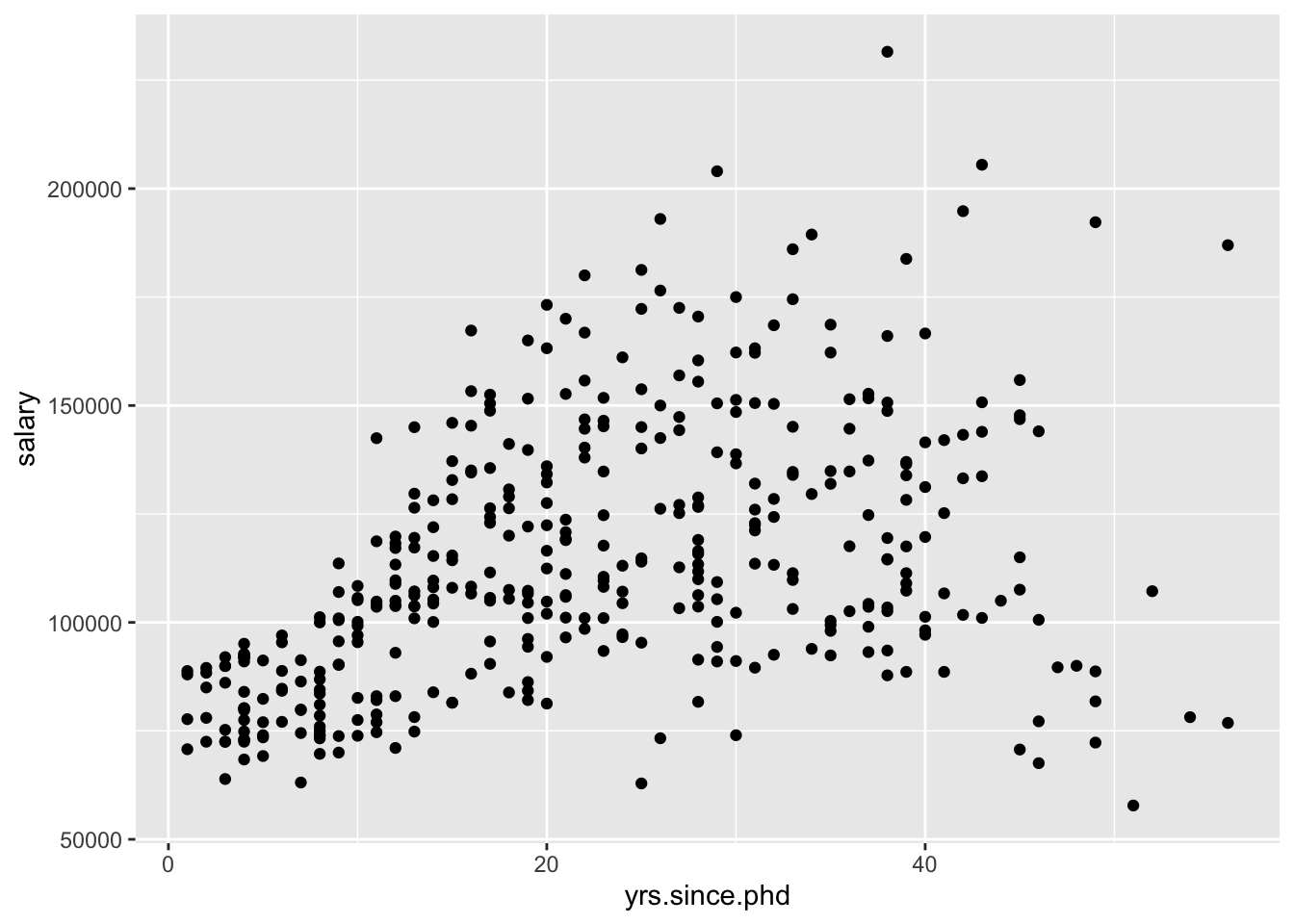
# ggplot(Salaries, aes(x = yrs.since.phd, y = salary)) +
# geom_text(aes(label = discipline))
# practice: make a scatterplot of salary vs. yrs.service- Side-by-side plots of 1 quantitative variable vs. 1 categorical
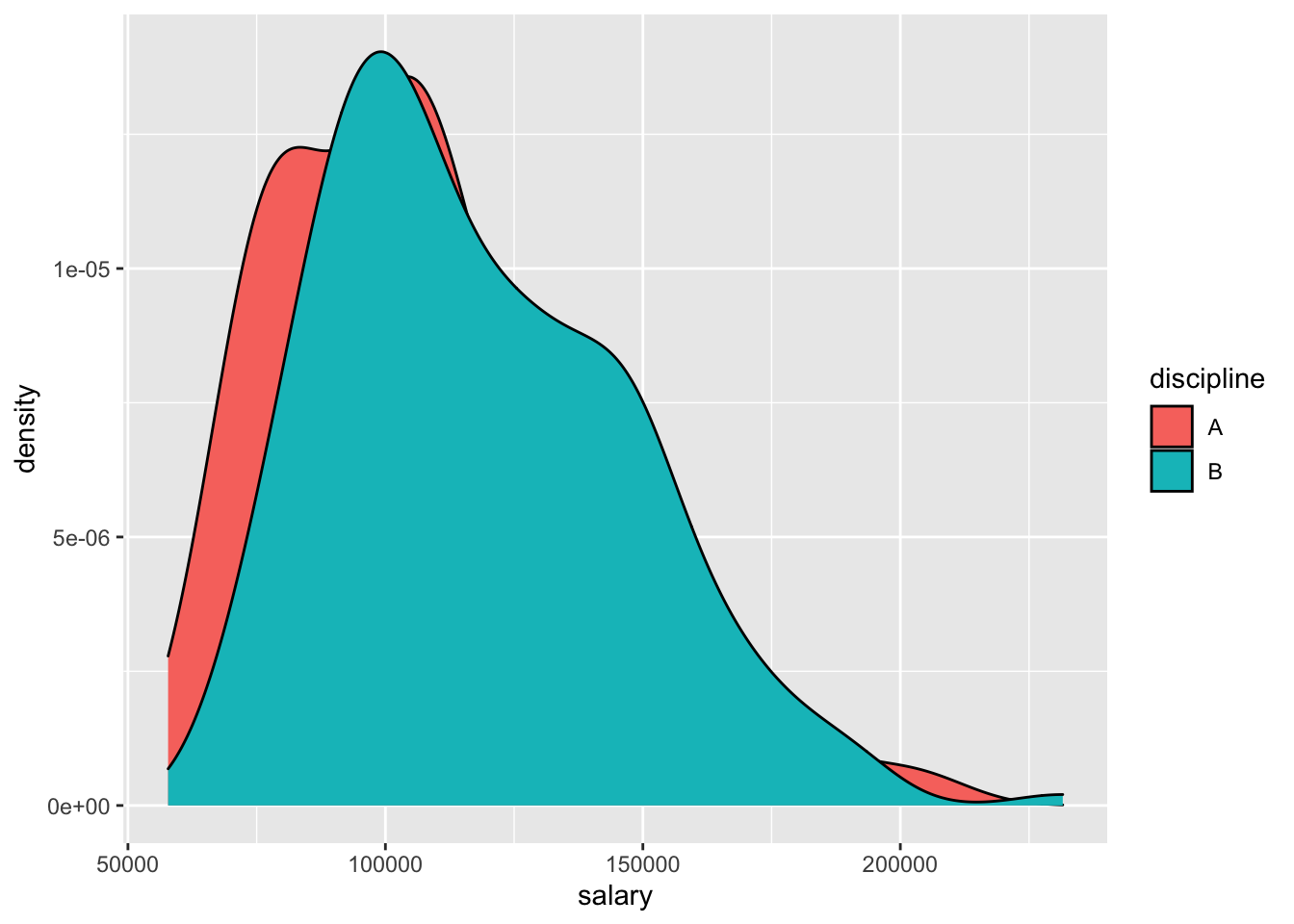
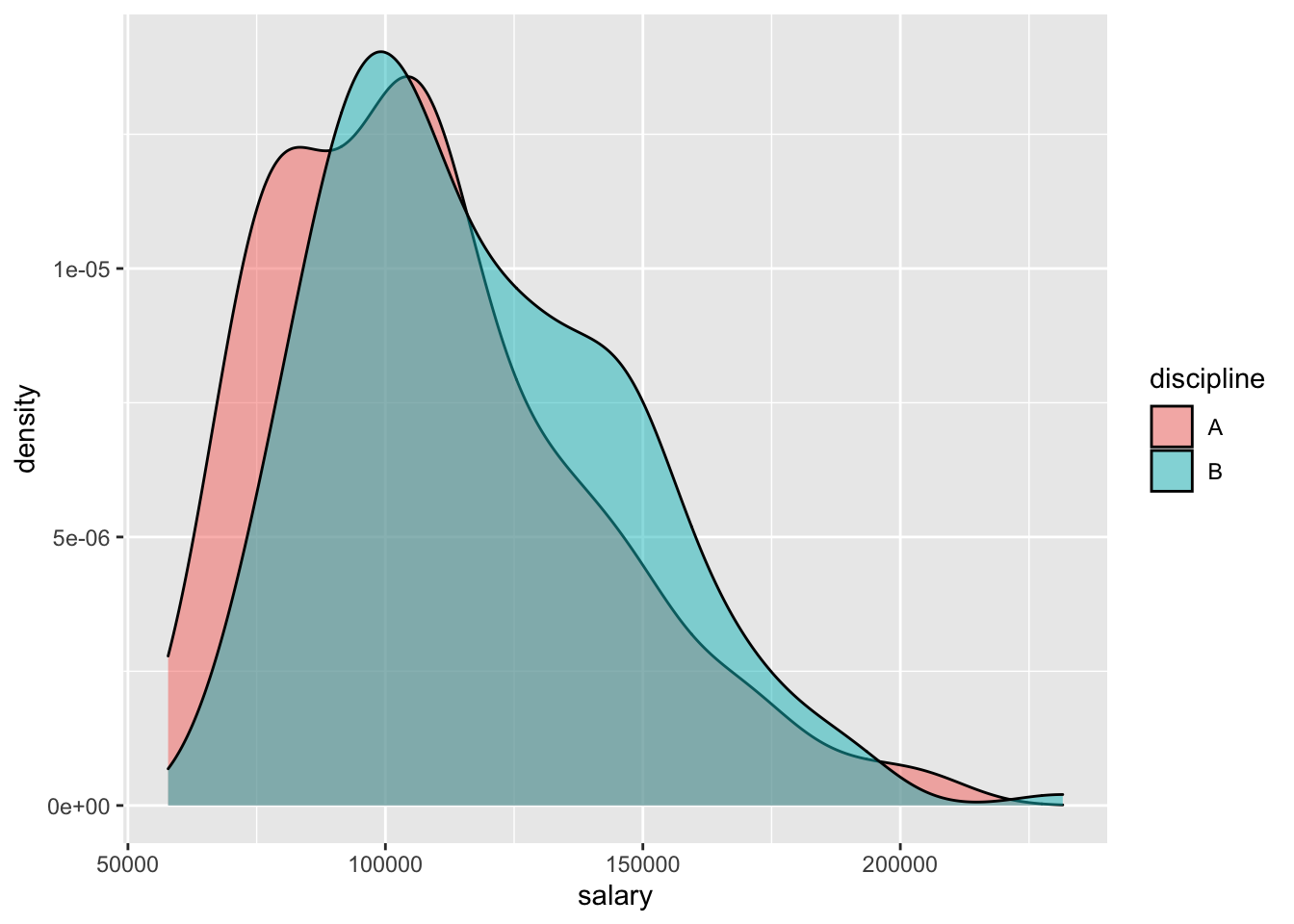
# ggplot(Salaries, aes(x = salary)) +
# geom_density() +
# facet_wrap(~discipline)
# practice: make side-by-side boxplots of salary by rank- Scatterplots of 2 quantitative variables plus a categorical
If yrs.since.phd and discpline both explain some variability in salary, why not look at all the variables at once?
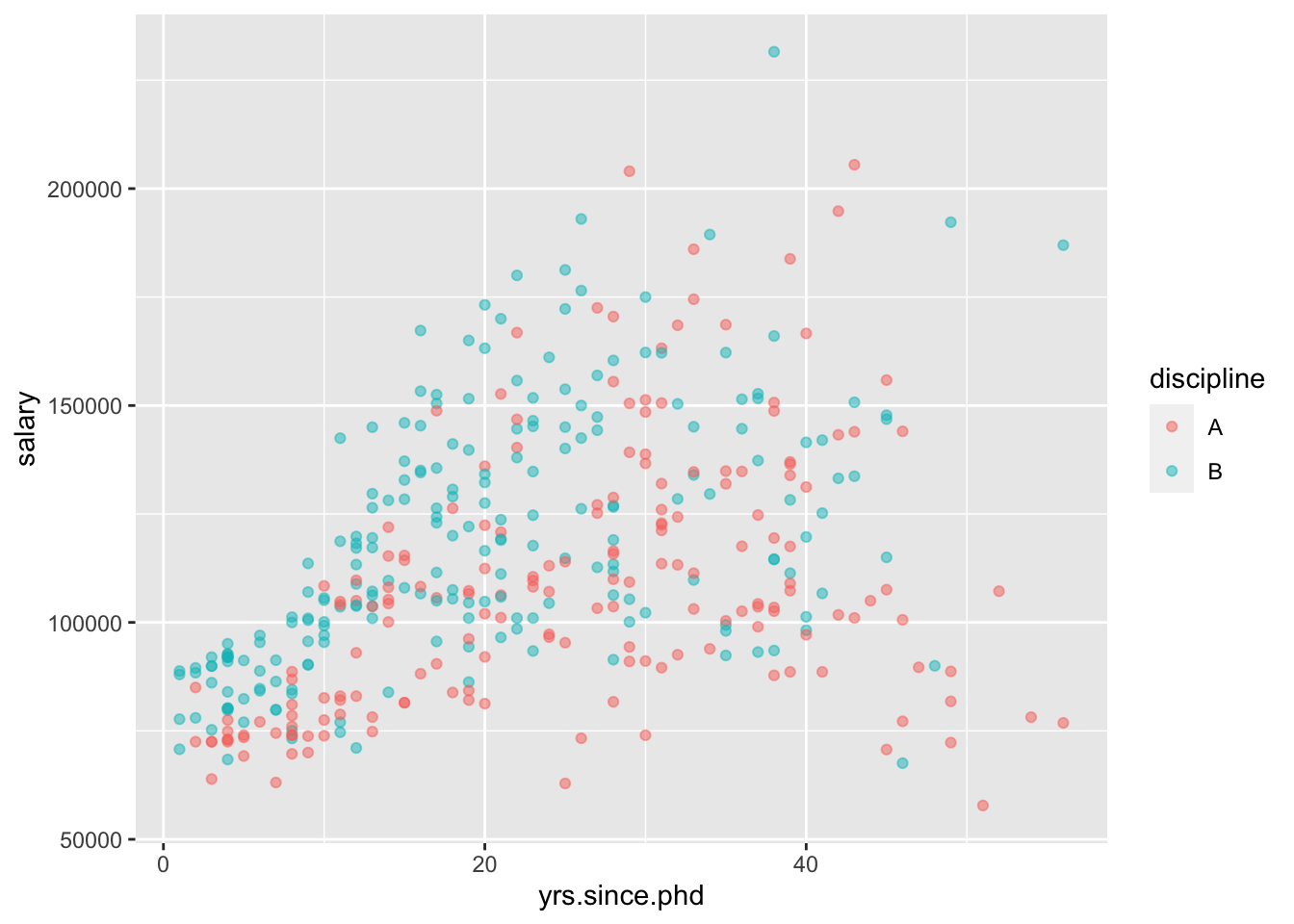
- Plots of 3 quantitative variables
Think back to our sketches. How might we include information about yrs.service in our plots?
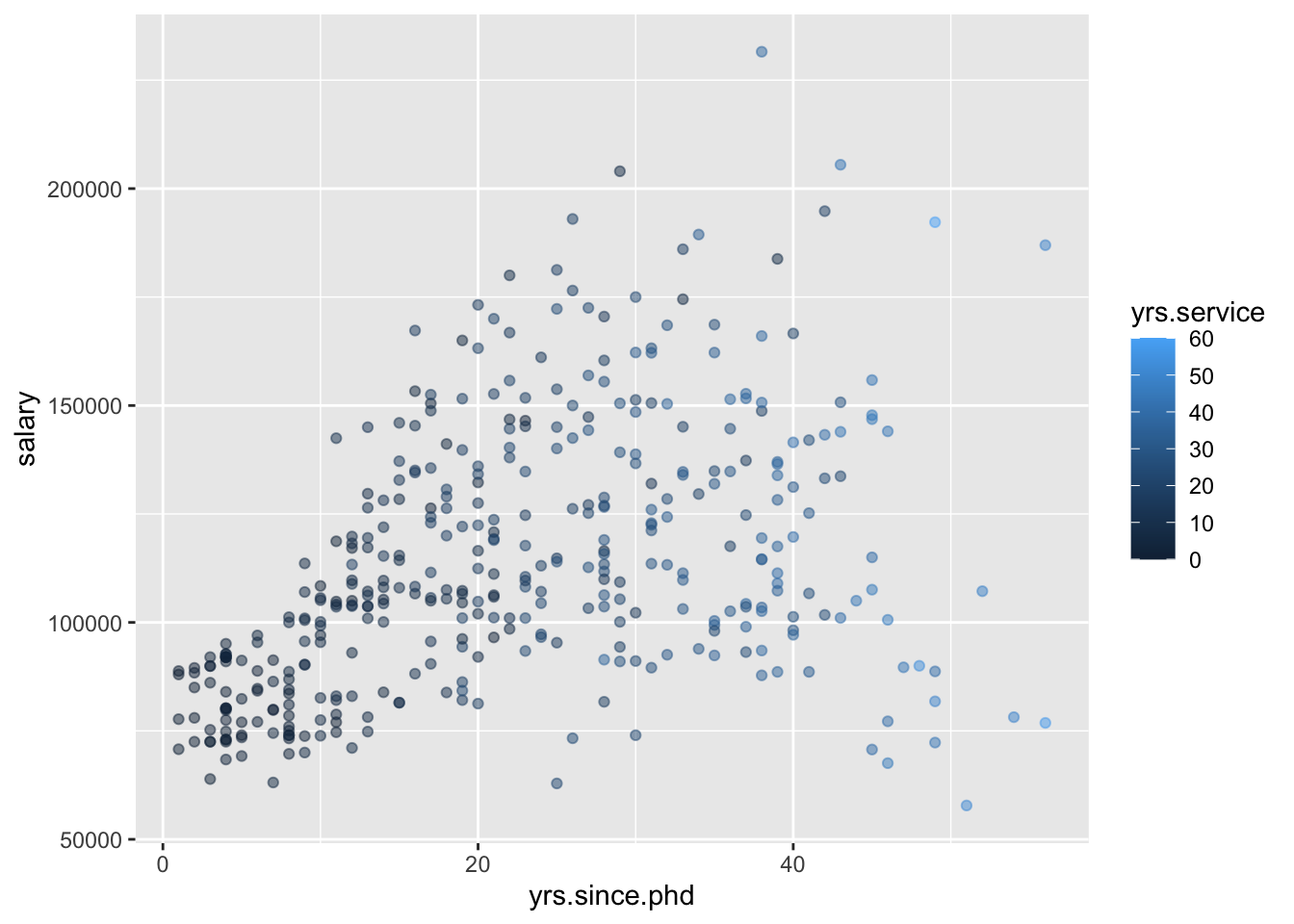
8.2 Modeling: choose, fit, assess, use
As we look at these pictures, we begin to build mental models in our head. Statistical models are much like mental models (they help us generalize and make predictions) but of course, more rigorous. No matter which model we are using, we will use the CFAU framework,
- Choose
- Fit
- Assess
- Use
We’re going to begin with linear models, which are one of the simplest ways to model.
8.3 Linear models
A linear model is a supervised learning method, in that we will use ground truth to help us fit the model, as well as assess how good our model is.
With regression, your input is
\[ x = (x_1, x_2, \dots, x_k) \] and the output is a quantitative \(y\). We can “model” \(y\) by \(x\) by finding
\[ y = f(x) + \epsilon \]
where \(f(x)\) is some function of \(x\), and \(\epsilon\) are errors/residuals. For simple linear regression, \(f(x)\) will be the equation of a line.
Specifically, we use the notation
\[ Y = \beta_0 + \beta_1\cdot X + \epsilon, \, \epsilon \sim N(0, \sigma_\epsilon) \] although we are primarily concerned with the fitted model,
\[ \hat{Y} = \hat{\beta_0} + \hat{\beta_1}\cdot X \] Notice that there is no \(\epsilon\) in the fitted model. If we want, we can calculate
\[ \epsilon_i = y_i - \hat{y}_i \] for each data point, to get an idea of how “off” the model is.
The coefficients \(\hat{\beta_0}\) and \(\hat{\beta_1}\) are useful for interpreting the model.
\(\hat{\beta_0}\) is the intercept coefficient, the average \(Y\) value when \(X=0\)
\(\hat{\beta_1}\) is the slope, the amount that we predict \(Y\) to change for a 1-unit change in \(X\)
We can use our model to predict \(y\) for new values of \(y\), explain variability in \(y\), and describe relationships between \(y\) and \(x\).
8.4 Linear models in R
The syntax for linear models is different than the tidyverse syntax, and instead is more similar to the syntax for lattice graphics.
The general framework is
goal ( y ~ x , data = mydata )
We’ll use it for modeling.
Given how the data looked in the scatterplot we saw above, it seems reasonable to choose a simple linear regression model. We can then fit it using R.
We’re using the assignment operator to store the results of our function into a named object.
I’m using the assignment operator <-, but you can also use =. As with many things, I’ll try to be consistent, but I often switch between the two.
The reason to use the assignment operator here is because we might want to do things with our model output later. If you want to skip the assignment operator, try just running lm(salary ~ yrs.since.phd, data = Salaries) in your console to see what happens.
Now, we want to move on to assessing and using our model. Typically, this means we want to look at the model output (the fitted coefficients, etc). If we run summary() on our model object we can look at the output.
##
## Call:
## lm(formula = salary ~ yrs.since.phd, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -84171 -19432 -2858 16086 102383
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 91718.7 2765.8 33.162 <2e-16 ***
## yrs.since.phd 985.3 107.4 9.177 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 27530 on 395 degrees of freedom
## Multiple R-squared: 0.1758, Adjusted R-squared: 0.1737
## F-statistic: 84.23 on 1 and 395 DF, p-value: < 2.2e-16The p-values are quite significant, which might lead us to assess that the model is pretty effective.
We could visualize the model using ggplot2, by adding an additional geom_,
ggplot(Salaries, aes(x = yrs.since.phd, y = salary)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE)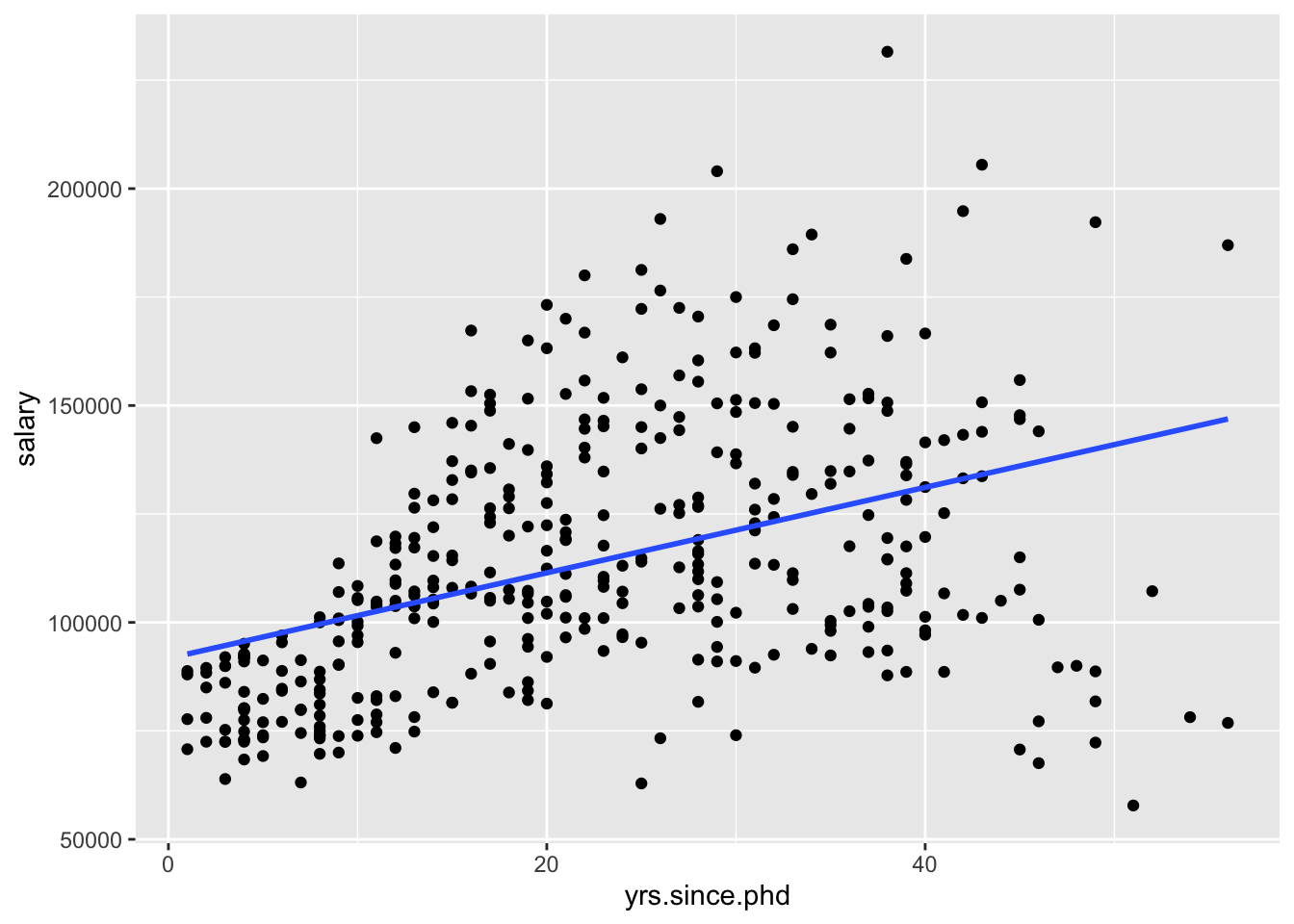
geom_smooth can use other methods, but we want a linear model. And we’ll talk about standard errors eventually, but for now we’ll turn them off.
We can also use the model for description. To write this model out in our mathematical notation,
\[ \widehat{\texttt{salary}} = 91718.7 + 985.3\cdot \texttt{yrs.since.phd} \]
- What would this model predict for the salary of a professor who had just finished their PhD?
## [1] 91718.7We would predict $91,719.
- What does the model predict for a professor who has had their PhD for 51 years?
## [1] 141969We would predict $141,969.
- What was the observed salary value for the person with 51 years of experience?
## salary
## 1 57800- What is the residual?
## [1] -84169To assess further, we can compare it to a “null” model, where we don’t use a predictor (Instead, we just use the mean as the model for every observation).
We can run two models and visualize them to compare! The first is the average, and the second is the least squares regression line. We can think of the latter as a the null model where \(\beta_1 = 0\) and \(\hat{y} = \bar{y}\). Which model do you think is better?
##
## Call:
## lm(formula = salary ~ 1, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -55906 -22706 -6406 20479 117839
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 113706 1520 74.8 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 30290 on 396 degrees of freedomp1 <- ggplot(Salaries, aes(x = yrs.since.phd, y = salary)) + geom_point()
p1 + geom_abline(slope = 0, intercept = 113706)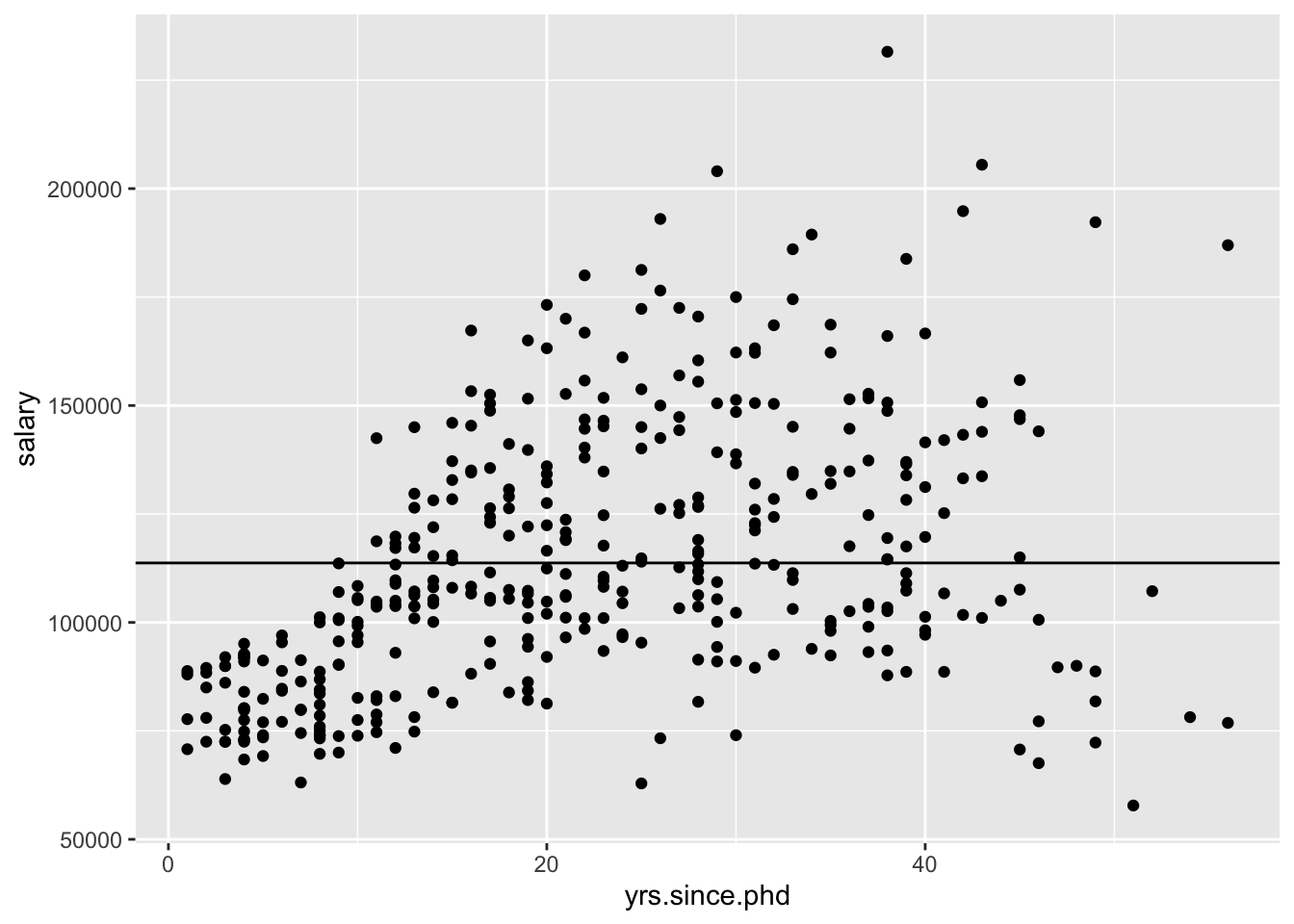
p1 + geom_smooth(method = lm, se = FALSE, color ="red") + geom_abline(slope = 0, intercept = 113706, color = "blue")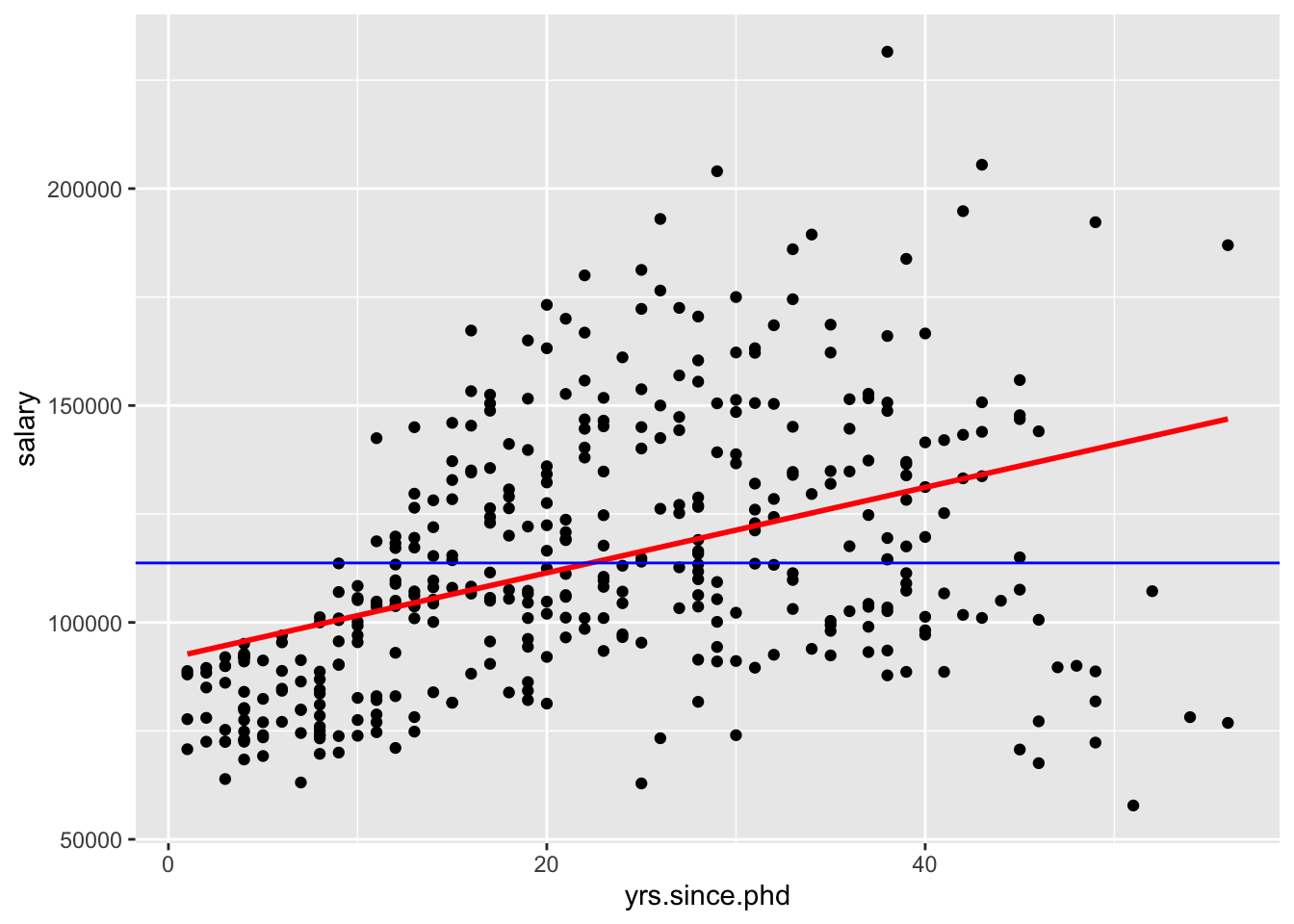
Let’s interpret the slope and intercept:
“For a 1-unit increase in x, we would expect to see a \(\hat{\beta}_1\)-unit increase/decrease in y.”
“For a 1-year increase in years since PhD, we would expect to see a 985-dollar increase in salary.”
8.4.1 Linear model with one categorical predictor
We thought that rank probably explained some of the variability in salary as well. We could make another simple linear regression model using just rank as a predictor,
##
## Call:
## lm(formula = salary ~ rank, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -68972 -16376 -1580 11755 104773
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 80776 2887 27.976 < 2e-16 ***
## rankAssocProf 13100 4131 3.171 0.00164 **
## rankProf 45996 3230 14.238 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 23630 on 394 degrees of freedom
## Multiple R-squared: 0.3943, Adjusted R-squared: 0.3912
## F-statistic: 128.2 on 2 and 394 DF, p-value: < 2.2e-16What is the estimated model formula?
\[ \widehat{salary} = 80776 + 13100\cdot rankAssocProf + 45996\cdot rankProf \]
What?! It seems like something is missing. This is because R splits categorical predictors up into a reference group (the first alphabetically) and indicators for the other groups. Here, Assistant Professors are the reference group and and
\[ \text{rankAssocProf} = \left\{ \begin{array}{ll} 1 \text{ if AssocProf} \\ 0 \text{ otherwise} \end{array} \right. \text{ and } \text{rankProf} = \left\{ \begin{array}{ll} 1 \text{ if Prof} \\ 0 \text{ otherwise} \end{array} \right. \]
In other words, rank is turned into 3 “dummy variables”:
\[\begin{eqnarray*} \begin{pmatrix} Prof \\ Prof\\ AsstProf\\ Prof\\ Prof\\ AssocProf \end{pmatrix} \rightarrow AsstProf = \begin{pmatrix} 0 \\ 0 \\ 1 \\ 0 \\ 0 \\ 0 \end{pmatrix}, AssocProf = \begin{pmatrix} 0 \\ 0 \\ 0 \\ 0 \\ 0 \\ 1 \end{pmatrix}, Prof = \begin{pmatrix} 1 \\ 1 \\ 0 \\ 1 \\ 1 \\ 0 \end{pmatrix} \end{eqnarray*}\]
Since these sum to 1, we only need to put 2 into our model and leave the other out as a reference level. With these ideas in mind, interpret all coefficients in your model. HINT: Plug in 0’s and 1’s to obtain 3 separate models for the different ranks
\[ \widehat{salary}_{AssistProf} = 80776 \\ \widehat{salary}_{AssocProf} = 80776 + 13100 = 93876 \\ \widehat{salary}_{Prof} = 80776 + 45996 = 126772 \]
How are the model coefficients related to the salaries across rank? (That is, what’s the dual meaning of these coefficients?)
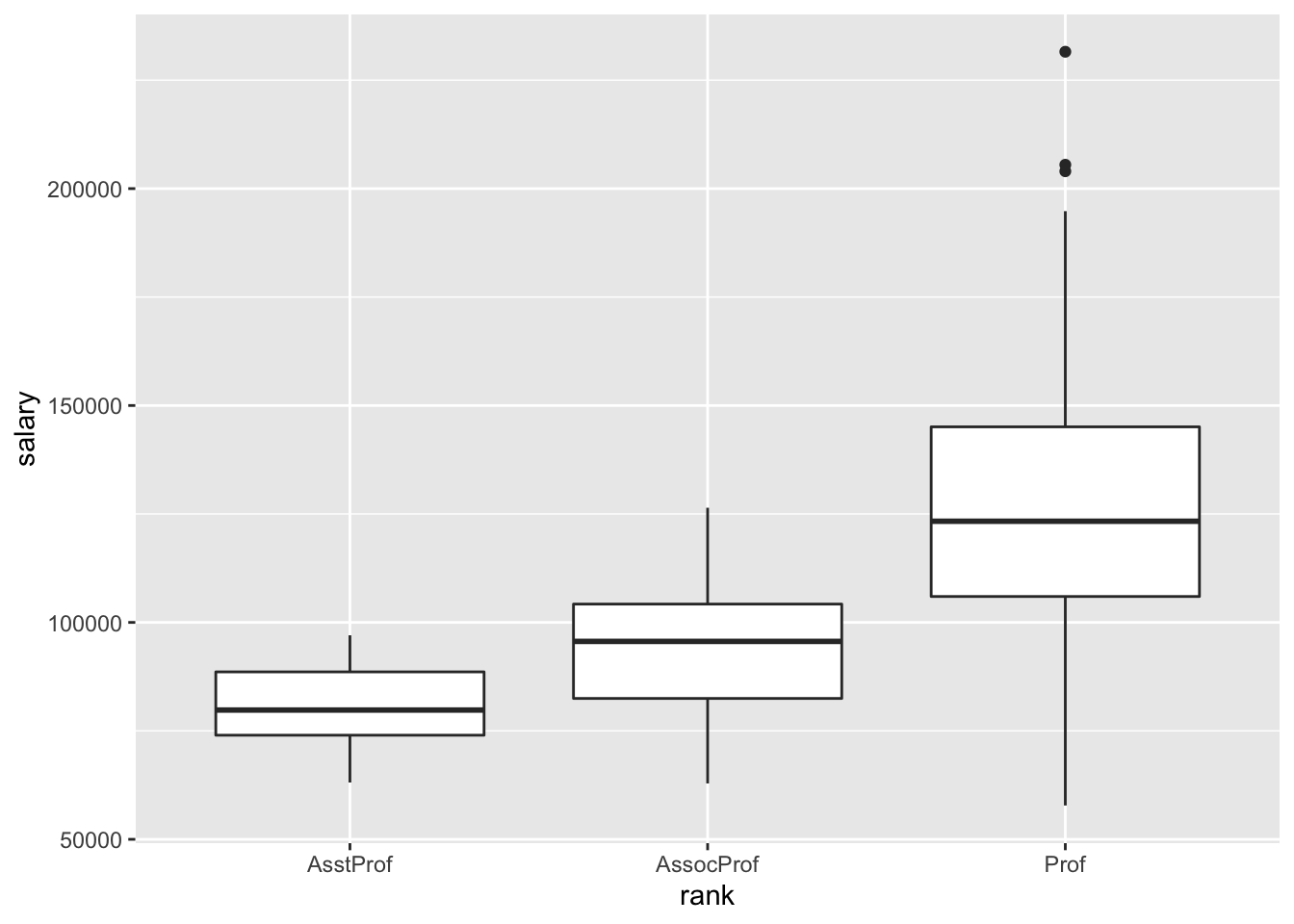
8.5 Multiple linear regression
Once we move beyond a single predict, we are no longer doing “simple” linear regression, and are instead doing “multiple” linear regression.
one quantitative response variable, more than one explanatory variable \[ Y = \beta_0 + \beta_1 \cdot X_1 + \beta_2 \cdot X_2 + \cdots + \beta_p \cdot X_p + \epsilon, \text{ where } \epsilon \sim N(0, \sigma_\epsilon) \]
Parallel slopes
More specifically, consider the case where X_1 is quantitative, but X_2 is an indicator variable. Then,
\[\begin{eqnarray*} \hat{Y} |_{ X_1, X_2 = 0} &= \hat{\beta}_0 + \hat{\beta}_1 \cdot X_1 \\ \hat{Y} |_{ X_1, X_2 = 1} &= \hat{\beta}_0 + \hat{\beta}_1 \cdot X_1 + \hat{\beta}_2 \cdot 1 \\ &= \left( \hat{\beta}_0 + \hat{\beta}_2 \right) + \hat{\beta}_1 \cdot X_1 \end{eqnarray*}\] This is called a parallel slopes model. (Why?)
Let’s consider both yrs.since.phd and rank in our model at once. Let’s build our model,
##
## Call:
## lm(formula = salary ~ rank + yrs.since.phd, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -67148 -16683 -1323 11835 105552
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 81186.23 2964.17 27.389 < 2e-16 ***
## rankAssocProf 13932.18 4345.76 3.206 0.00146 **
## rankProf 47860.42 4412.63 10.846 < 2e-16 ***
## yrs.since.phd -80.37 129.47 -0.621 0.53510
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 23650 on 393 degrees of freedom
## Multiple R-squared: 0.3948, Adjusted R-squared: 0.3902
## F-statistic: 85.47 on 3 and 393 DF, p-value: < 2.2e-16It’s more challenging to visualize this model, because ggplot2 by default won’t create this “parallel slopes” model. However, there is a specialty geom_ from the moderndive package that will help us. (I don’t think I told you to install this package, but you could get it with install.packages("moderndive") if you wanted to follow along very closely.)
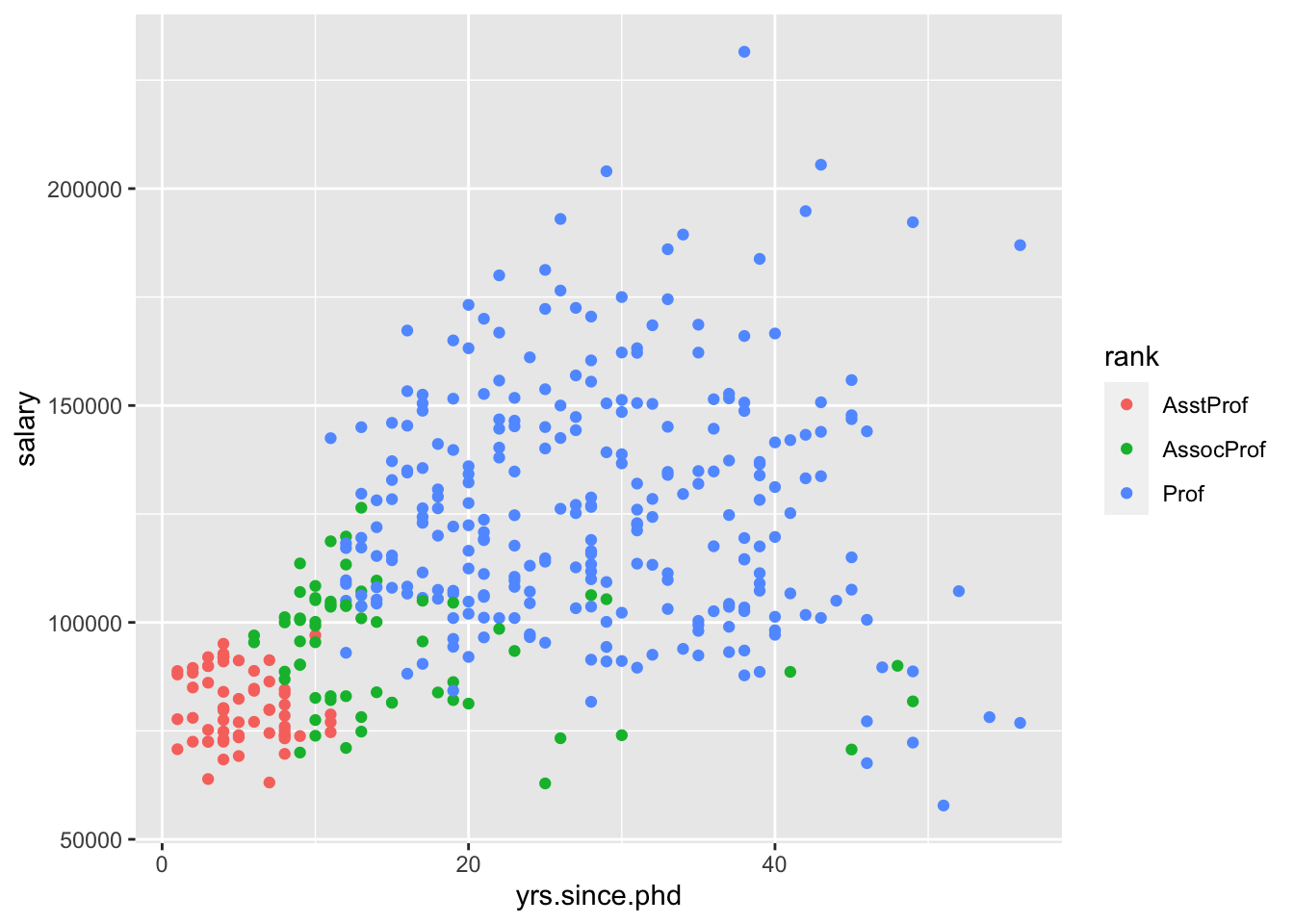
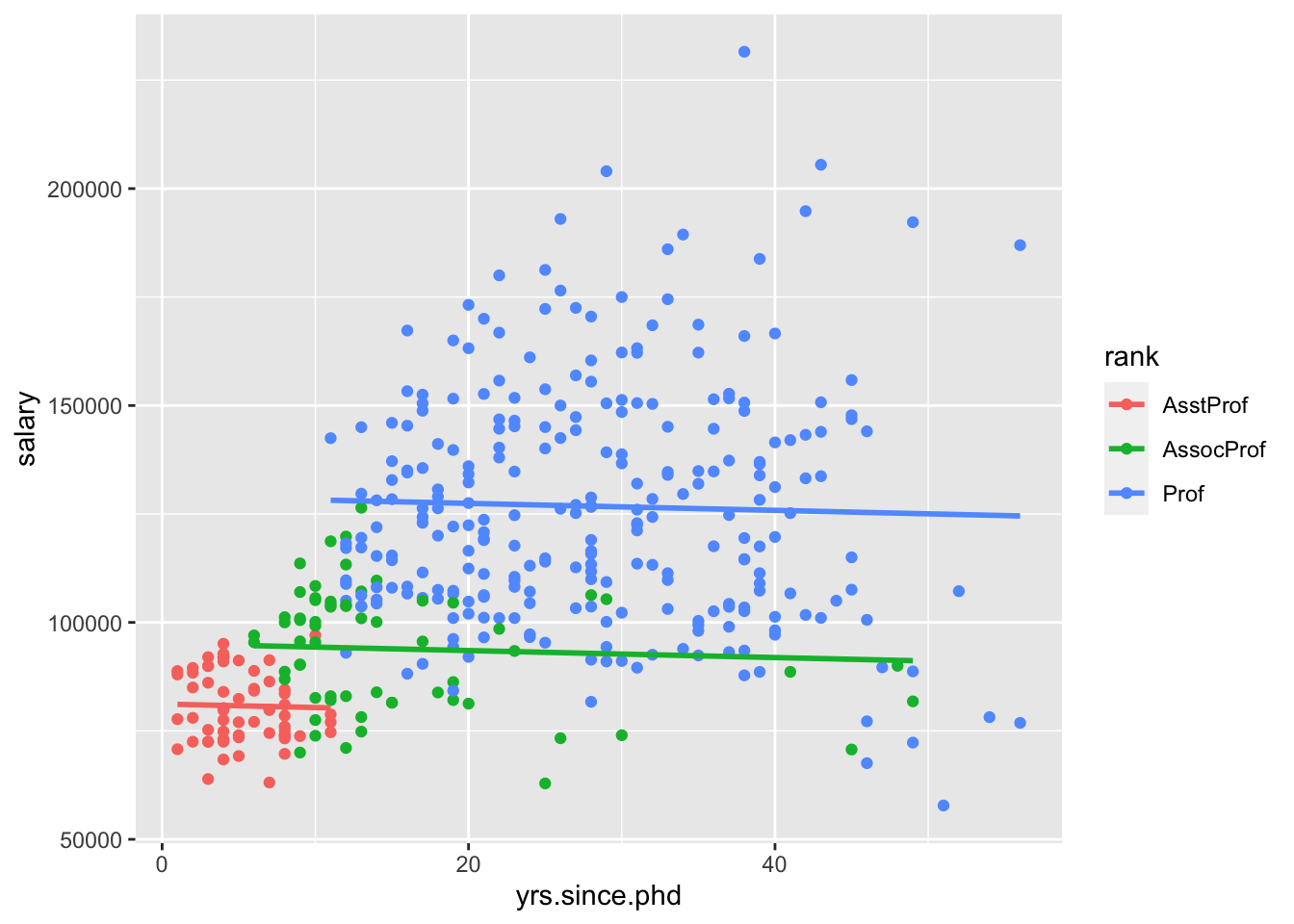
Interpret all the model coefficients by applying the techniques you learned above. These interpretations will differ from those in m1 and m2 – coefficients are defined / interpreted differently depending upon what other predictors are in the model. HINT:
- First write out the estimated model formula: \[ \widehat{salary} = 81186 - 80 \cdot yrs.since.phd + 13932 \cdot rankAssocProf + 47860 \cdot rankProf \]
- Then, notice that the presence of a categorical variable results in the separation of the model formula by group. To this end, plug in 0’s and 1’s to obtain 3 separate model formulas for the ranks: \[ \widehat{salary}_{AssistProf} = 81186 -80 \cdot yrs.since.phd. \\ \widehat{salary}_{AssocProf} = 95118 -80 \cdot yrs.since.phd. \\ \widehat{salary}_{Prof} = 1029046 -80 \cdot yrs.since.phd. \] Is there anything that strikes you as odd about this model? Do you have any concerns about the variables we are including?
Let’s try another model with a categorical variable and a quantitative variable. This time, let’s use discipline as our categorical variable.
##
## Call:
## lm(formula = salary ~ yrs.since.phd + discipline, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -79837 -16607 -4373 15679 93098
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 80158.3 3328.3 24.083 < 2e-16 ***
## yrs.since.phd 1118.5 105.8 10.576 < 2e-16 ***
## disciplineB 15784.2 2733.3 5.775 1.56e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 26470 on 394 degrees of freedom
## Multiple R-squared: 0.2401, Adjusted R-squared: 0.2362
## F-statistic: 62.24 on 2 and 394 DF, p-value: < 2.2e-16Again, I’ll visualize that relationship for you,
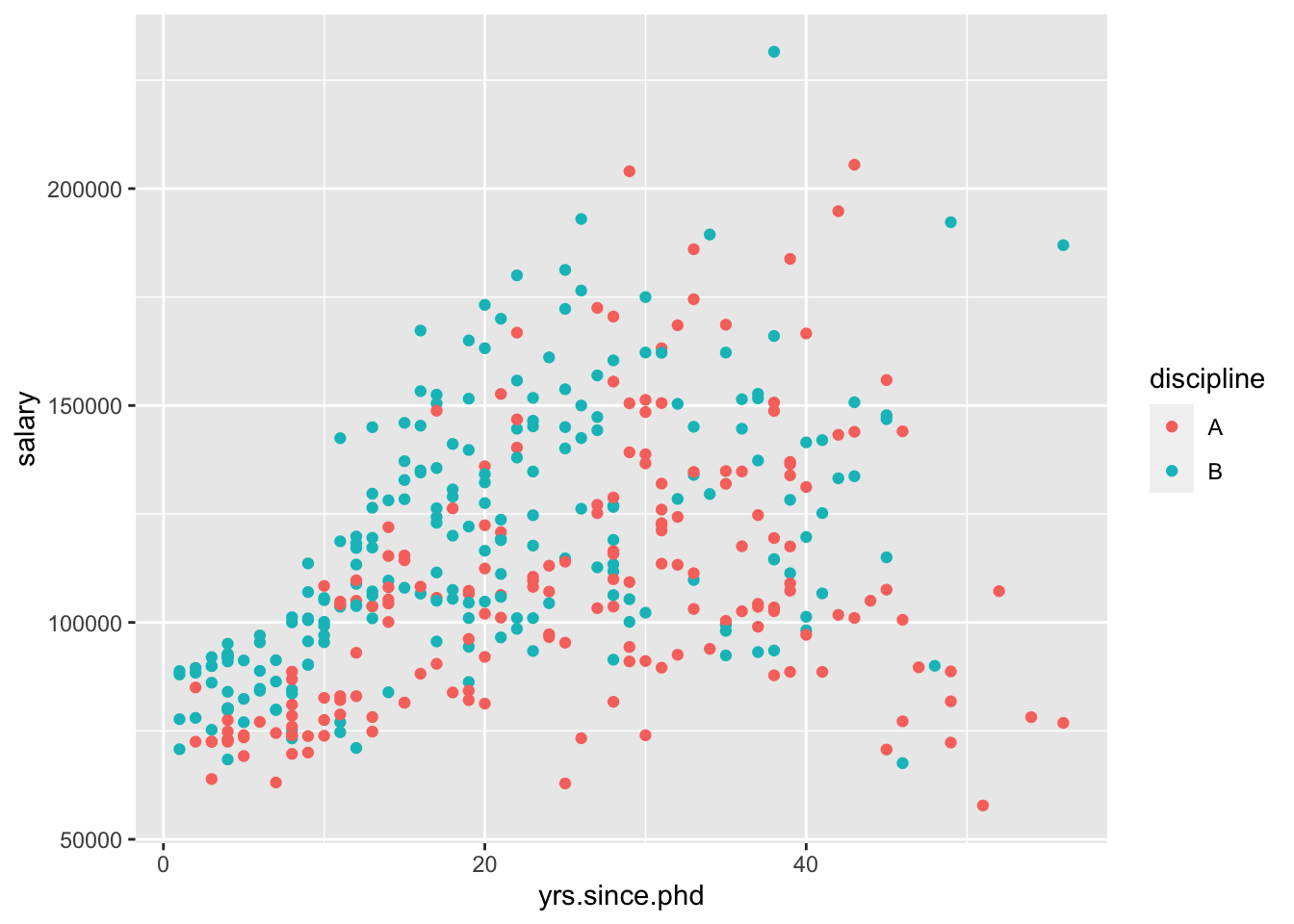
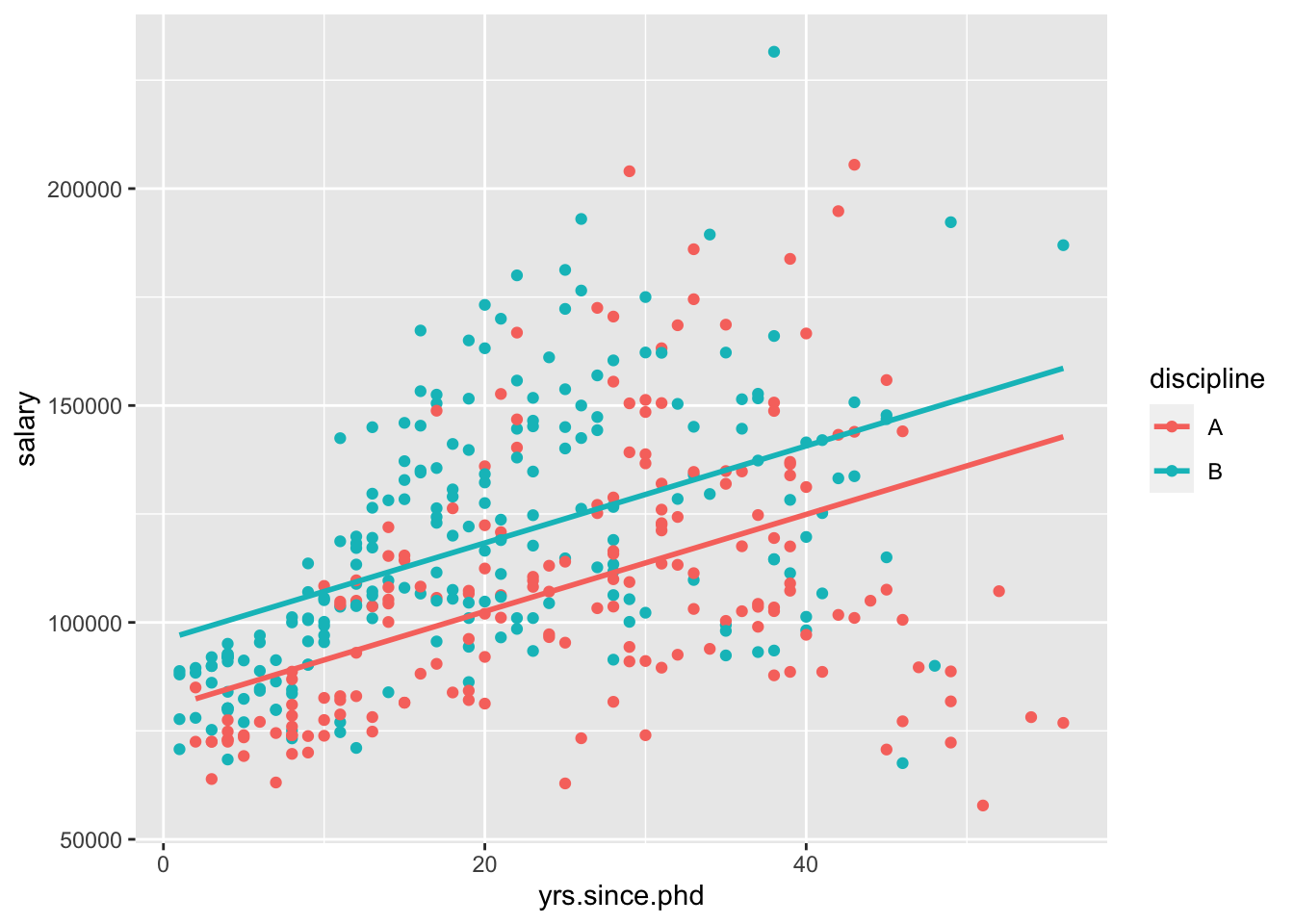
“For a 1-year increase in years since PhD, we would expect a 1118-dollar increase in salary, holding the discipline constant.”
“Compared to discipline A, we would expect people in discipline B to be making 15784 dollars more, holding years of experience constant.”
This model assumes that the relationship between yrs.since.phd and salary is the same between disciplines. However, we could include an interaction between the two variables,
##
## Call:
## lm(formula = salary ~ yrs.since.phd + discipline + yrs.since.phd *
## discipline, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -84580 -16974 -3620 15733 92072
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 84845.4 4283.9 19.806 < 2e-16 ***
## yrs.since.phd 933.9 150.0 6.225 1.24e-09 ***
## disciplineB 7530.0 5492.2 1.371 0.1711
## yrs.since.phd:disciplineB 365.3 211.0 1.731 0.0842 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 26400 on 393 degrees of freedom
## Multiple R-squared: 0.2458, Adjusted R-squared: 0.2401
## F-statistic: 42.7 on 3 and 393 DF, p-value: < 2.2e-16Now, we can visualize this using standard ggplot2 geom_s,
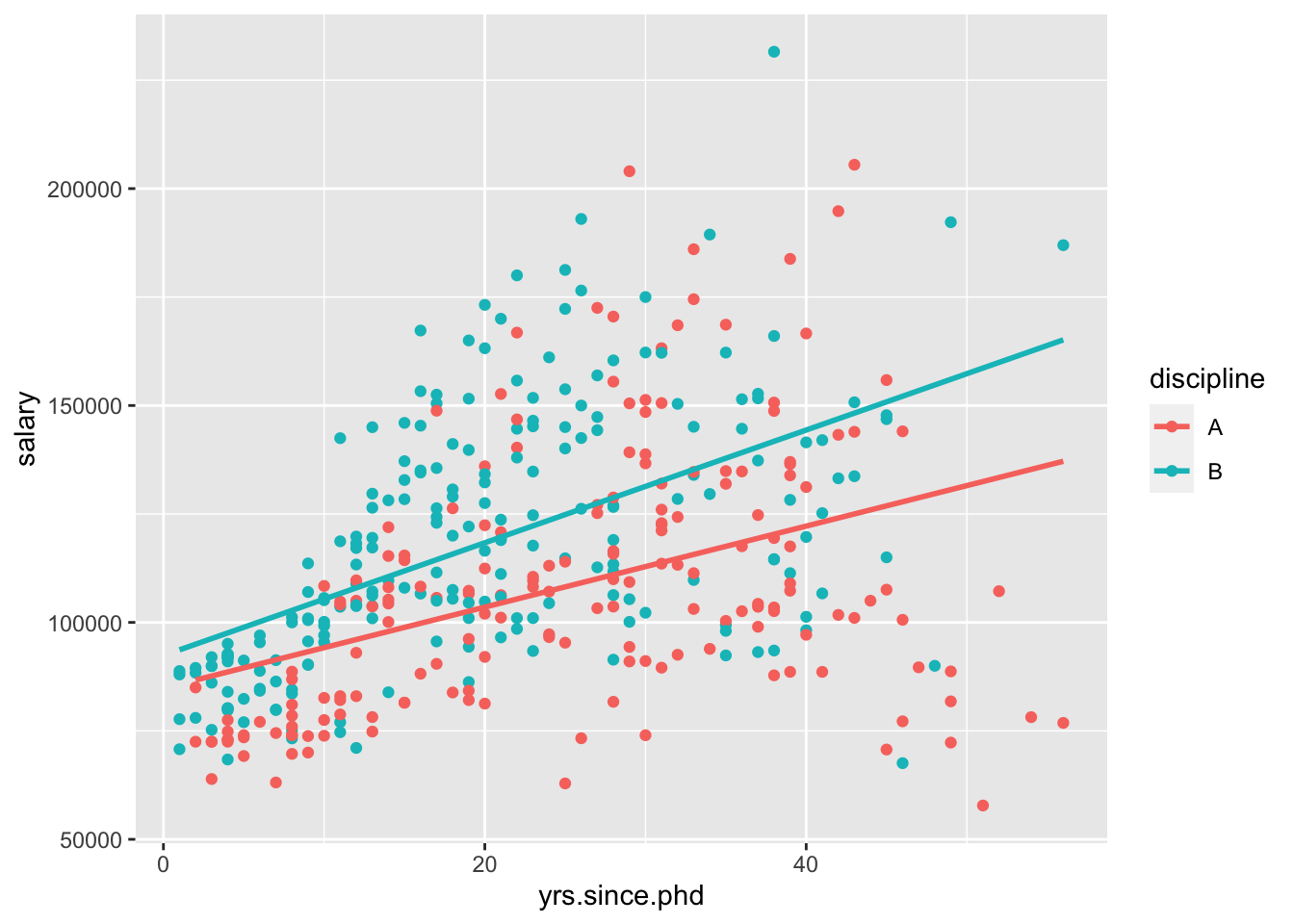
\[ \widehat{salary}_{A} = 84845 + 934\cdot yrs.since.phd \\ \widehat{salary}_{B} = 84845 + 934\cdot yrs.since.phd + 7530 + 365\cdot yrs.since.phd \\ \widehat{salary}_{B} = (84845+ 7530) + (934 + 365)\cdot yrs.since.phd \\ \widehat{salary}_{B} = 92375 + 1299\cdot yrs.since.phd \]
Which of those two models do you prefer?
8.5.1 Models with 2 quantitative predictors
Let’s include both yrs.since.phd and yrs.service in one model.
##
## Call:
## lm(formula = salary ~ yrs.since.phd + yrs.service, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -79735 -19823 -2617 15149 106149
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 89912.2 2843.6 31.620 < 2e-16 ***
## yrs.since.phd 1562.9 256.8 6.086 2.75e-09 ***
## yrs.service -629.1 254.5 -2.472 0.0138 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 27360 on 394 degrees of freedom
## Multiple R-squared: 0.1883, Adjusted R-squared: 0.1842
## F-statistic: 45.71 on 2 and 394 DF, p-value: < 2.2e-16If we were going to visualize this model, what would it look like?
Interpret the yrs.service coefficient.
“For a 1-year increase in years of service, we would expect a $629 decrease in salary, holding years since PhD constant.”
8.5.2 Models with 2 categorical predictors
We could fit a model with rank and sex as our two predictors,
8.5.3 Lots of variables
You don’t have to be limited to just two variables! Multiple regression models can contain as many variables as you (as the analyst) find appropriate.
m8 <- lm(salary ~ yrs.since.phd + yrs.service + rank + discipline + sex + yrs.service*discipline, data = Salaries)
summary(m8)##
## Call:
## lm(formula = salary ~ yrs.since.phd + yrs.service + rank + discipline +
## sex + yrs.service * discipline, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -71931 -13185 -1896 10086 94613
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 70095.1 4885.5 14.348 < 2e-16 ***
## yrs.since.phd 554.0 239.7 2.311 0.02136 *
## yrs.service -687.7 226.7 -3.033 0.00258 **
## rankAssocProf 12166.2 4132.9 2.944 0.00344 **
## rankProf 43744.8 4249.5 10.294 < 2e-16 ***
## disciplineB 6990.5 3907.6 1.789 0.07440 .
## sexMale 5223.0 3840.6 1.360 0.17463
## yrs.service:disciplineB 420.0 177.4 2.367 0.01841 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 22410 on 389 degrees of freedom
## Multiple R-squared: 0.4624, Adjusted R-squared: 0.4527
## F-statistic: 47.8 on 7 and 389 DF, p-value: < 2.2e-168.6 Homework
Before we meet next time, I’d like you to try some visualization and modeling on your own.
- Start a new RMarkdown document
- Load the following packages at the top of your Rmd:
dplyr,ggplot2,fivethirtyeight,skimr - When interpreting visualizations, models, etc, be sure to do so in a contextually meaningful way.
- This homework is a resource for you. Record all work that is useful for your current learning & future reference. Further, try your best, but don’t stay up all night trying to finish all of the exercises! We’ll discuss any questions / material you didn’t get to tomorrow.
For this assignment, you will need an additional package I forgot to tell you to install before! So, run the following code in your Console:
##
## The downloaded binary packages are in
## /var/folders/m7/f7bf48x11fdgvv8wy155qyyhhxrxdr/T//RtmpAbQysS/downloaded_packagesDo not include this code in your RMarkdown document, otherwise you will re-download the package every time you knit!
Once the package has downloaded, you can load the package and load the data
 Horst, Hill, and Gorman (2020)
Horst, Hill, and Gorman (2020)
You should read about the data in the documentation,
Let response variable \(Y\) be the length in mm of a penguin’s flipper and \(X\) be their body mass in grams. Then the (population) linear regression model of \(Y\) vs \(X\)
is
\[ Y=\beta_0+\beta_1\cdot X+ \epsilon \]
Note that the \(\beta_i\) represent the population coefficients. We can’t measure all penguins, thus don’t know the “true” values of the \(\beta_i\). Rather, we can use sample data to estimate the\(\beta_i\) by \(\hat{\beta}_i\).
That is, the sample estimate of the population model trend is
\[ Y=\hat{\beta}_0+\hat{\beta}_1\cdot X \] In choosing the “best” estimates \(\hat{\beta}_i\), we’re looking for the coefficients that best describe the relationship between \(Y\) and \(X\) among the sample subjects. In the visual below, we can see that the red line does a better job at capturing this relationship than the blue line does:
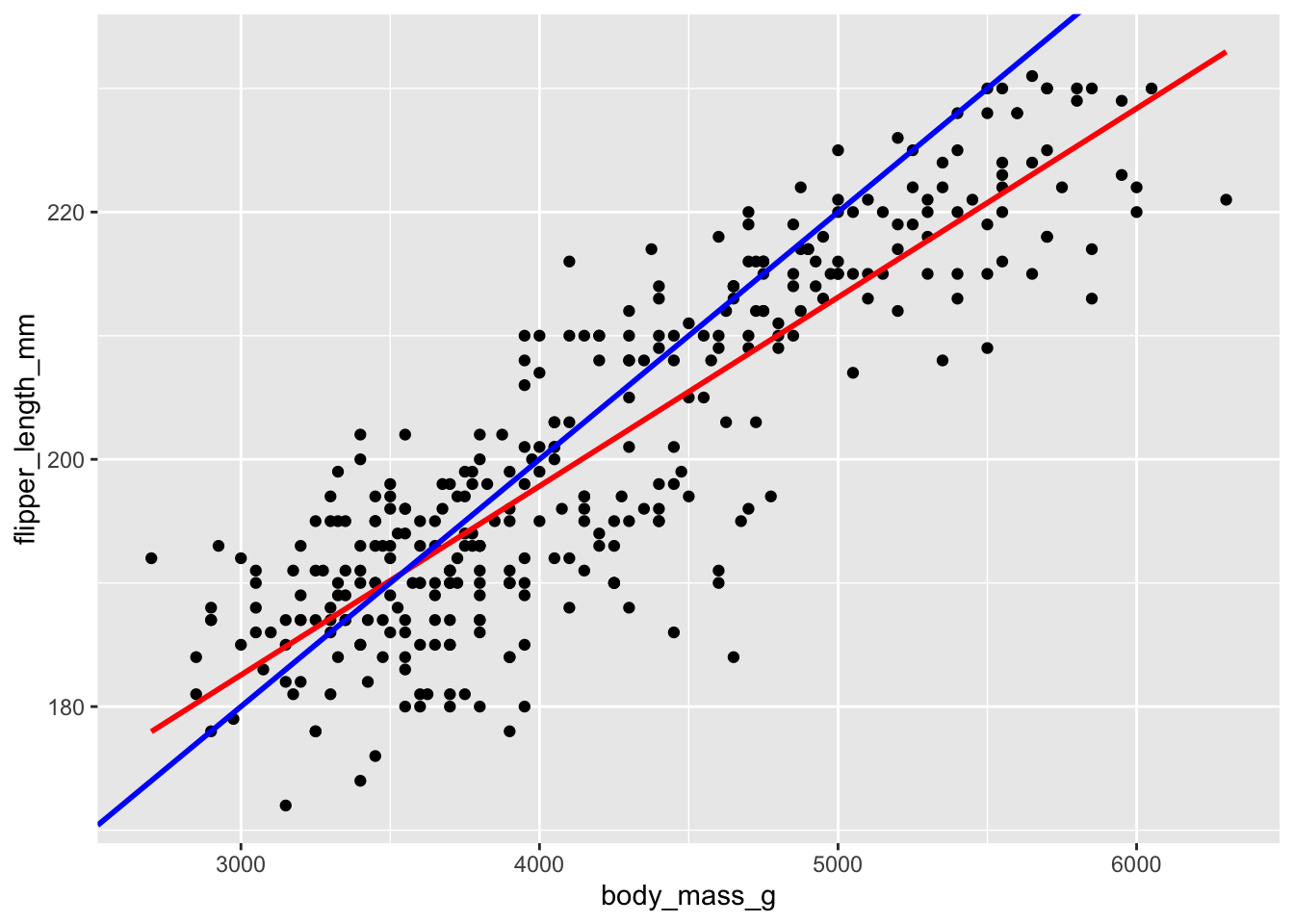
Mainly, on average, the individual points fall closer to the red line than the blue line. The distance between an individual observation and its model value (prediction) is called a residual.
\[ \epsilon_i = y_i - \hat{y}_i \] Let’s use our sample data to estimate the population model.
- Fit a simple linear regression, and look at the summary:
##
## Call:
## lm(formula = flipper_length_mm ~ body_mass_g, data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -23.7626 -4.9138 0.9891 5.1166 16.6392
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.367e+02 1.997e+00 68.47 <2e-16 ***
## body_mass_g 1.528e-02 4.668e-04 32.72 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 6.913 on 340 degrees of freedom
## (2 observations deleted due to missingness)
## Multiple R-squared: 0.759, Adjusted R-squared: 0.7583
## F-statistic: 1071 on 1 and 340 DF, p-value: < 2.2e-16What is the equation of the fitted model?
Interpret the slope and the intercept.
Make predictions for the flipper length of the following two penguins:
## # A tibble: 2 x 4
## species island flipper_length_mm body_mass_g
## <fct> <fct> <int> <int>
## 1 Adelie Torgersen 181 3750
## 2 Gentoo Biscoe 210 4200What are the residuals for these two penguins?
penguin_modis anlm“object.” Not only does it contain info about the model coefficients, it contains the numerical values of the residuals (residuals) and model predictions (fitted.values) for each penguin in the data set:
## [1] "lm"## [1] "coefficients" "residuals" "effects" "rank"
## [5] "fitted.values" "assign" "qr" "df.residual"
## [9] "na.action" "xlevels" "call" "terms"
## [13] "model"You can print out all the residuals in the data frame by doing something like,
## 1 2 3 5 6 7
## -13.01424280 -8.77803858 8.62371500 3.56853188 -2.48665124 -11.10475335
## 8 9 10 11 12 13
## -13.14446474 3.18663399 -11.65220061 -1.14008078 -13.25044702 -3.61248922
## 14 15 16 17 18 19
## -3.77803858 -5.94358795 -8.25044702 5.56853188 -8.47117951 -3.52197867
## 20 21 22 23 24 25
## -6.88840483 -14.66767234 -11.72285546 -5.77803858 -12.06942592 -14.77803858
## 26 27 28 29 30 31
## -7.77803858 -7.95905968 1.38751078 -12.84869344 -17.06942592 -8.37628500
## 32 33 34 35 36 37
## -18.30563014 0.85991922 -12.30563014 7.47802133 -4.12460905 -7.06942592
## 38 39 40 41 42 43
## -10.95905968 -6.14008078 -23.76256685 -2.84869344 -1.30563014 1.91510234
## 44 45 46 47 48 49
## -7.94358795 2.44269390 -16.99877107 -7.04957023 -3.17540821 0.56853188
## 50 51 52 53 54 55
## -9.12460905 -4.19526390 -14.41599639 0.56853188 1.40298251 5.97028546
## 56 57 58 59 60 61
## -2.25044702 -4.95905968 -1.77803858 0.73408124 -0.01424280 0.15130656
## 62 63 64 65 66 67
## -8.94358795 -6.72285546 -6.59701749 3.73408124 -5.06942592 7.09612344
## 68 69 70 71 72 73
## -11.36081327 6.67889812 -6.70738373 -1.72285546 -6.30563014 5.04094032
## 74 75 76 77 78 79
## -3.12460905 -3.25044702 -6.65220061 -2.25044702 -12.30563014 -3.95905968
## 80 81 82 83 84 85
## -2.83322170 3.38751078 -12.52636263 -7.77803858 -7.88840483 3.09612344
## 86 87 88 89 90 91
## 3.04094032 -4.77803858 -1.19526390 -8.06942592 -1.72285546 11.04094032
## 92 93 94 95 96 97
## 2.58400361 -3.66767234 -18.70738373 -0.14008078 5.58400361 -3.25044702
## 98 99 100 101 102 103
## -7.17979217 -3.02971454 -7.36081327 -1.63234491 -5.90826052 -0.70299977
## 104 105 106 107 108 109
## -11.65220061 11.58838757 -6.95905968 4.98575720 -6.30563014 -4.23059133
## 110 111 112 113 114 115
## -12.67205630 2.84006353 -15.99877107 7.38751078 -5.03409850 -5.30563014
## 116 117 118 119 120 121
## -2.97891538 6.97028546 4.60385931 1.09612344 1.47802133 2.15130656
## 122 123 124 125 126 127
## 7.80473610 -13.43146812 6.07626775 2.67889812 1.16677830 4.24181711
## 128 129 130 131 132 133
## -7.41599639 7.67889812 12.16677830 2.47802133 6.80473610 2.80473610
## 134 135 136 137 138 139
## -6.08928162 -2.04957023 -6.30563014 5.76940867 2.54867619 -3.66767234
## 140 141 142 143 144 145
## -8.65220061 4.33232766 -2.81336601 4.67889812 -3.63234491 9.44269390
## 146 147 148 149 150 151
## -7.48665124 -11.65220061 -5.81336601 5.56853188 -1.01424280 -6.25044702
## 152 153 154 155 156 157
## 3.16677830 5.52882049 6.19772176 5.29261627 -5.80227824 -4.21950356
## 158 159 160 161 162 163
## 3.76502471 0.94604581 2.83567957 5.05641205 -0.40052465 6.23743315
## 164 165 166 167 168 169
## -5.51089090 6.23743315 -13.09366558 9.11159517 -9.09366558 9.87539095
## 170 171 172 173 174 175
## -11.96782760 -1.05395419 3.54429222 -5.80227824 1.89086269 9.05641205
## 176 177 178 179 180 181
## 1.12706691 1.89086269 0.36327113 16.63918673 -8.03848246 3.00122893
## 182 183 184 185 186 187
## -1.51089090 5.07188379 0.47363737 -6.87293309 0.85115130 4.59947535
## 188 189 190 191 192 193
## 0.78049644 0.65465847 2.07188379 4.82020783 -10.45570778 10.93057408
## 194 195 196 197 198 199
## 1.19772176 7.58400361 6.70984159 0.48910910 5.41845425 9.11159517
## 200 201 202 203 204 205
## 5.78049644 -1.63672887 -2.69191200 -0.81774997 2.30808800 6.05641205
## 206 207 208 209 210 211
## 11.89086269 5.41845425 6.12706691 5.58400361 6.89086269 3.29261627
## 212 213 214 215 216 217
## 2.48910910 7.11159517 3.30808800 10.05641205 7.96151754 10.47363737
## 218 219 220 221 222 223
## 6.19772176 6.23743315 3.67013020 11.47363737 1.48910910 6.70984159
## 224 225 226 227 228 229
## 7.89086269 6.36327113 0.83567957 7.47363737 4.67013020 2.00122893
## 230 231 232 233 234 235
## -8.38505292 5.70984159 -4.62125714 4.61933104 1.01670066 3.09173948
## 236 237 238 239 240 241
## 5.54429222 2.70984159 5.72531332 11.00122893 0.30808800 0.80035214
## 242 243 244 245 246 247
## 8.48910910 5.65465847 8.78049644 2.70984159 0.96151754 3.18225003
## 248 249 250 251 252 253
## 9.83567957 4.03655636 10.80035214 -4.38066896 8.07188379 8.18225003
## 254 255 256 257 258 259
## 5.72531332 2.27276058 7.25290488 7.09173948 -5.74709512 1.47363737
## 260 261 262 263 264 265
## -1.74709512 1.38312682 -11.74709512 2.89086269 1.37874286 5.23743315
## 266 267 268 269 270 271
## 9.25290488 13.43830994 3.90633442 5.80035214 -6.38505292 2.03655636
## 273 274 275 276 277 278
## 4.18225003 -2.56607402 -4.16432043 -6.21950356 1.80473610 -0.30563014
## 279 280 281 282 283 284
## 0.51334876 -2.57716179 3.36765509 0.93057408 -8.37628500 2.98575720
## 285 286 287 288 289 290
## -5.12460905 4.74955298 -1.77803858 -0.39614069 -8.25044702 2.40298251
## 291 292 293 294 295 296
## -1.34095757 2.40298251 9.85991922 -12.25044702 0.56853188 -8.94358795
## 297 298 299 300 301 302
## -10.72285546 2.33232766 5.97028546 -1.77803858 7.85991922 -3.12460905
## 303 304 305 306 307 308
## 11.33232766 5.22196142 -2.25044702 -1.23497529 1.38751078 -1.41599639
## 309 310 311 312 313 314
## -0.90387656 3.63918673 3.27714454 2.69436986 -0.54183436 -0.05395419
## 315 316 317 318 319 320
## 14.02546859 -0.47117951 12.93057408 -5.48665124 5.04094032 5.80473610
## 321 322 323 324 325 326
## 3.13145087 -3.70738373 1.33232766 9.58400361 0.62371500 5.13145087
## 327 328 329 330 331 332
## 11.47802133 3.93057408 1.27714454 4.40298251 -0.90387656 7.56853188
## 333 334 335 336 337 338
## 4.62371500 4.40298251 7.22196142 3.42283821 8.93057408 -3.48665124
## 339 340 341 342 343 344
## 2.51334876 9.16677830 13.33232766 -1.39614069 10.63918673 3.60385931- See if you can confirm that the mean residual equals 0. This property always holds for regression models!
## Min. 1st Qu. Median Mean 3rd Qu. Max.
## -23.7626 -4.9138 0.9891 0.0000 5.1166 16.6392## [1] -4.416659e-16- Calculate the standard deviation of the residuals. Where does this information appear in
summary(penguinmod)(within rounding error)? SIDENOTE: The Residual standard error is calculated slightly differently than the standard deviation of residuals. Let \(\epsilon_i\) denote a residual for case \(i\), \(n\) as sample size, and \(k\) the number of coefficients in our model (here we have 2), then the standard deviation of residuals are
\[ \frac{\sum_{i=1}^n \epsilon_i^2}{n-1} \] whereas the residual standard error is
\[ \frac{\sum_{i=1}^n \epsilon_i^2}{n-k} \]
## [1] 6.903249Now that you have thought a lot about a simple linear regression model, explore some more variables. Try making plots to see if you can find other variables that help explain some variability in flipper_length_mm.
Then, try out a variety of models. Try using more than one variable, making parallel slopes, including interaction effects, etc. You can name your models as you go, but I am most interested in your “final” favorite model.
- Write down your favorite model,
favmod <- lm(flipper_length_mm ~ species + bill_depth_mm + body_mass_g + sex,
data = penguins)
summary(favmod)##
## Call:
## lm(formula = flipper_length_mm ~ species + bill_depth_mm + body_mass_g +
## sex, data = penguins)
##
## Residuals:
## Min 1Q Median 3Q Max
## -14.358 -3.185 0.009 3.036 17.782
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.497e+02 6.372e+00 23.494 < 2e-16 ***
## speciesChinstrap 5.491e+00 7.781e-01 7.057 1.02e-11 ***
## speciesGentoo 2.227e+01 2.131e+00 10.453 < 2e-16 ***
## bill_depth_mm 9.786e-01 3.639e-01 2.689 0.00753 **
## body_mass_g 5.859e-03 9.572e-04 6.121 2.66e-09 ***
## sexmale 1.466e+00 9.259e-01 1.584 0.11426
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.295 on 327 degrees of freedom
## (11 observations deleted due to missingness)
## Multiple R-squared: 0.8594, Adjusted R-squared: 0.8573
## F-statistic: 399.8 on 5 and 327 DF, p-value: < 2.2e-16Why is this your preferred model? What is good about it?
How would you visualize your model object?
## Warning: Removed 2 rows containing missing values (geom_point).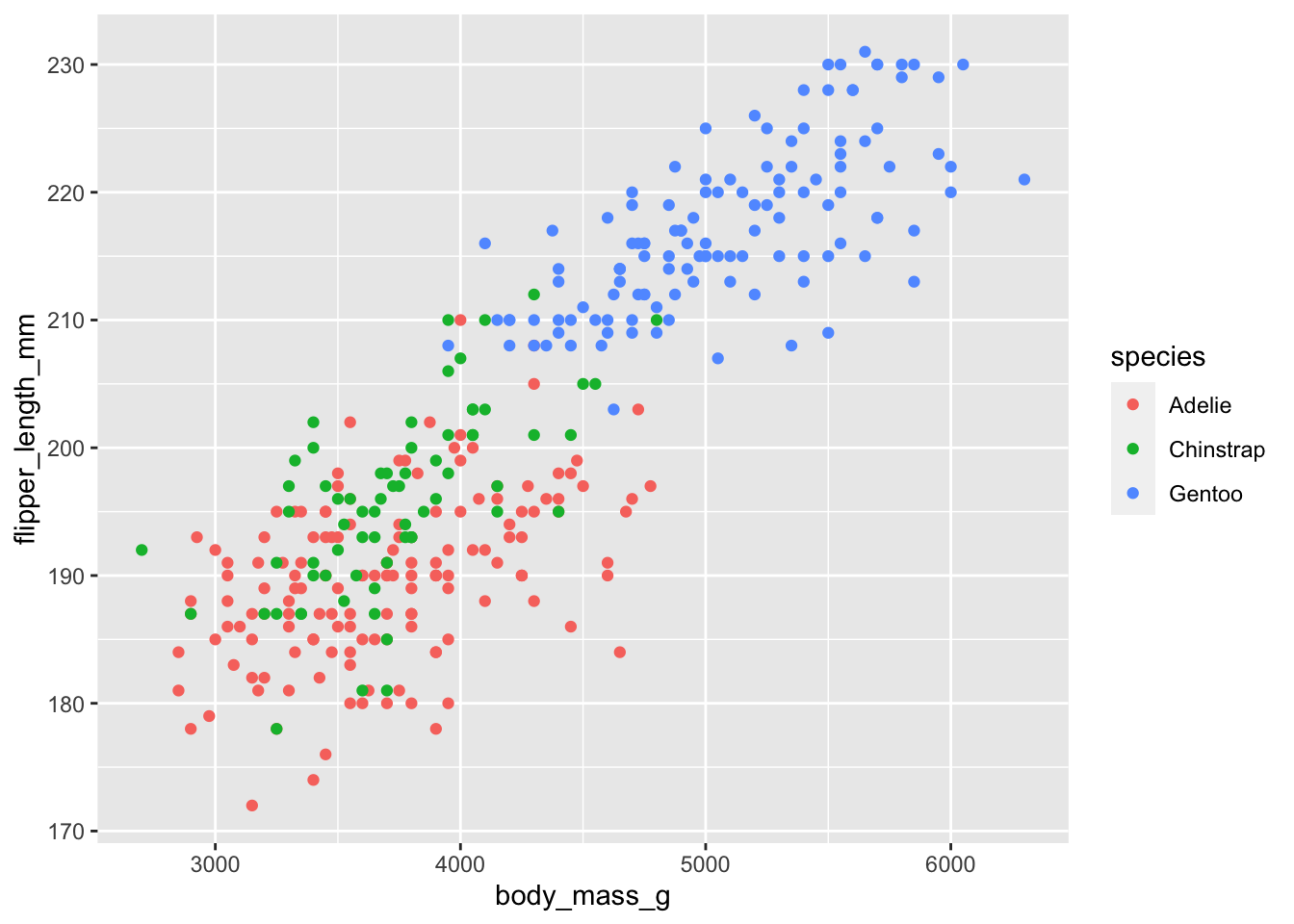
References
Horst, Allison Marie, Alison Presmanes Hill, and Kristen B Gorman. 2020. Palmerpenguins: Palmer Archipelago (Antarctica) Penguin Data. https://doi.org/10.5281/zenodo.3960218.
Wickham, Hadley, and Garrett Grolemund. 2017. R for Data Science. O’Reilly. https://r4ds.had.co.nz.