Chapter 9 Data wrangling for model assessment
[This “chapter” is essentially the same as 3, but with some of the notes I wrote in the document as we went.]
We’ll talk about the dplyr package, and assessing models.
Much of this code was adapted from Master the Tidyverse
9.1 dplyr verbs
We have already seen some dplyr verbs, but we are going to need more as we move on. So, let’s spend some time focusing on them. Here are the main verbs we will cover:
| verb | action | example |
|---|---|---|
select() |
take a subset of columns | select(x,y), select(-x) |
filter() |
take a subset of rows | filter(x == __, y > __) |
arrange() |
reorder the rows | arrange(x), arrange(desc(x)) |
summarize() |
a many-to-one or many-to-few summary | summarize(mean(x), median(y)) |
group_by() |
group the rows by a specified column | group_by(x) %>% something() |
mutate() |
a many-to-many operation that creates a new variable | mutate(x = ___, y = ___) |
We’ll use the babynames package to play with the verbs, so let’s begin by loading that dataset.
We could skim the data, to learn something about it:
| Name | babynames |
| Number of rows | 1924665 |
| Number of columns | 5 |
| _______________________ | |
| Column type frequency: | |
| character | 2 |
| numeric | 3 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| sex | 0 | 1 | 1 | 1 | 0 | 2 | 0 |
| name | 0 | 1 | 2 | 15 | 0 | 97310 | 0 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| year | 0 | 1 | 1974.85 | 34.03 | 1880 | 1951 | 1985 | 2003 | 2017.00 | ▁▂▃▅▇ |
| n | 0 | 1 | 180.87 | 1533.34 | 5 | 7 | 12 | 32 | 99686.00 | ▇▁▁▁▁ |
| prop | 0 | 1 | 0.00 | 0.00 | 0 | 0 | 0 | 0 | 0.08 | ▇▁▁▁▁ |
9.1.1 select()
## # A tibble: 1,924,665 x 2
## name prop
## <chr> <dbl>
## 1 Mary 0.0724
## 2 Anna 0.0267
## 3 Emma 0.0205
## 4 Elizabeth 0.0199
## 5 Minnie 0.0179
## 6 Margaret 0.0162
## 7 Ida 0.0151
## 8 Alice 0.0145
## 9 Bertha 0.0135
## 10 Sarah 0.0132
## # … with 1,924,655 more rows- Alter the code to select just the
ncolumn
## # A tibble: 1,924,665 x 1
## n
## <int>
## 1 7065
## 2 2604
## 3 2003
## 4 1939
## 5 1746
## 6 1578
## 7 1472
## 8 1414
## 9 1320
## 10 1288
## # … with 1,924,655 more rows9.1.1.1 select() helpers
:select range of columns,select(storms, storm:pressure)-select every column butselect(storms, -c(storm, pressure))starts_with()select columns that start with…select(storms, starts_with("w"))ends_with()select columns that end with…select(storms, ends_with("e"))- …and more! Check out the Data Transformation cheatsheet
9.1.2 filter()
extract rows that meet logical criteria
## # A tibble: 169 x 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1880 F Amelia 221 0.00226
## 2 1881 F Amelia 235 0.00238
## 3 1882 F Amelia 252 0.00218
## 4 1883 F Amelia 262 0.00218
## 5 1884 F Amelia 315 0.00229
## 6 1885 F Amelia 298 0.00210
## 7 1886 F Amelia 326 0.00212
## 8 1887 F Amelia 344 0.00221
## 9 1888 F Amelia 358 0.00189
## 10 1889 F Amelia 346 0.00183
## # … with 159 more rowsNotice I’m using ==, which tests if things are equal. In R, = sets something. There are other logical comparisons you can use
| x < y | less than |
| x > y | greater than |
| x == y | equal to |
| x <= y | less than or equal to |
| x >= y | greater than or equal to |
| x != y | not equal to |
| x %in% y | group membership |
| is.na(x) | is NA |
| !is.na(x) | is not NA |
- Now, see if you can use the logical operators to manipulate our code to show:
- All of the names where
propis greater than or equal to 0.08 - All of the children named “Sea”
- All of the names that have a missing value for
n(Hint: this should return an empty data set).
Common mistakes:
- using
=instead of== - forgetting quotes
We can also filter rows that match every logical criteria,
## # A tibble: 1 x 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1880 F Amelia 221 0.00226For this, you need to use Boolean operators
| a & b | and |
| a | b | or |
| xor(a, b) | exactly or |
| !a | not |
| a %in% c(a, b) | one of (in) |
- Use Boolean operators to alter the code below to return only the rows that contain:
- Girls named Sea
- Names that were used by exactly 5 or 6 children in 1880
- Names that are one of Acura, Lexus, or Yugo
## # A tibble: 57 x 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1990 F Lexus 36 0.0000175
## 2 1990 M Lexus 12 0.00000558
## 3 1991 F Lexus 102 0.0000502
## 4 1991 M Lexus 16 0.00000755
## 5 1992 F Lexus 193 0.0000963
## 6 1992 M Lexus 25 0.0000119
## 7 1993 F Lexus 285 0.000145
## 8 1993 M Lexus 30 0.0000145
## 9 1994 F Lexus 381 0.000195
## 10 1994 F Acura 6 0.00000308
## # … with 47 more rows9.1.3 arrange()
Orders rows from smallest to largest values
- Arrange babynames by n. Add prop as a second (tie breaking) variable to arrange by.
## # A tibble: 1,924,665 x 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1880 F Adelle 5 0.0000512
## 2 1880 F Adina 5 0.0000512
## 3 1880 F Adrienne 5 0.0000512
## 4 1880 F Albertine 5 0.0000512
## 5 1880 F Alys 5 0.0000512
## 6 1880 F Ana 5 0.0000512
## 7 1880 F Araminta 5 0.0000512
## 8 1880 F Arthur 5 0.0000512
## 9 1880 F Birtha 5 0.0000512
## 10 1880 F Bulah 5 0.0000512
## # … with 1,924,655 more rows- Can you tell what the smallest value of
nis? Any guesses why?
Another helpful function is desc(), which changes the ordering to largest smallest,
## # A tibble: 1,924,665 x 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1947 F Linda 99686 0.0548
## 2 1948 F Linda 96209 0.0552
## 3 1947 M James 94756 0.0510
## 4 1957 M Michael 92695 0.0424
## 5 1947 M Robert 91642 0.0493
## 6 1949 F Linda 91016 0.0518
## 7 1956 M Michael 90620 0.0423
## 8 1958 M Michael 90520 0.0420
## 9 1948 M James 88588 0.0497
## 10 1954 M Michael 88514 0.0428
## # … with 1,924,655 more rows- Use
desc()to find the names with the highestprop.
## # A tibble: 1,924,665 x 5
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1880 M John 9655 0.0815
## 2 1881 M John 8769 0.0810
## 3 1880 M William 9532 0.0805
## 4 1883 M John 8894 0.0791
## 5 1881 M William 8524 0.0787
## 6 1882 M John 9557 0.0783
## 7 1884 M John 9388 0.0765
## 8 1882 M William 9298 0.0762
## 9 1886 M John 9026 0.0758
## 10 1885 M John 8756 0.0755
## # … with 1,924,655 more rows- Use
desc()to find the names with the highestn.
9.2 %>%, the pipe
I %>%
— Hadley Wickham (@hadleywickham) February 11, 2021
tumble(out_of = "bed") %>%
stumble(to = "the kitchen") %>%
pour(who = "myself", unit = "cup", what = "ambition") %>%
yawn() %>%
stretch() %>%
try(come_to_live()) https://t.co/plnA2dZdJ4
In other words, you can nest functions together in R, much like
\[ f(g(x)) \]
but, once you go beyond a function or two, that becomes hard to read.
try(come_to_life(stretch(yawn(pour(stumble(tumble(I, out_of = "bed"), to = "the kitchen"), who = "myself", unit = "cup", what = "ambition")))))The pipe allows you to unnest your functions, and pass data along a pipeline.
I %>%
tumble(out_of = "bed") %>%
stumble(to = "the kitchen") %>%
pour(who = "myself", unit = "cup", what = "ambition") %>%
yawn() %>%
stretch() %>%
try(come_to_life())(Those examples are not valid R code!)
We could see this with a more real-life example:
## # A tibble: 14,024 x 2
## name n
## <chr> <int>
## 1 Noah 19613
## 2 Liam 18355
## 3 Mason 16610
## 4 Jacob 15938
## 5 William 15889
## 6 Ethan 15069
## 7 James 14799
## 8 Alexander 14531
## 9 Michael 14413
## 10 Benjamin 13692
## # … with 14,014 more rowsWhat does this code do?
## # A tibble: 14,024 x 2
## name n
## <chr> <int>
## 1 Noah 19613
## 2 Liam 18355
## 3 Mason 16610
## 4 Jacob 15938
## 5 William 15889
## 6 Ethan 15069
## 7 James 14799
## 8 Alexander 14531
## 9 Michael 14413
## 10 Benjamin 13692
## # … with 14,014 more rowsWhat does this code do?
babynames %>%
filter(year == 2015, sex == "M") %>%
arrange(desc(n)) %>%
lm(prop~year, data = .) # . passes data in a different spot##
## Call:
## lm(formula = prop ~ year, data = .)
##
## Coefficients:
## (Intercept) year
## 6.681e-05 NAWe pronounce the pipe, %>%, as “then.”
[Side note: many languages use | as the pipe, but that means “or” or “given” in R, depending on the syntax.]
- Use
%>%to write a sequence of functions that
- Filter babynames to just the girls that were born in 2015
- Select the
nameandncolumns - Arrange the results so that the most popular names are near the top.
library(ggplot2)
babynames %>%
filter(name %in% c("Amelia", "Richard", "Sofia")) %>%
ggplot(aes(x=year, y=n, color = name)) +
geom_line() +
facet_wrap(~sex)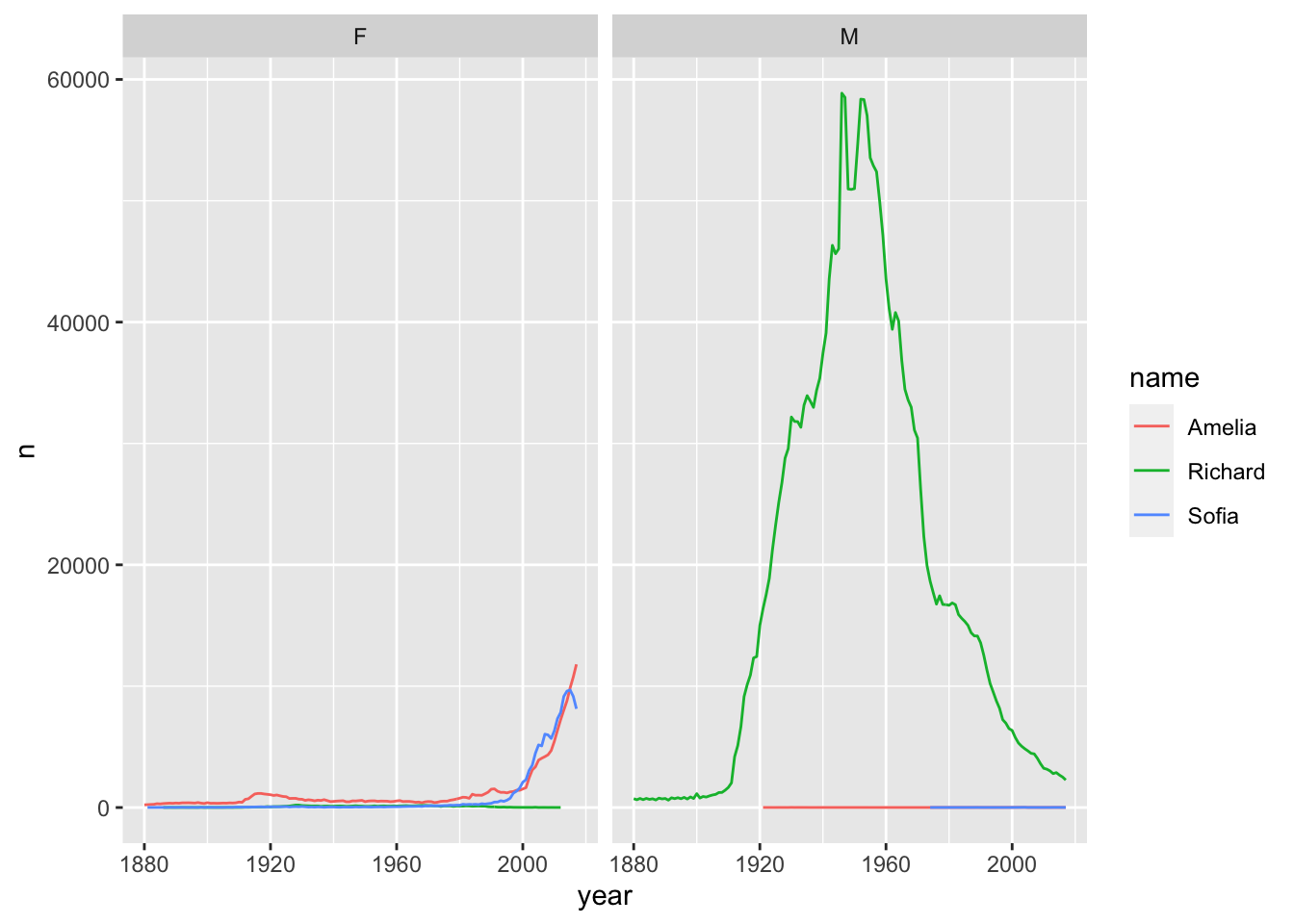
- [Combining
dplyrknowledge withggplot2!]
- Trim
babynamesto just the rows that contain a particular name and sex. This could be your name/sex or that of a friend or famous person. - Trim the result to just the columns that you’ll need for the plot
- Plot the results as a line graph with
yearon the x axis andpropon the y axis
[Hint: “trim” here is a colloquial word, you will need to translate it to the appropriate dplyr verb in each case.]
9.3 Modeling conditions
9.3.1 Least squares
When R finds the line of best fit, it is minimizing the sum of the squared residuals,
\[ SSE = \sum_{i=1}^n (y_i - \hat{y_i})^2 \] in order for the model to be appropriate, a number of conditions must be met.
- Linearity
- Independence
- Normality
- Equality of variance
These conditions are mainly related to the distribution of the residuals.
| Assumption | Consequence | Diagnostic | Solution |
|---|
Independence | inaccurate inference | common sense/context | use a different technique/ don’t model \(E(\epsilon)=0\) | lack of model fit | plot of residuals vs. fitted values | transform \(x\) and/or \(y\) \(Var(\epsilon)=\sigma^2\) | inaccurate inference | plot of residuals v. fitted values | transform \(y\) \(\epsilon\sim N(\mu, \sigma)\) | if extreme, inaccurate inference | QQ plot | transform \(y\)
We would like to be able to work with the residuals from our models to assess whether the conditions are met, as well as to determine which model explains the most variability.
We would like to be able to work with the model objects we created yesterday using dplyr verbs, but model objects are untidy objects. This is where the broom package comes in! broom helps you tidy up your models. Its two most useful functions are augment and tidy.
Let’s re-create our simple linear regression model from before (again, I’m hoping this isn’t just hanging out in your Environment already!).
Let’s augment() that model.
Look at the new object in your environment. What is it like?
One parameter to augment() is data=. Let’s try again with that,
What’s different about that object?
We could use this augmented version of our dataset to do things like look for the largest residuals.
- Use a
dplyrverb to find the rows where we over-predicted the most.
## # A tibble: 397 x 12
## rank discipline yrs.since.phd yrs.service sex salary .fitted .resid
## <fct> <fct> <int> <int> <fct> <int> <dbl> <dbl>
## 1 Prof B 38 38 Male 231545 129162. 102383.
## 2 Prof A 51 51 Male 57800 141971. -84171.
## 3 Prof A 29 7 Male 204000 120294. 83706.
## 4 Prof B 26 19 Male 193000 117338. 75662.
## 5 Prof A 43 43 Male 205500 134088. 71412.
## 6 Prof A 56 57 Male 76840 146898. -70058.
## 7 Prof B 46 45 Male 67559 137044. -69485.
## 8 Prof A 49 43 Male 72300 140000. -67700.
## 9 Prof A 54 49 Male 78162 144927. -66765.
## 10 Prof B 22 9 Male 180000 113396. 66604.
## # … with 387 more rows, and 4 more variables: .hat <dbl>, .sigma <dbl>,
## # .cooksd <dbl>, .std.resid <dbl>## # A tibble: 221 x 12
## rank discipline yrs.since.phd yrs.service sex salary .fitted .resid
## <fct> <fct> <int> <int> <fct> <int> <dbl> <dbl>
## 1 Asst… B 4 3 Male 79750 95660. -15910.
## 2 Prof B 45 39 Male 115000 136059. -21059.
## 3 Asso… B 6 6 Male 97000 97631. -631.
## 4 Asst… B 7 2 Male 79800 98616. -18816.
## 5 Asst… B 1 1 Male 77700 92704. -15004.
## 6 Asst… B 2 0 Male 78000 93689. -15689.
## 7 Prof B 20 18 Male 104800 111426. -6626.
## 8 Prof B 19 20 Male 101000 110440. -9440.
## 9 Prof A 38 34 Male 103450 129162. -25712.
## 10 Prof A 37 23 Male 124750 128176. -3426.
## # … with 211 more rows, and 4 more variables: .hat <dbl>, .sigma <dbl>,
## # .cooksd <dbl>, .std.resid <dbl>We could also use this dataset to plot our residuals, to see if they conform to our conditions. One way to see residual plots is to use the convenience function plot() on our original model object.
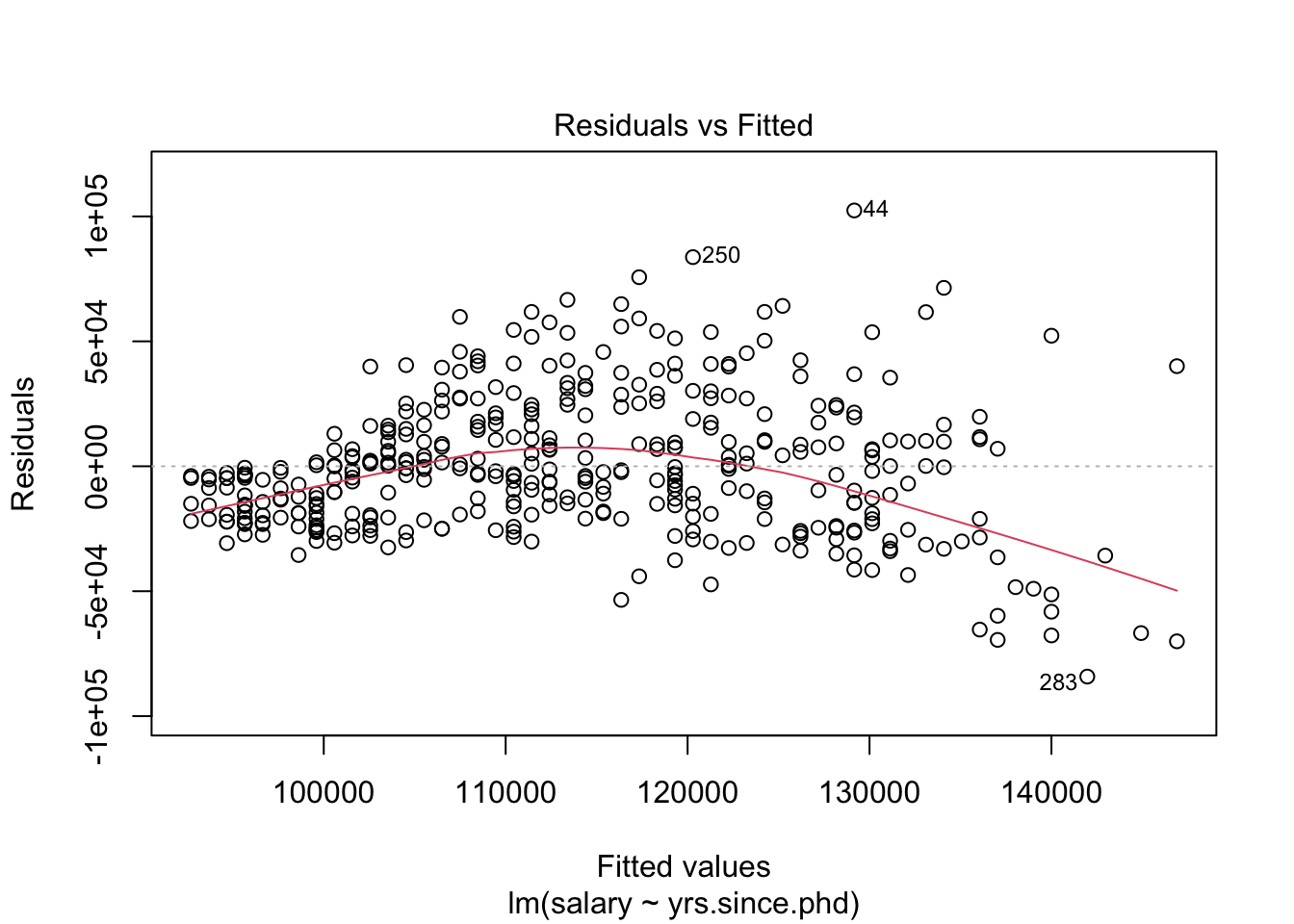
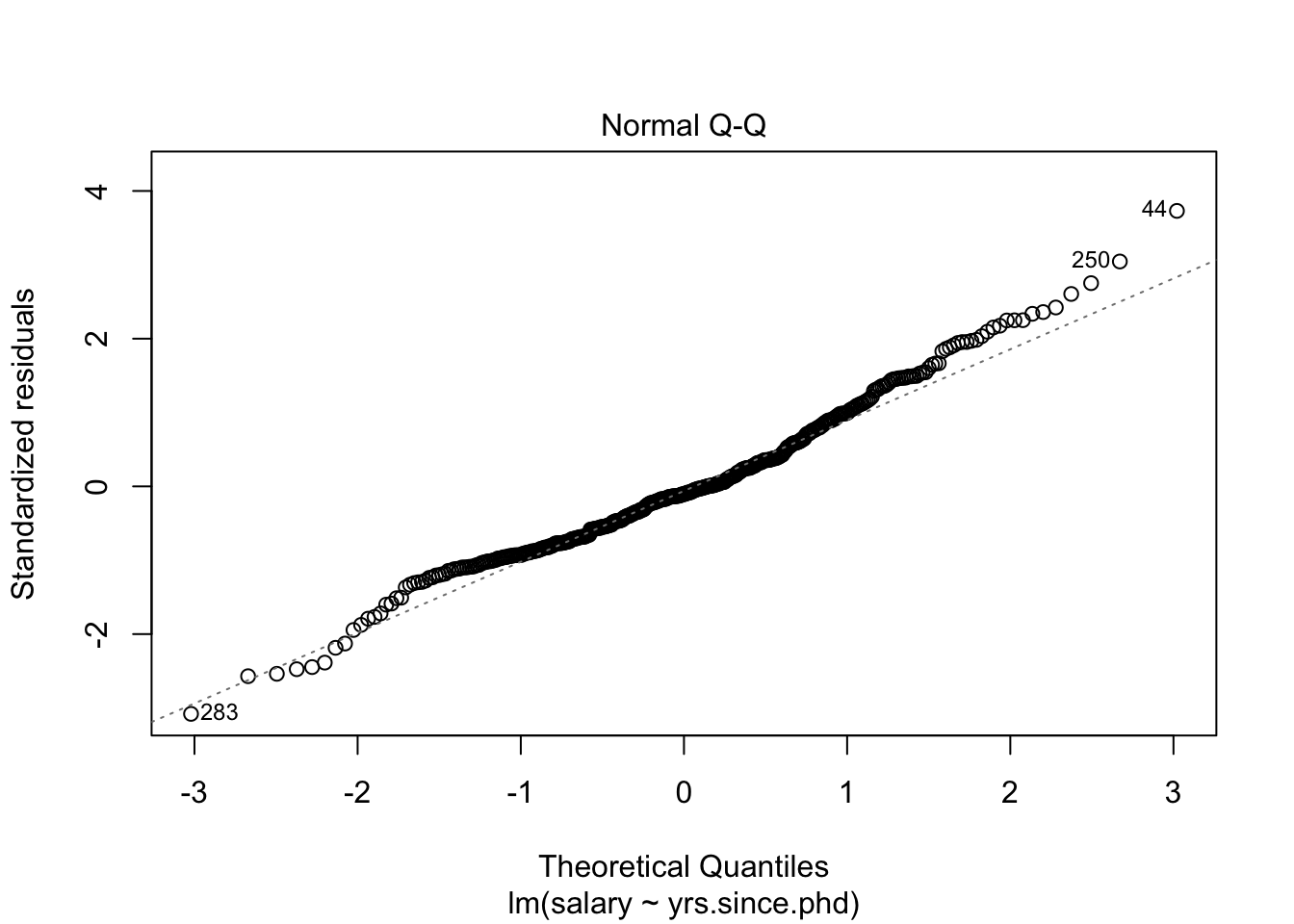
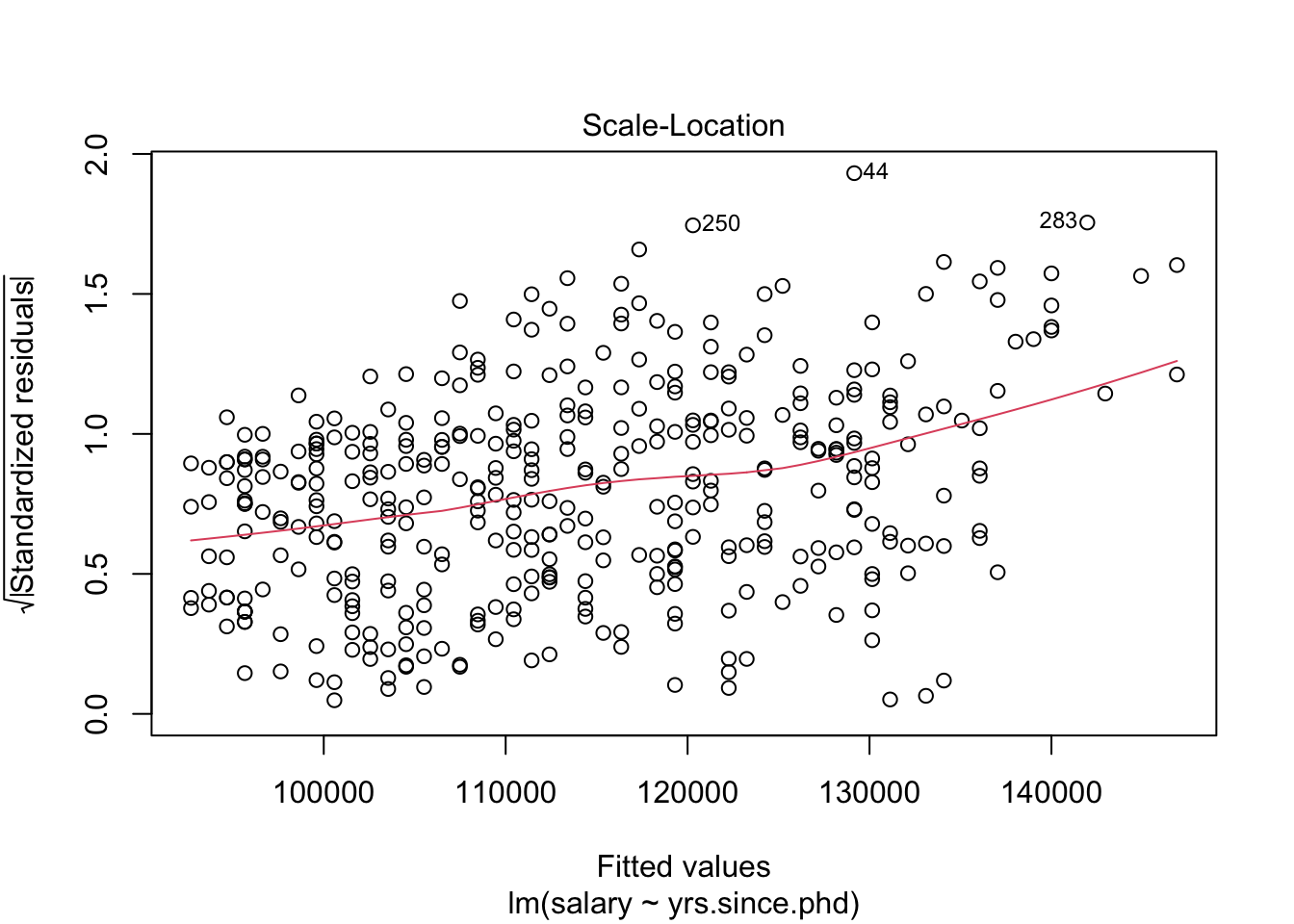
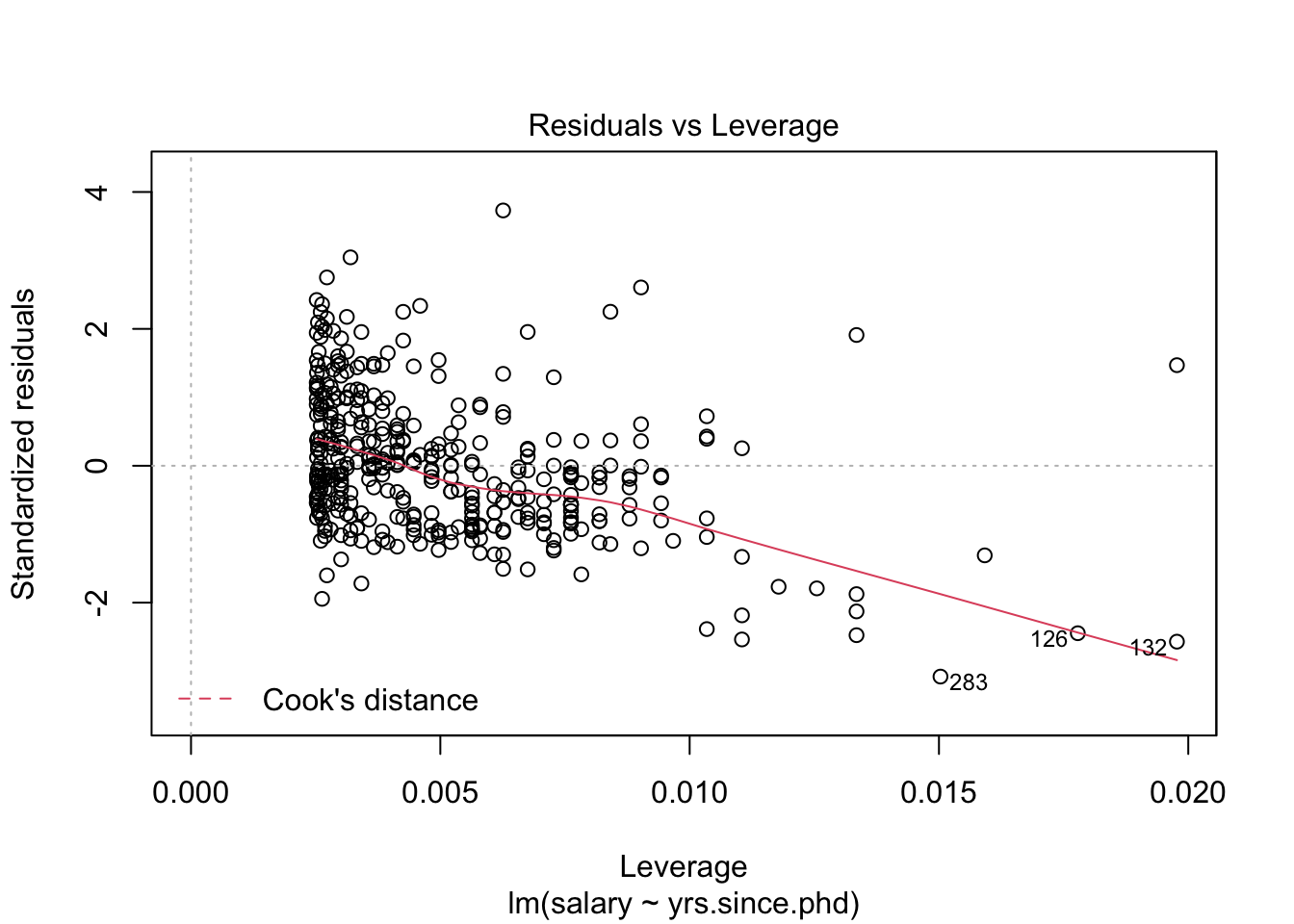
But, a more flexible approach is to create our own residual plots. The augmented data allows us to do this!
ggplot(m1_augmented, aes(x=.fitted, y=.resid)) +
geom_point() +
geom_smooth(method = "loess", se=FALSE, formula = "y~x")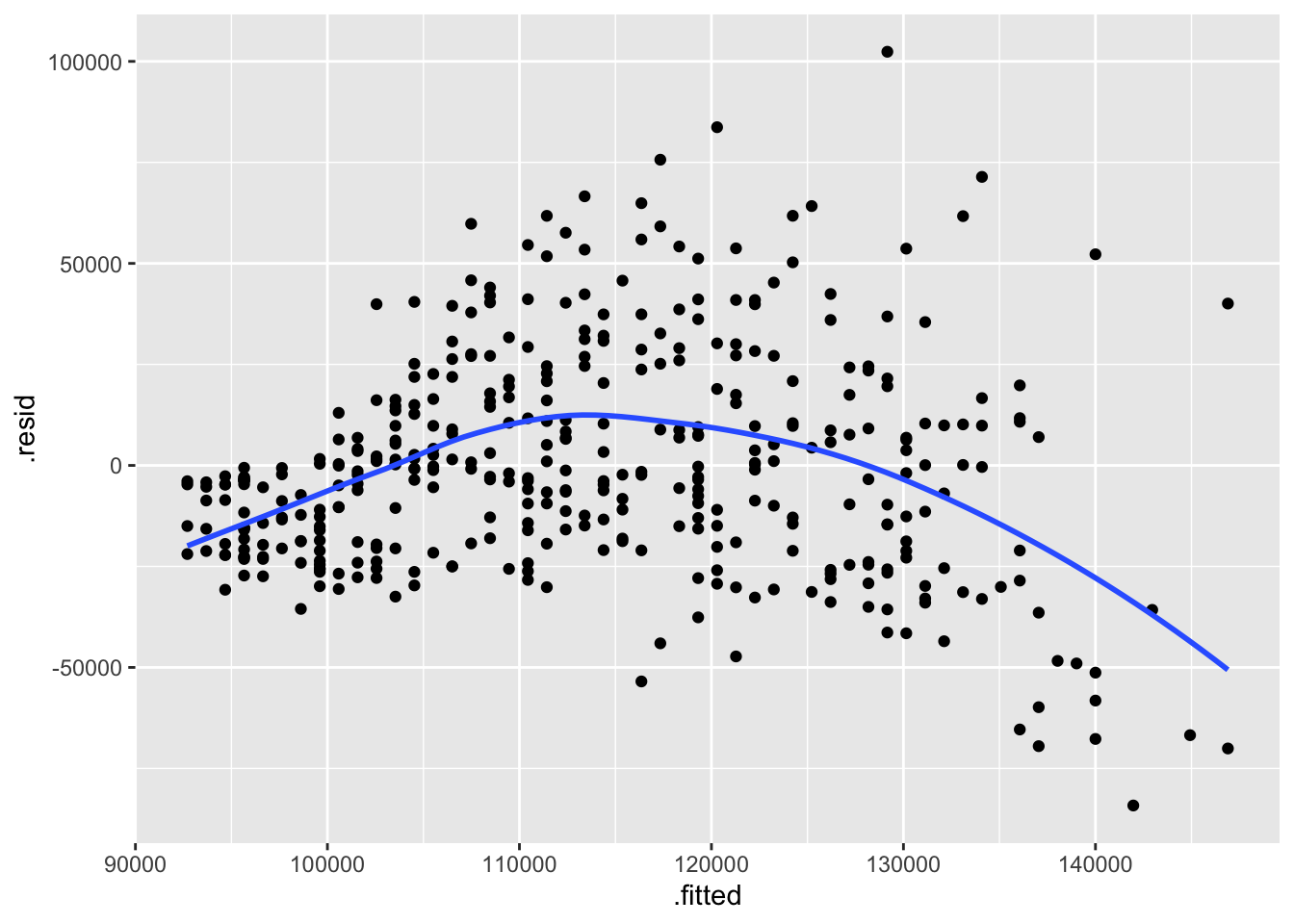
Residual v. fitted plot. Use it to check linearity and equality of variance.
ggplot(Salaries, aes(x=yrs.since.phd, y = salary)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE)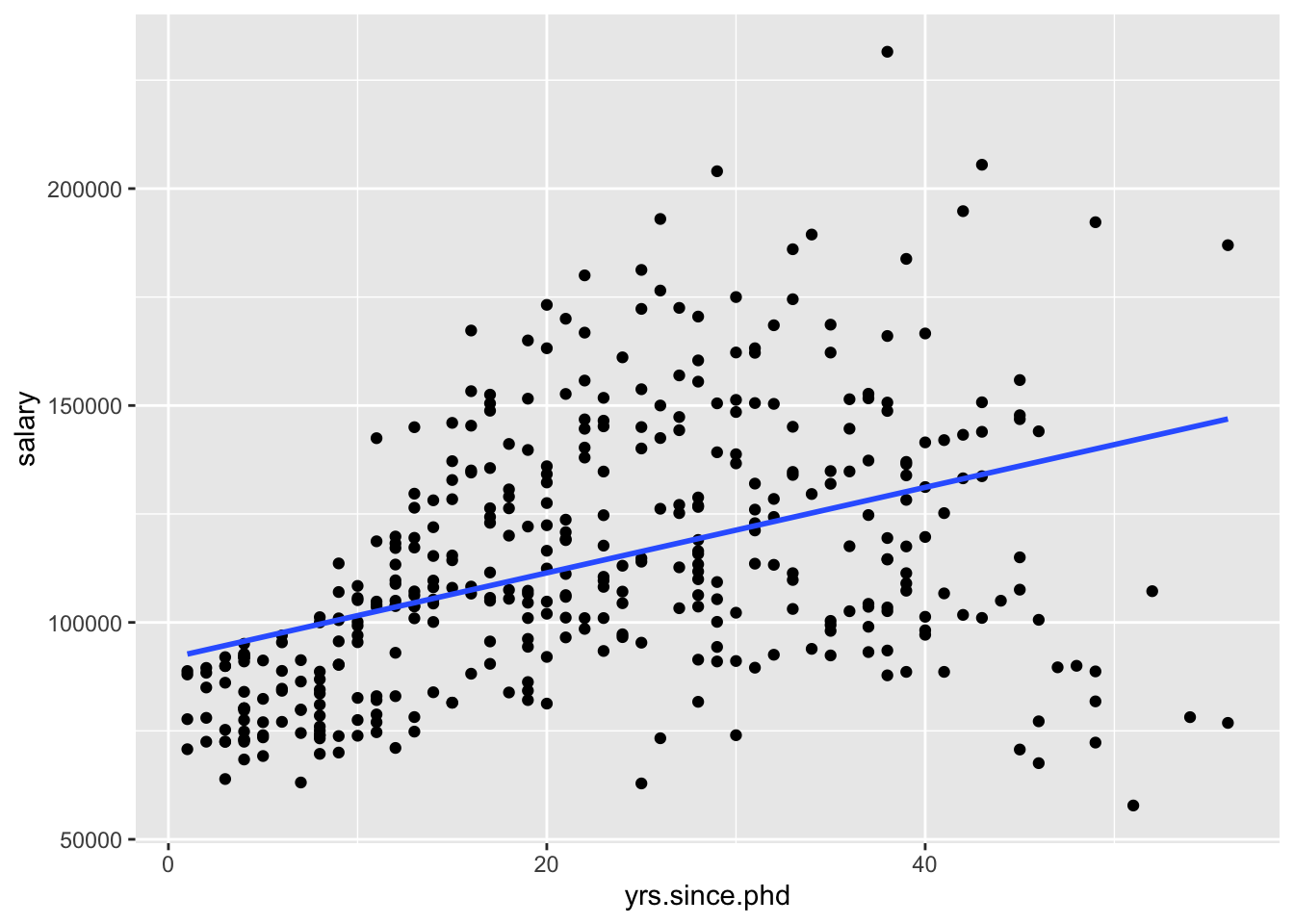
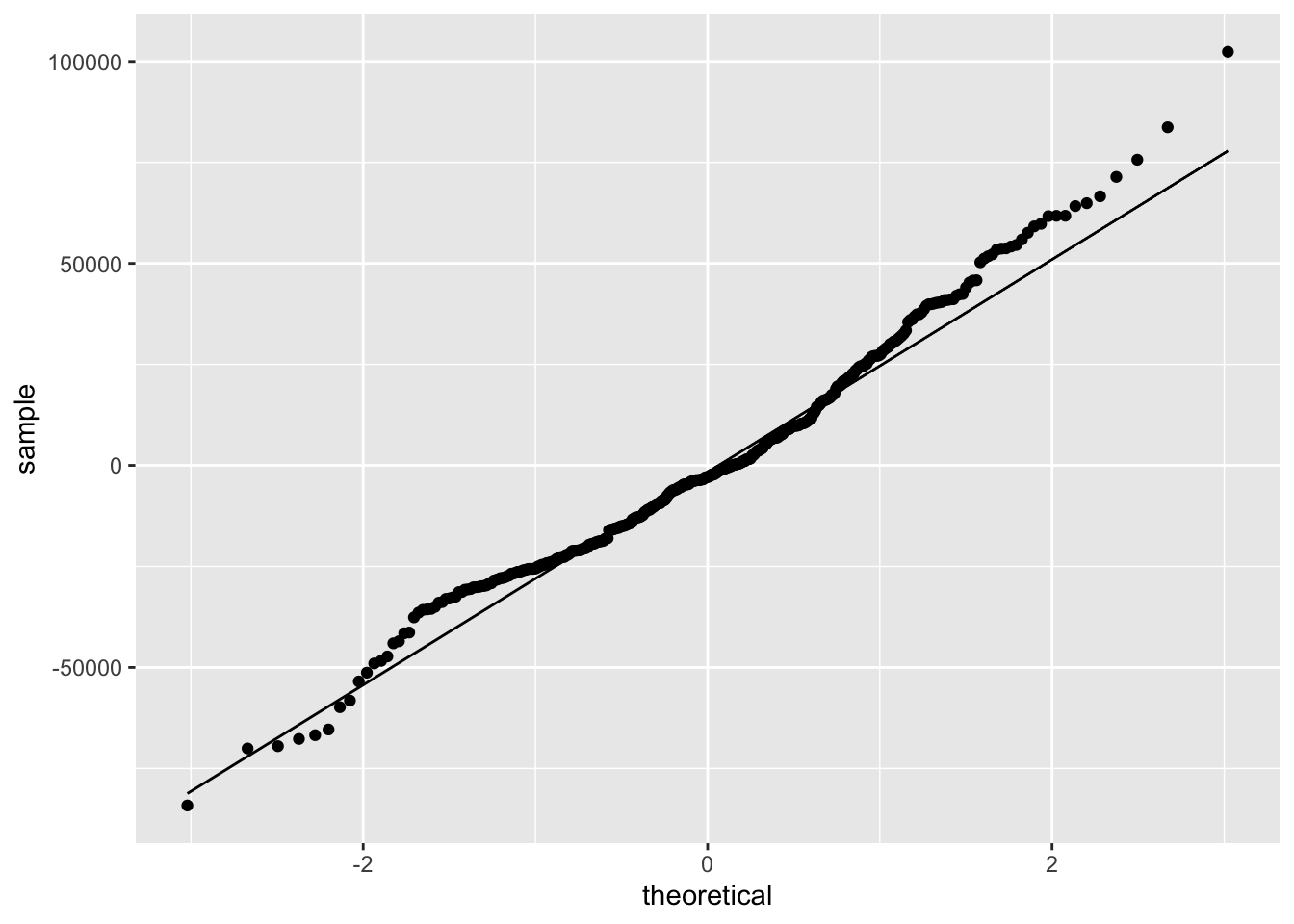
One benefit to making our own residual plots is we can do things like color by a different variable, or facet the plot, to see if there is unexplained variability.
Try coloring the residual v. fitted plot by each of the categorical variables. Which categorical variable do you think explains the most additional variability? How can you tell?
ggplot(m1_augmented, aes(x=.fitted, y=.resid)) +
geom_point(aes(color = sex)) +
geom_smooth(method = "loess", se=FALSE, formula = "y~x")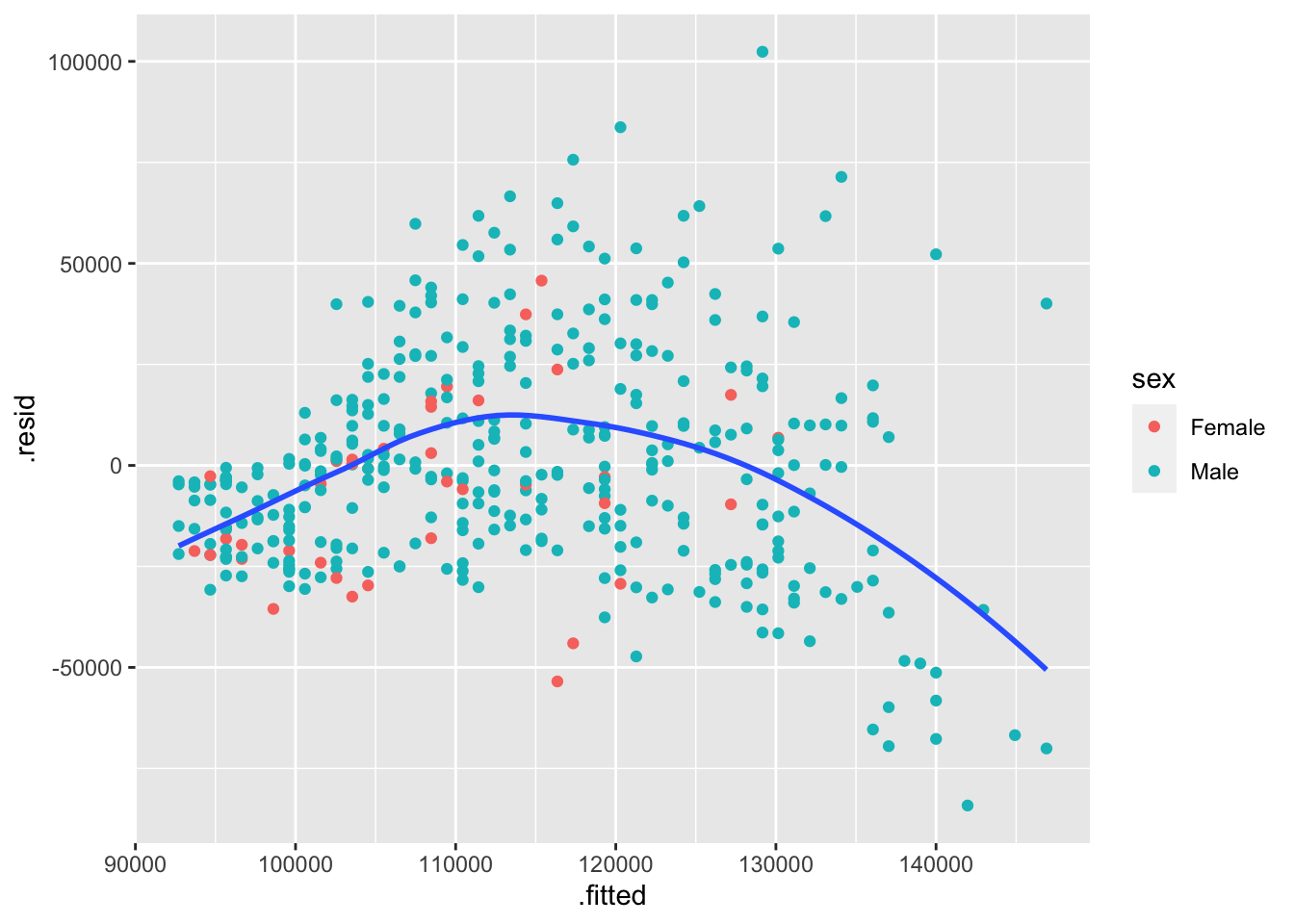
m2 <- lm(log(salary) ~ yrs.since.phd + discipline,
data = Salaries)
m2_augmented <- augment(m2, data=Salaries)
ggplot(m2_augmented, aes(x=.fitted, y=.resid)) +
geom_point(aes(color = discipline)) +
geom_smooth(method = "loess", se=FALSE, formula = "y~x")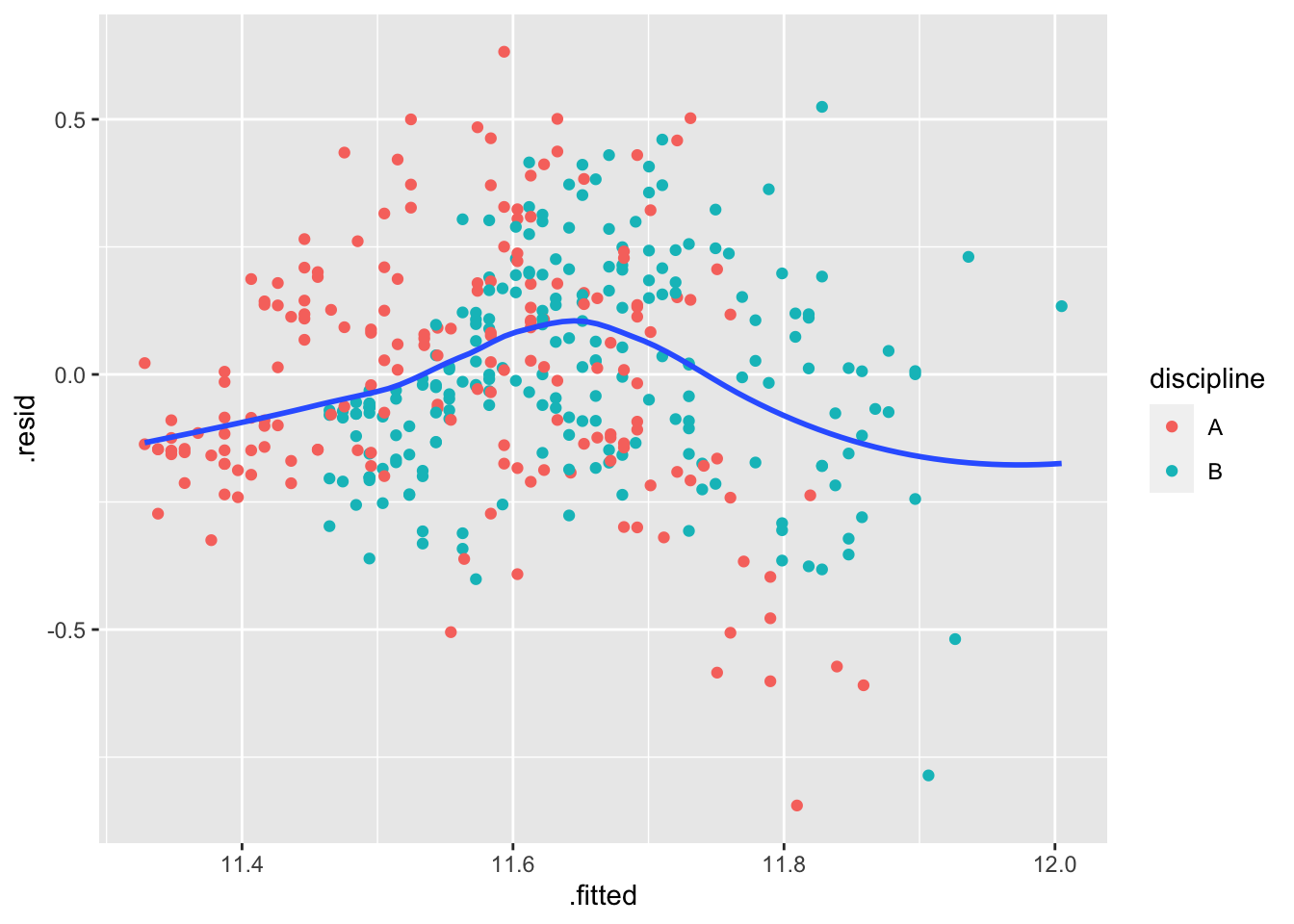
9.4 More dplyr verbs
So far we have learned about filter, select and arrange. Now we want to go into the verbs that modify the data in some way. First, summarize
9.4.1 summarize()
[Note: both the British and American spellings are accepted! I use summarize() most of the time, but summarise() also works.]
This can be thought of as a many-to-one operation. We are moving from many rows of data, and condensing down to just one.
## # A tibble: 1 x 2
## total max
## <int> <int>
## 1 348120517 99686- Use
summarize()to compute three statistics about the data:
- The first (minimum) year in the dataset
- The last (maximum) year in the dataset
- The total number of children represented in the data
There are a few useful helper functions for summarize(),
n(), which counts the number of rows in a dataset or groupn_distinct(), which counts number of distinct values in a variable
Right now, n() doesn’t seem that useful
## # A tibble: 1 x 1
## n
## <int>
## 1 1924665n_distinct() might seem better,
## # A tibble: 1 x 1
## nname
## <int>
## 1 97310But, these become even more useful when combined with…
9.4.2 group_by()
The group_by() function just groups cases by a common value of a particular variable.
## # A tibble: 1,924,665 x 5
## # Groups: sex [2]
## year sex name n prop
## <dbl> <chr> <chr> <int> <dbl>
## 1 1880 F Mary 7065 0.0724
## 2 1880 F Anna 2604 0.0267
## 3 1880 F Emma 2003 0.0205
## 4 1880 F Elizabeth 1939 0.0199
## 5 1880 F Minnie 1746 0.0179
## 6 1880 F Margaret 1578 0.0162
## 7 1880 F Ida 1472 0.0151
## 8 1880 F Alice 1414 0.0145
## 9 1880 F Bertha 1320 0.0135
## 10 1880 F Sarah 1288 0.0132
## # … with 1,924,655 more rowsWhen combined with other dplyr verbs, it can be very useful!
## # A tibble: 2 x 2
## sex total
## * <chr> <int>
## 1 F 172371079
## 2 M 1757494389.4.3 mutate()
Our final single-table verb is mutate(). I think of mutate() as a many-to-many transformation. It adds additional columns (variables) to the data.
## # A tibble: 1,924,665 x 6
## year sex name n prop percent
## <dbl> <chr> <chr> <int> <dbl> <dbl>
## 1 1880 F Mary 7065 0.0724 7.24
## 2 1880 F Anna 2604 0.0267 2.67
## 3 1880 F Emma 2003 0.0205 2.05
## 4 1880 F Elizabeth 1939 0.0199 1.99
## 5 1880 F Minnie 1746 0.0179 1.79
## 6 1880 F Margaret 1578 0.0162 1.62
## 7 1880 F Ida 1472 0.0151 1.51
## 8 1880 F Alice 1414 0.0145 1.45
## 9 1880 F Bertha 1320 0.0135 1.35
## 10 1880 F Sarah 1288 0.0132 1.32
## # … with 1,924,655 more rows9.5 More model analysis
Since we have the residuals in m1_augmented, we can use that to compute the sum of squared residuals.
## # A tibble: 1 x 1
## SSE
## <dbl>
## 1 299448839521.Notice that I’m naming my summary statistic, so I could use it later as a variable name.
We can think of partitioning the variability in our response variable as follows,
\[ SST = SSM + SSE \]
where
\[\begin{eqnarray*} SST &=& \sum_{i=1}^n (y_i-\bar{y})^2 \\ SSM &=& \sum_{i=1}^n (\bar{y} - \hat{y})^2 \\ SSE &=& \sum_{i=1}^n (y_i -\hat{y})^2 \end{eqnarray*}\]
Let’s find the other two sums of squares
m1_augmented %>%
mutate(meansalary = mean(salary)) %>%
select(salary, .fitted, .resid, meansalary) %>%
summarize(SSE = sum(.resid^2),
SSM = sum((meansalary - .fitted)^2),
SST = sum((salary - meansalary)^2))## # A tibble: 1 x 3
## SSE SSM SST
## <dbl> <dbl> <dbl>
## 1 299448839521. 63851803040. 363300642561.We don’t have a nice way to interpret those sums of squares, but we can use them to calculate the \(R^2\) value,
\[ R^2 = 1 - \frac{SSE}{SST} = \frac{SSM}{SST} \]
m1_augmented %>%
mutate(meansalary = mean(salary)) %>%
summarize(SSE = sum(.resid^2),
SSM = sum((meansalary - .fitted)^2),
SST = sum((salary - meansalary)^2)) %>%
summarize(R2 = 1 - SSE/SST)## # A tibble: 1 x 1
## R2
## <dbl>
## 1 0.176We can use the \(R^2\) value to assess the model. The larger the \(R^2\) value, the more variability we can explain using the model.
Unfortunately, \(R^2\) always increases as you add predictors, so it is not a good statistic for comparing between models. Instead, we should use adjusted \(R^2\)
\[ R^2_{adj} = 1- \frac{SSE/(n-1)}{SST/(n-k-1)} \]
The adjusted \(R^2\) doesn’t have a nice interpretation, but it can be used to compare between models.
The \(R^2\) and \(R^2_{adj}\) values are given by the model summary table.
##
## Call:
## lm(formula = salary ~ yrs.since.phd, data = Salaries)
##
## Residuals:
## Min 1Q Median 3Q Max
## -84171 -19432 -2858 16086 102383
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 91718.7 2765.8 33.162 <2e-16 ***
## yrs.since.phd 985.3 107.4 9.177 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 27530 on 395 degrees of freedom
## Multiple R-squared: 0.1758, Adjusted R-squared: 0.1737
## F-statistic: 84.23 on 1 and 395 DF, p-value: < 2.2e-16We can also use the tidy function from broom to tidy up the model coefficients,
## # A tibble: 2 x 5
## term estimate std.error statistic p.value
## <chr> <dbl> <dbl> <dbl> <dbl>
## 1 (Intercept) 91719. 2766. 33.2 3.33e-116
## 2 yrs.since.phd 985. 107. 9.18 2.50e- 18and glance to look at model summaries,
## # A tibble: 1 x 12
## r.squared adj.r.squared sigma statistic p.value df logLik AIC BIC
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 0.176 0.174 27534. 84.2 2.50e-18 1 -4621. 9248. 9260.
## # … with 3 more variables: deviance <dbl>, df.residual <int>, nobs <int>Try re-making a few more of our models from yesterday, and glanceing to see which one has the highest adjusted \(R^2\).
library(palmerpenguins)
data("penguins")
library(broom)
favmod <- lm(flipper_length_mm ~ species + bill_depth_mm + body_mass_g + sex, data = penguins)
favmod_augment <- augment(favmod) # works## Error: Assigned data `predict(x, na.action = na.pass, ...) %>% unname()` must be compatible with existing data.
## x Existing data has 344 rows.
## x Assigned data has 333 rows.
## ℹ Only vectors of size 1 are recycled.I didn’t know how to fix this, so I had to look at documentation!
It turns out it has to do with the na.action in lm.
favmod <- lm(flipper_length_mm ~ species + bill_depth_mm + body_mass_g + sex, data = penguins, na.action = "na.exclude")
favmod_augment <- augment(favmod, data = penguins) # works! Beyond \(R^2\), another useful statistic for assessing a model is the mean squared error, or the root mean squared error
\[
MSE = \frac{1}{n}\sum_{i=1}^n (y_i-\hat{y}_i)^2 \\
RMSE = \sqrt{MSE}
\]
Try using dplyr verbs to compure the RMSE.
9.6 Bonus: comment on theory!
Although we are trying to “minimize” the sum of squared residuals, we don’t have to use a simulation method. Regression is actually done using matrix operations.
Suppose we have a sample of \(n\) subjects. For subject \(i\in{1,...,n}\) let \(Y_i\) denote the observed response value and \((x_{i1},x_{i2},\dots,x_{ik})\) denote the observed values of the \(k\) predictors. Then we can collect our observed response values into a vector \(y\), our predictor values into a matrix \(X\), and our regression coefficients into a vector \(\beta\). Note that a column of 1s is included for an intercept term in \(X\):
\[\begin{eqnarray*}y= \begin{pmatrix} y_1 \\ y_2 \\ \vdots \\ y_n \end{pmatrix}, X = \begin{pmatrix} 1 & x_{11} x_{12} \dots x_{1k} \\ 1 & x_{21} & x_{22} & \dots x_{2k} \\ \vdots & \vdots & \vdots & \dots & \vdots \\ 1 & x_{n1} & x_{n2} & \dots & x_{nk} \end{pmatrix}, \text{ and } \beta = \begin{pmatrix} beta_1 \\ \beta_2 \\ \vdots \\ \beta_k \end{pmatrix} \end{eqnarray*}\]
Then we can express the model \(y_i=\beta_0+\beta_1 x_{i1}+\dots +\beta_{k}x_{ik}\) for \(i\in{1,\dots,n}\) using linear algebra:
\[ y=X\beta \] Further, let \(\hat{\beta}\) denote the vector of sample estimated \(\hat{beta}\), and \(\hat{y}\) denote the vector of predictions/model values:
\[ \hat{y}=X\hat{\beta} \] Thus the residual vector is \[ y−\hat{y}=X\beta−X\hat{\beta} \] and the sum of squared residuals is \[ (y−\hat{y})^T(y−\hat{y}) \] Challenge: Prove that the following formula for sample coefficients \(\beta\) are the least squares estimates of \(\beta\), ie. they minimize the sum of squared residuals: \[ \hat{\beta}=(X^TX)^{-1}X^Ty \]