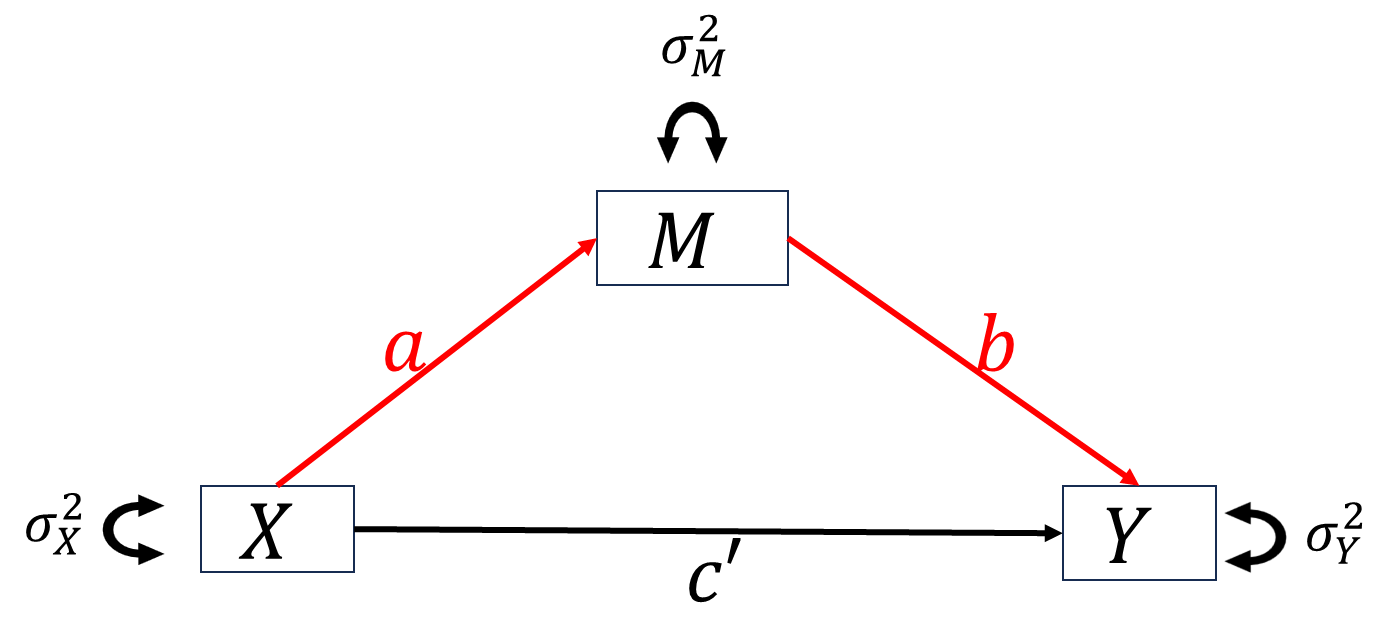
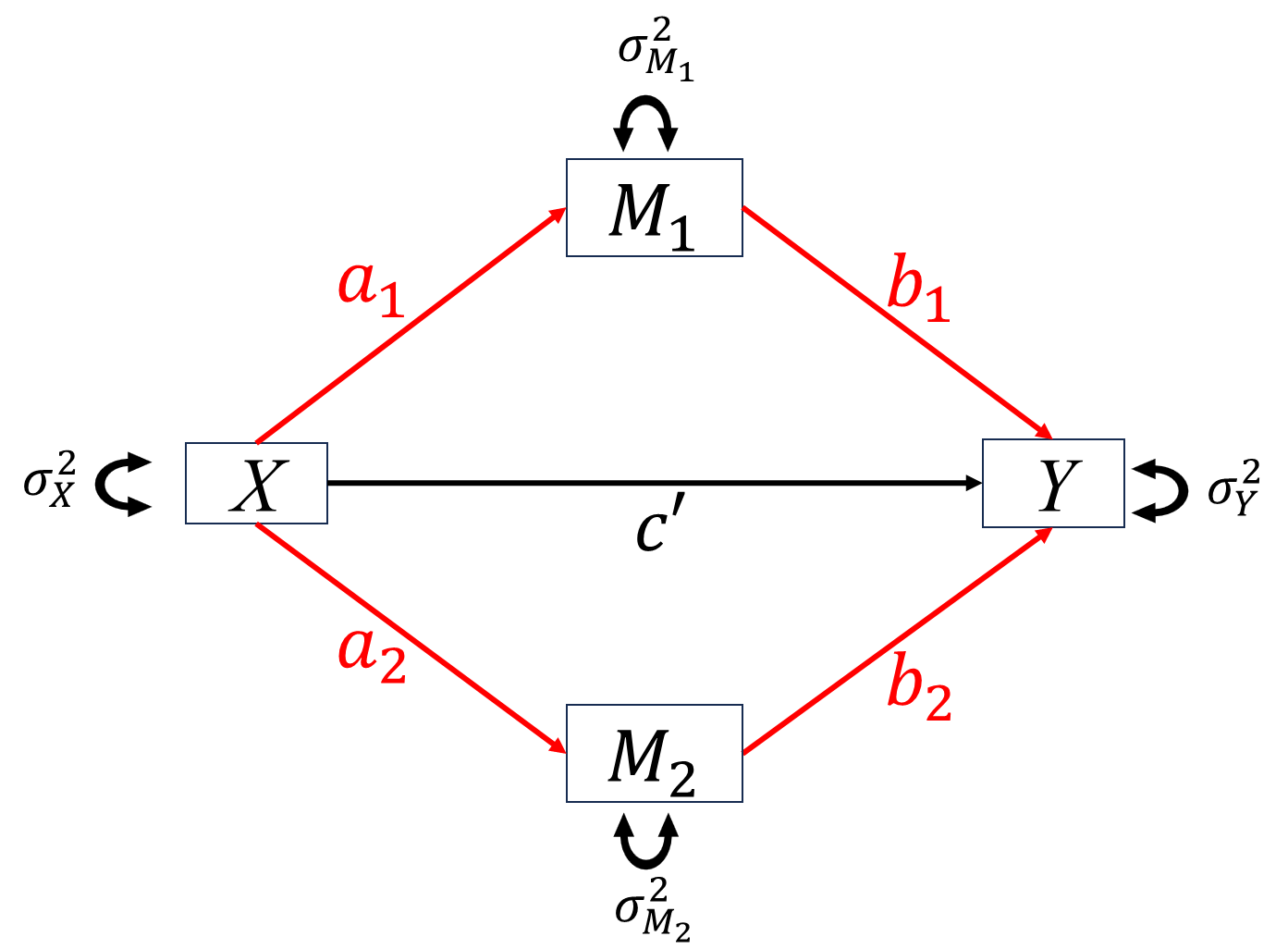
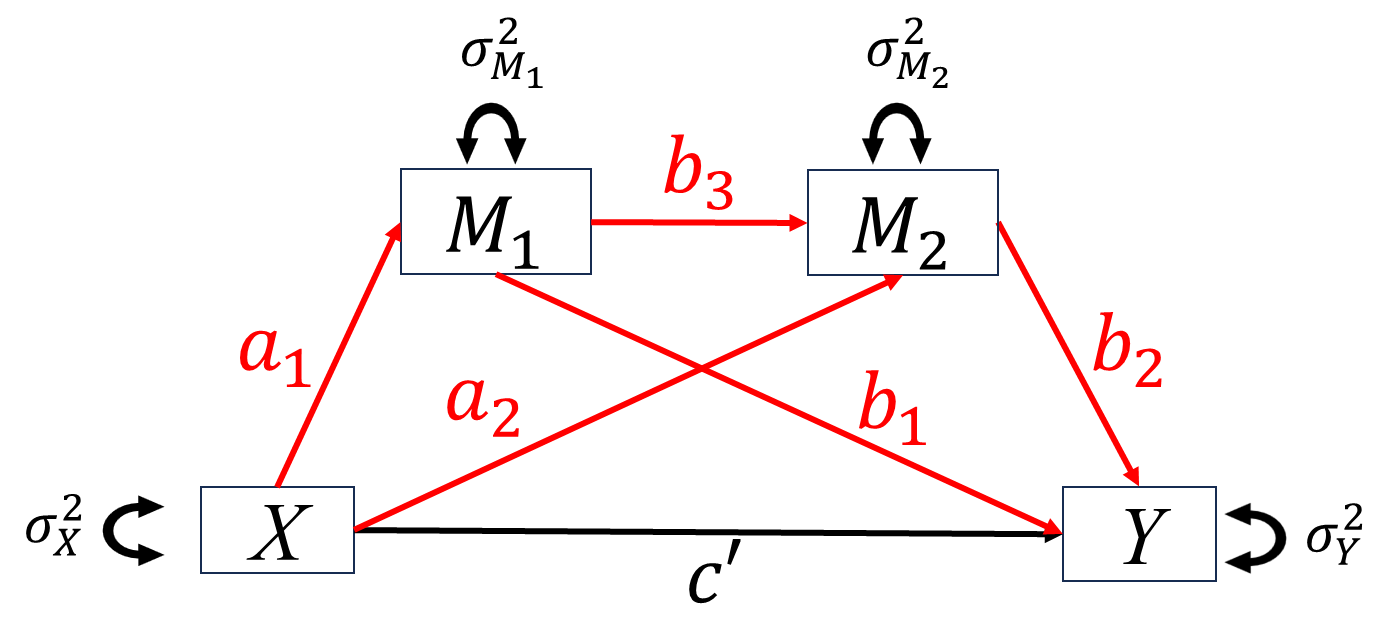
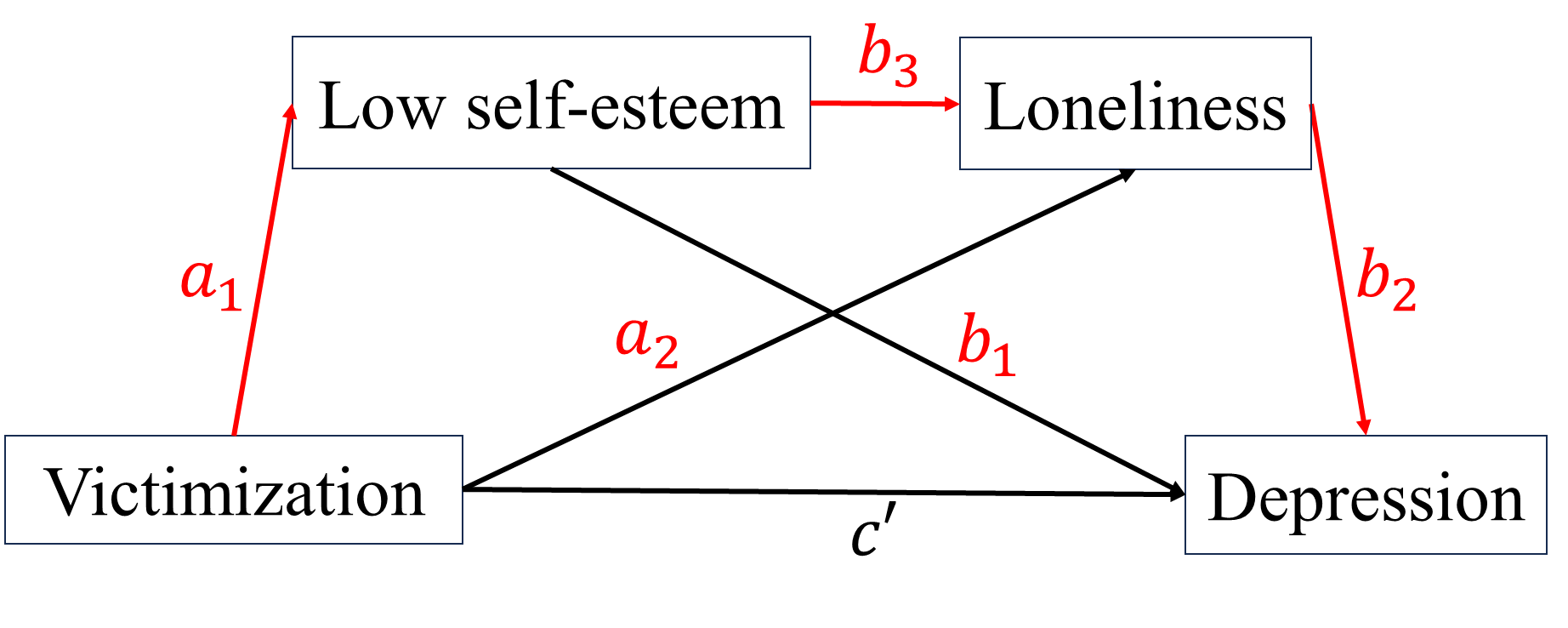
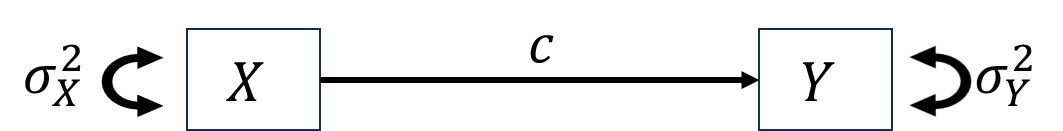When studying the relationship between independent variable \(X\) and dependent variable \(Y\), we want to know how \(X\) impact \(Y\). For this purpose, researchers proposed two types of basic “causal” mechanisms between \(X\) and \(Y\), mediation and moderation.
The simplest scenario: when everything is continuous
When \(x\) is categorical
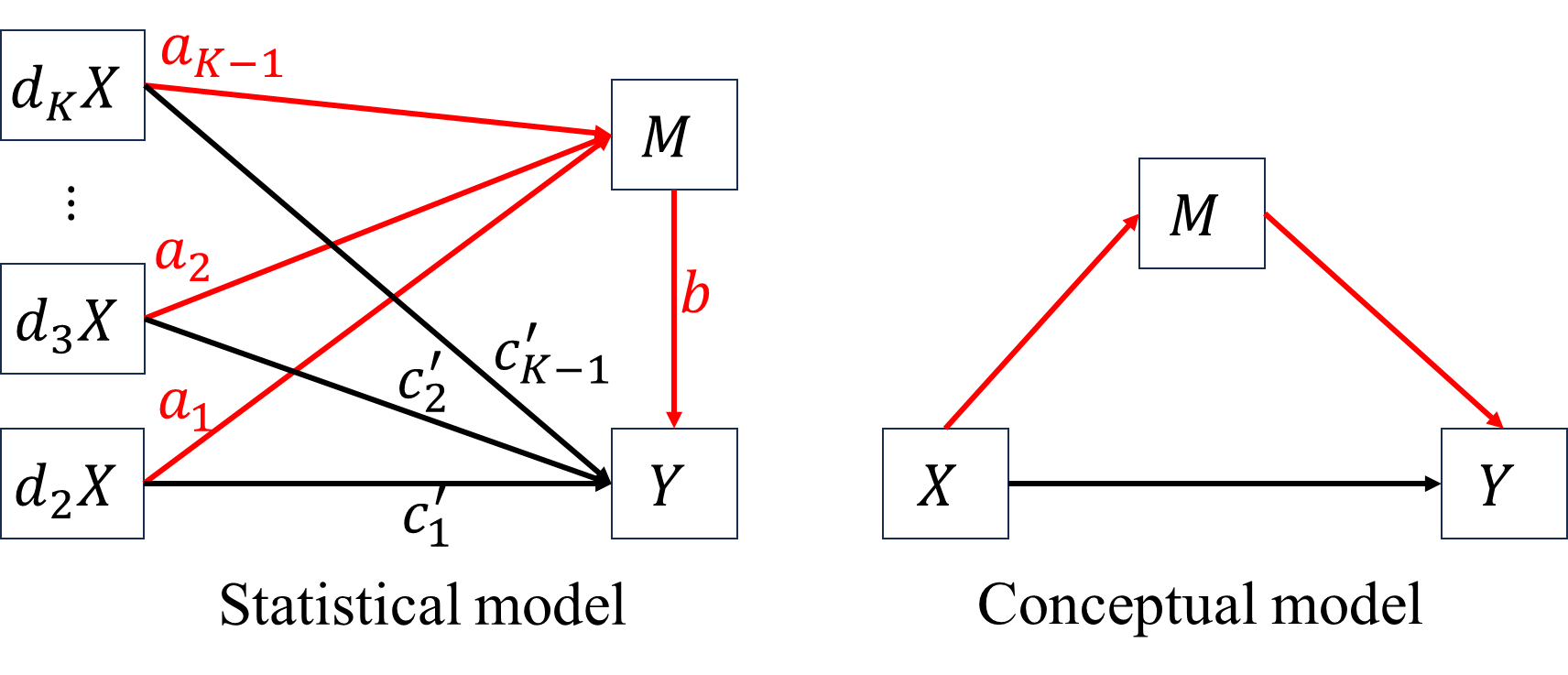
Suppose \(X\) is a K-level categorical variable and dummy coded, the 1st category of \(X\) is used as reference group. \[\begin{align*}
M&=a_0+a_1d2X+a_2d3X+\cdots+a_{K-1}dKX+\epsilon_M\\
Y&=c_0'+bM+c_1'd2X+c_2'd3X+\cdots+c_{K-1}'dKX+\epsilon_{Y}.
\end{align*}\] Combine these two equations we have \[\begin{align*}
Y&=\red a_0b\black+c_0'+(\red a_1b\black+c_1')d2X+(\red a_2b\black+c_2')d3X+\cdots+(\red a_{K-1}b\black+c_{K-1}')dKX+\\
&\quad\enspace b\epsilon_M+\epsilon_Y.
\end{align*}\] When \(X=1\), \(d2X=\cdots=dKX=0\), \[\begin{align*}
Y=\red a_0b\black+c_0'.
\end{align*}\] When \(X=2\), \(d2X=1\), \(d3X=\cdots=dKX=0\), \[\begin{align*}
Y=\red a_0b+a_1b\black+c_0'+c_1'.
\end{align*}\] When \(X=3\), \(d2X=0\), \(d3X=1\), \(d4X=\cdots=dKX=0\), \[\begin{align*}
Y=\red a_0b+a_2b\black+c_0'+c_2'.
\end{align*}\] When \(X=K\), \(d2X=d3X=\cdots=d_{K-1}X=0\), \(dKX=1\), \[\begin{align*}
Y=\red a_0b+a_{K-1}b\black+c_0'+c_{K-1}'.
\end{align*}\] Interested readers in more details about basic mediation analysis with categorical \(X\) are referred to Hayes & Preacher (2014).
When dependent variable(s) is(are) categorical
When either \(M\) or \(Y\) is, or both of them are categorical, the mediation analysis soon becomes much more complicated because logistic/probit regression involved, interested readers are referred to Breen et al. (2013).