7 Data Exploration
Loading packages
library(readr)
library(dplyr)
library(tidyr)
library(openair) # http://davidcarslaw.github.io/openair/
library(purrr)
library(lubridate)
library(ggplot2)
library(stringr)
library(knitr)
library(xts)
library(zoo)
library(gridExtra)
library(astsa)
library(rvest)
library(fpp2)
library(ranger)
library(broom)
library(RcppRoll)
library(reshape2)Data loading
air_data_2 <- readRDS("data_rds/air_data_2.rds")7.1 Trends exploration
We take a look to the general trend of several indicators through the last 18 years
# We calcule the yearly mean of the pollutants levels.
year_avgs <- air_data_2 %>% select(station_alias, date_time_utc, PM10, PM25, SO2, NO2, NO, O3, BEN, CO, MXIL, TOL) %>%
group_by(station_alias, year = year(date_time_utc)) %>%
summarise_all(funs(mean(., na.rm = TRUE))) %>%
select(-date_time_utc) # We drop this variable
# We export the years_avgs as a csv file
write_csv(year_avgs, "data_final_csvs/year_avgs.csv")
# We convert the table to long format
year_avgs_long <- gather(year_avgs, contaminante, value, 3:length(year_avgs)) %>%
filter(!(grepl('Constit', station_alias)
& year == '2006' &
contaminante %in% c('BEN', 'MXIL', 'TOL'))) %>%
# We filter this data because is only completed in 0.01%
filter(!(grepl('Constit', station_alias) &
year == '2008' & contaminante == 'PM25'))
# We filter this data because is only completed in 0.02%
# We present the data in a grid of graphs
ggplot(year_avgs_long, aes(x = year, y = value)) +
geom_line() +
facet_grid(contaminante~station_alias,scales="free_y") +
theme(axis.text = element_text(size = 6))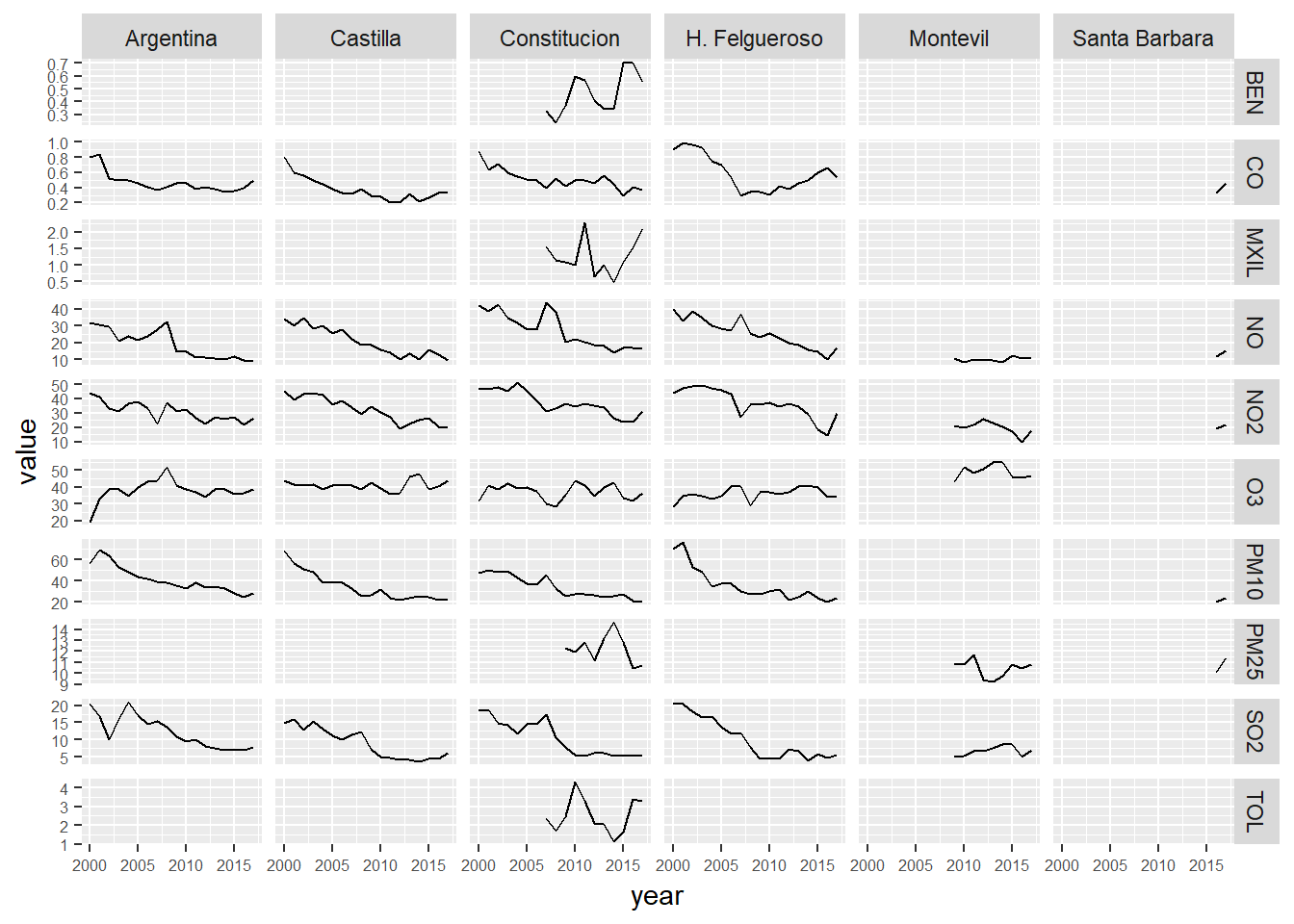
# We calcule the hourly mean of the pollutants levels.
hour_avgs <- air_data_2 %>% select(station_alias, hour, PM10, PM25, SO2, NO2, NO, O3, BEN, CO, MXIL, TOL) %>%
group_by(station_alias, hour) %>%
summarise_all(funs(mean(., na.rm = TRUE))) # quito ahora esta variable, porque no tiene sentido que salga su media.
# We convert the table to long format
hour_avgs_long <- gather(hour_avgs, contaminante, value, 3:length(hour_avgs))
# We present the data in a grid of graphs
ggplot(hour_avgs_long, aes(x = hour, y = value)) +
geom_line() +
facet_grid(contaminante~station_alias,scales="free_y") +
theme(axis.text = element_text(size = 6))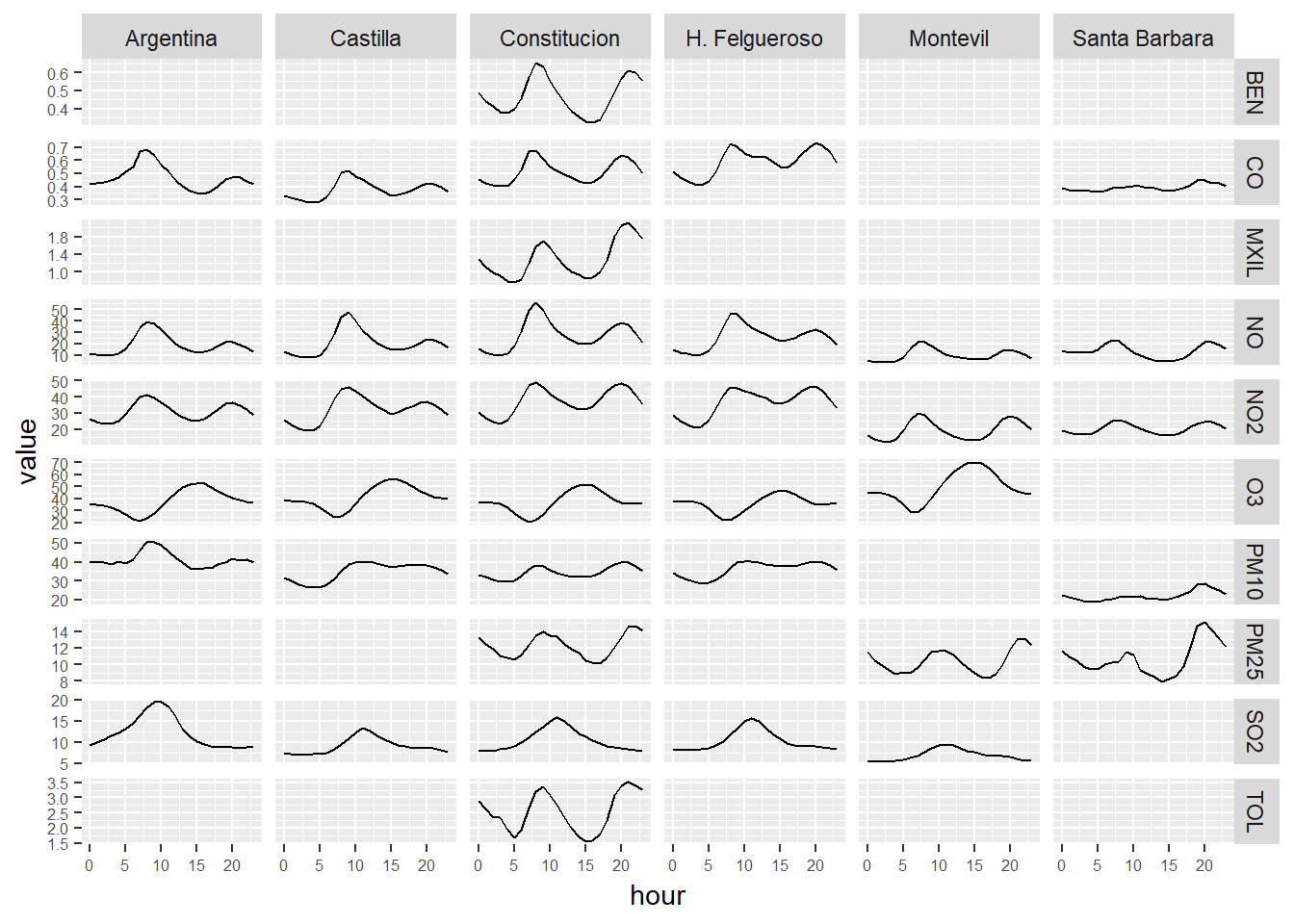
# We calcule the monthly mean of the pollutants levels.
month_avgs <- air_data_2 %>% select(station_alias, month, PM10, PM25, SO2, NO2, NO, O3, BEN, CO, MXIL, TOL) %>%
group_by(station_alias, month) %>%
summarise_all(funs(mean(., na.rm = TRUE))) # quito ahora esta variable, porque no tiene sentido que salga su media.
# We convert the table to long format
month_avgs_long <- gather(month_avgs, contaminante, value, 3:length(month_avgs))
# We present the data in a grid of graphs
ggplot(month_avgs_long, aes(x = month, y = value)) +
geom_line() +
facet_grid(contaminante~station_alias,scales="free_y") +
theme(axis.text = element_text(size = 6))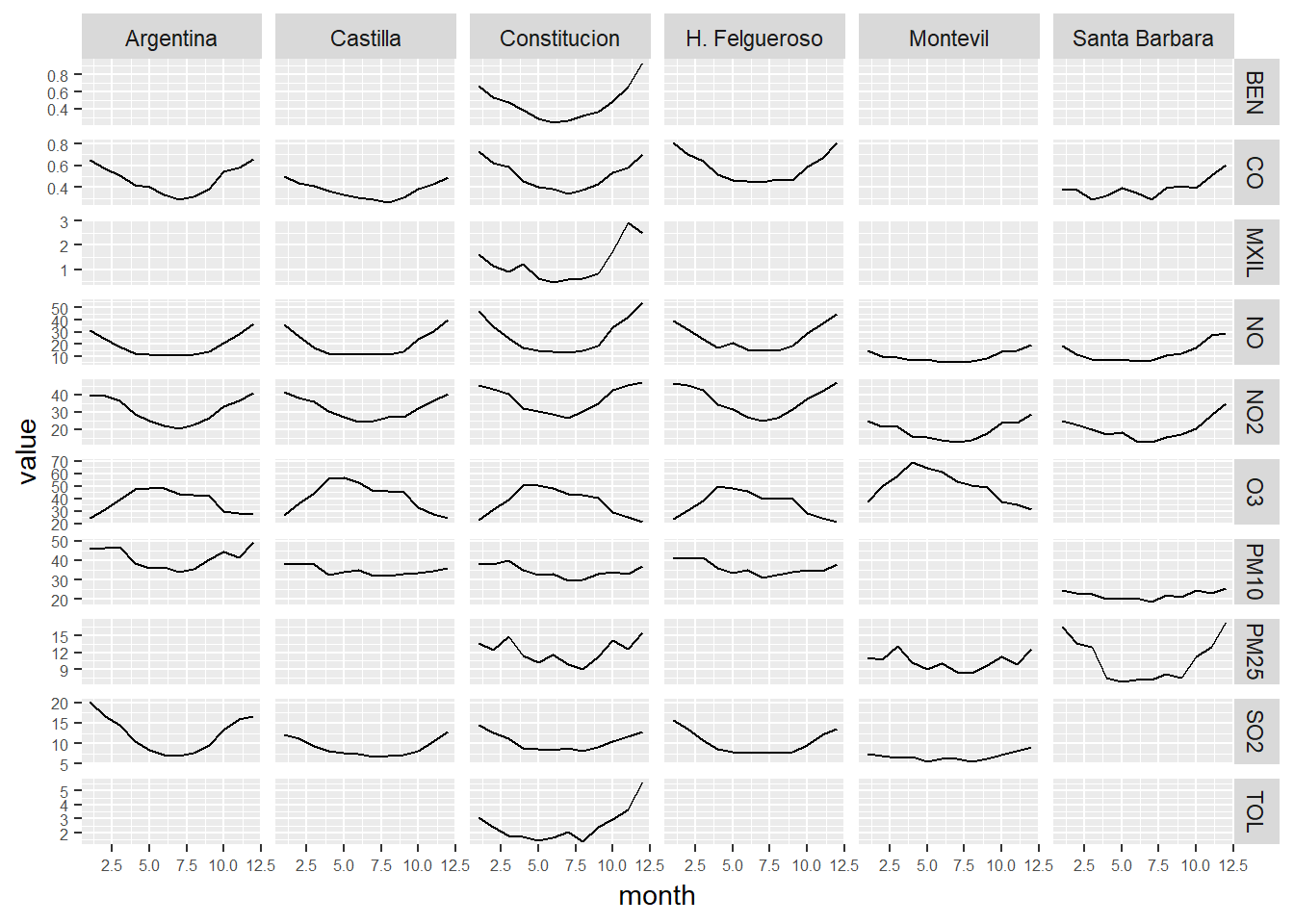
# We calcule the weekly mean of the pollutants levels.
week_day_avgs <- air_data_2 %>% select(station_alias, week_day, PM10, PM25, SO2, NO2, NO, O3, BEN, CO, MXIL, TOL) %>%
group_by(station_alias, week_day) %>%
summarise_all(funs(mean(., na.rm = TRUE))) # quito ahora esta variable, porque no tiene sentido que salga su media.
# We convert the table to long format
week_day_avgs_long <- gather(week_day_avgs, contaminante, value, 3:length(week_day_avgs))
# We present the data in a grid of graphs
ggplot(week_day_avgs_long, aes(x = week_day, y = value)) +
geom_line() +
facet_grid(contaminante~station_alias,scales="free_y") +
theme(axis.text = element_text(size = 6))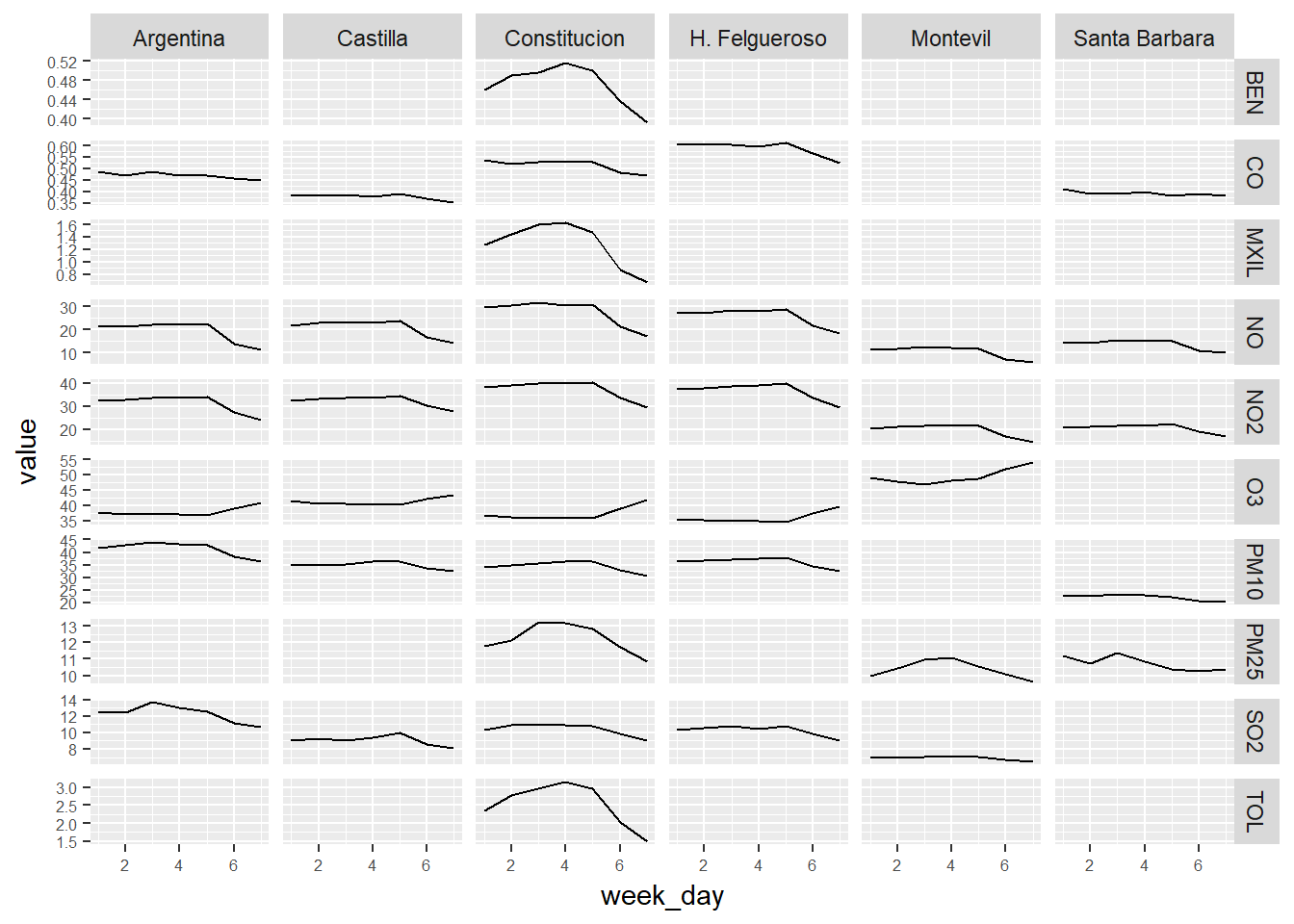
We take a look to the distributions
hist_table <- air_data_2 %>% filter(year(date_time_utc) == '2017') %>%
select(station_alias, PM10, PM25, SO2, NO2, NO, O3, BEN, CO, MXIL, TOL)
# We convert the table to long format
hist_table_long <- hist_table %>% gather(pollutant, value, 2:length(hist_table))
# We present the data in a grid of graphs
ggplot(hist_table_long, aes(x = value)) +
geom_histogram() +
xlim(c(0, 150)) +
facet_grid(pollutant~station_alias,scales="free_y") +
theme(axis.text = element_text(size = 6))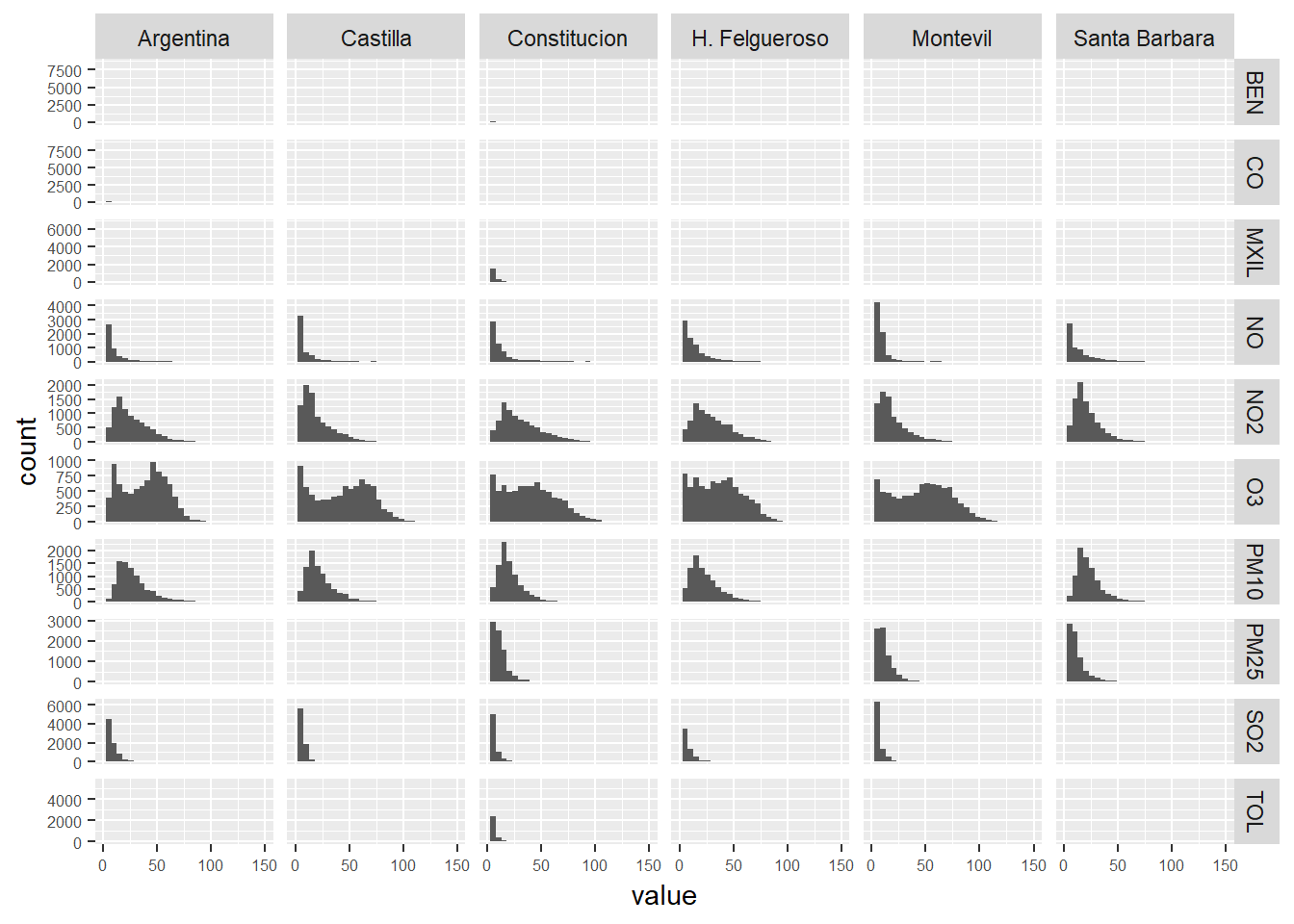
7.2 PM10 Constitucion Station
We create the dataset pm10 with PM10 values from the Constitución Station and we execute a summary
pm10 <- air_data_2 %>% filter(station == '1') %>%
select(date_time_utc, PM10)
summary(pm10)## date_time_utc PM10
## Min. :2000-01-01 00:00:00 Min. : 0.00
## 1st Qu.:2004-06-30 23:15:00 1st Qu.: 19.00
## Median :2009-01-02 00:30:00 Median : 29.00
## Mean :2008-12-31 18:50:45 Mean : 34.39
## 3rd Qu.:2013-07-02 23:45:00 3rd Qu.: 44.00
## Max. :2017-12-31 23:00:00 Max. :888.00
## NA's :310625% of the values are between 44.00 and 888.00. 888.00 is a value really extreme. How many extreme values (outliers) do we have in this series? We plot all the values to visualise this:
ggplot(pm10, aes(x = date_time_utc, y = PM10)) +
geom_point(alpha = 0.1)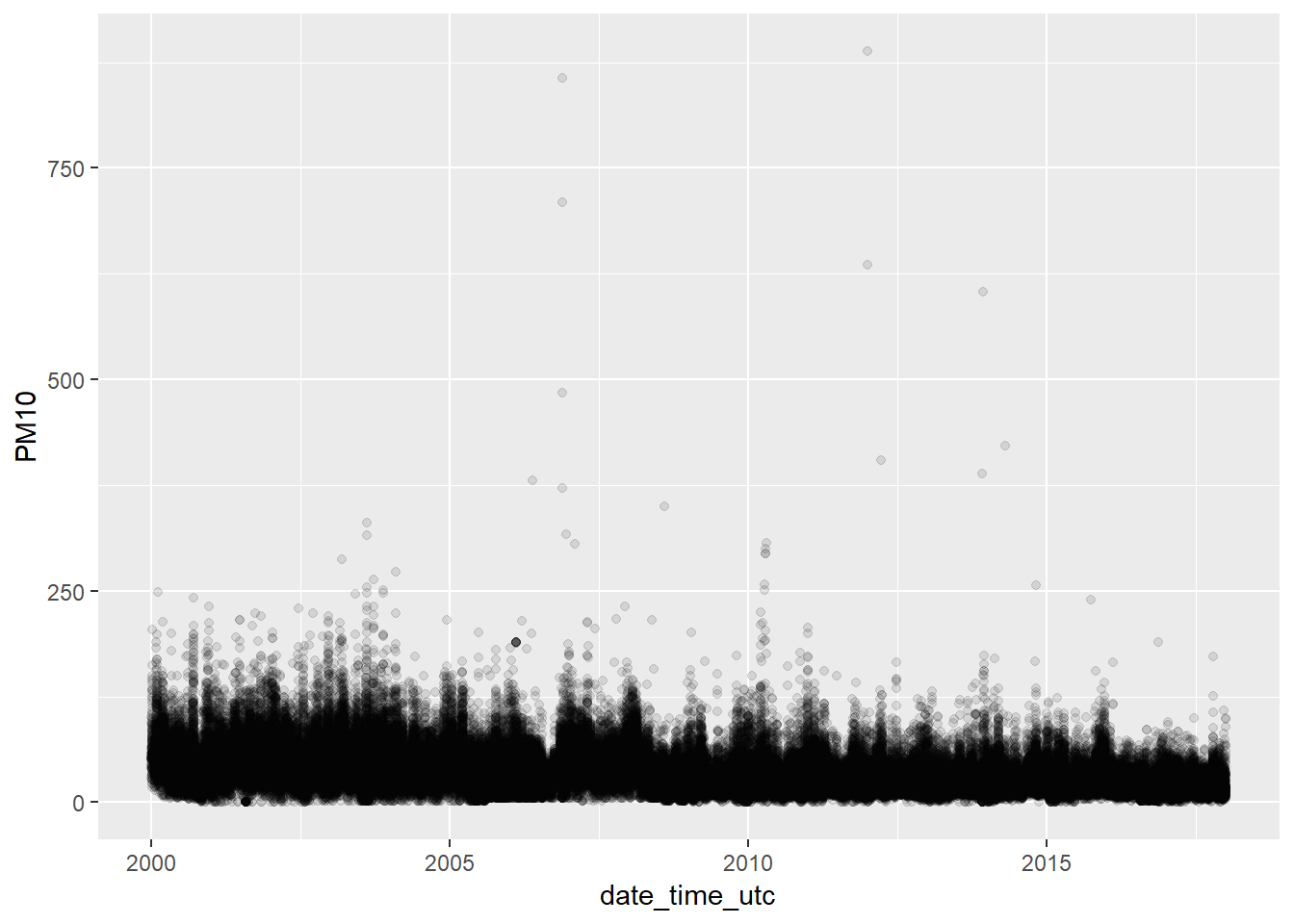 We have very few values greater than 250. So, it doesn’t seem we have a problem with the outliers.
We have very few values greater than 250. So, it doesn’t seem we have a problem with the outliers.
We filter out the most extreme values to improve the visualization, but
pm10_no_outliers <- pm10 %>% filter(PM10 <= 300)
ggplot(pm10_no_outliers, aes(x = date_time_utc, y = PM10)) +
geom_point(alpha = 0.1)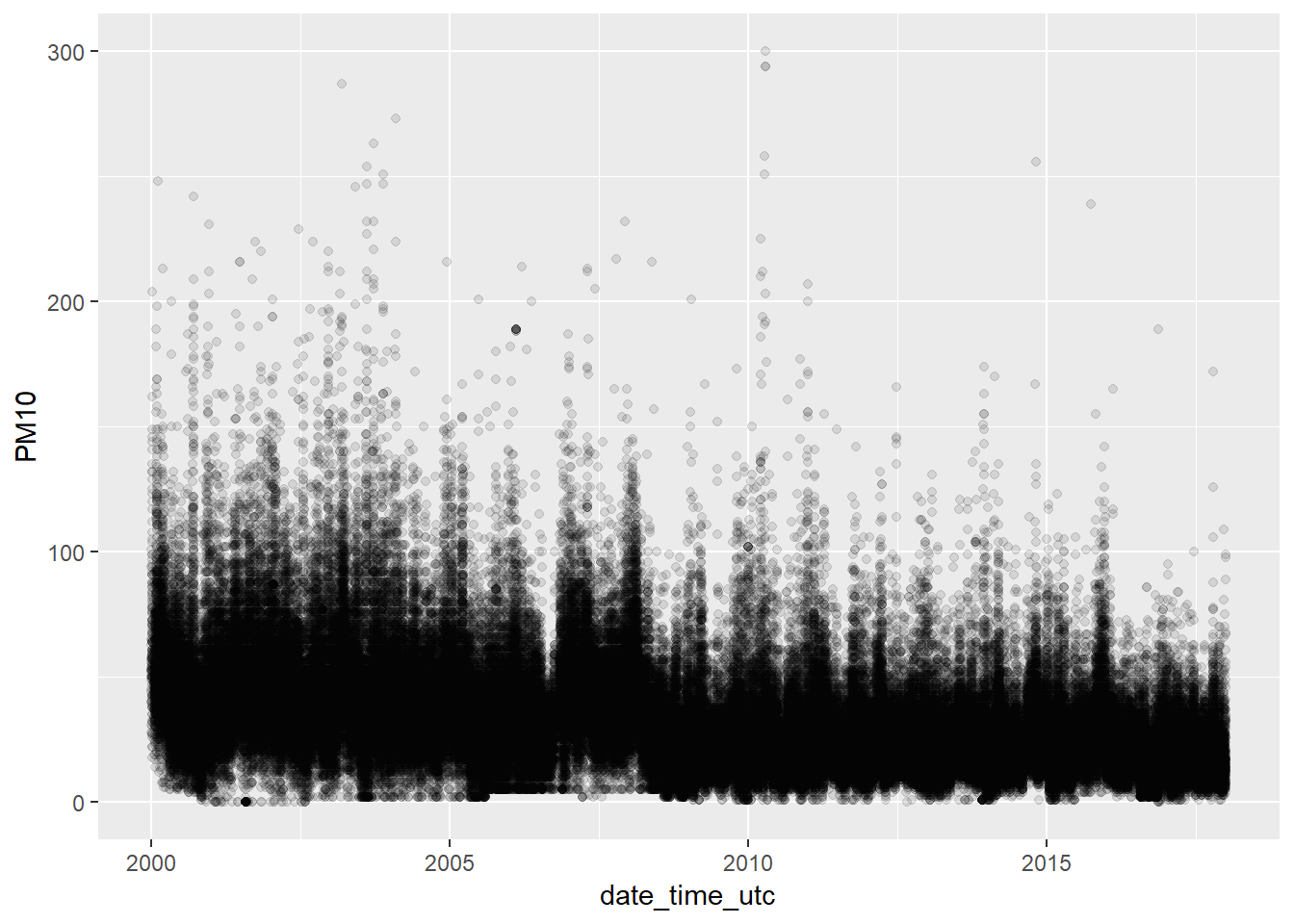
ggplot(pm10, aes(x = date_time_utc, y = PM10)) +
geom_point(alpha = 0.1, size = 0.2) +
scale_y_continuous(trans='log2')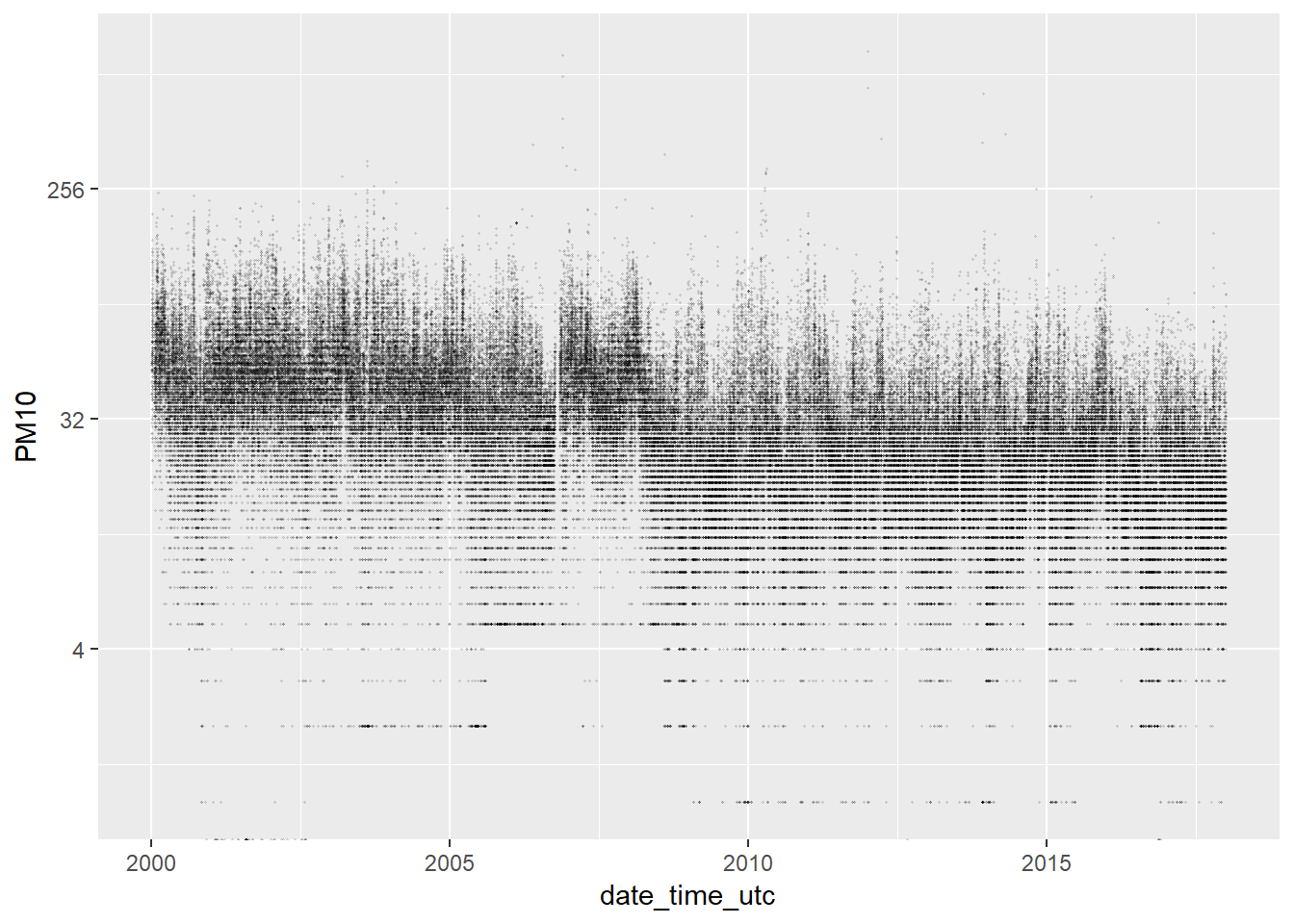
PM10_2017 <- pm10 %>% filter(year(date_time_utc) == '2017')
ggplot(PM10_2017, aes(x = date_time_utc, y = PM10)) +
geom_point(alpha = 0.1, size = 1) +
scale_y_continuous(trans='log2')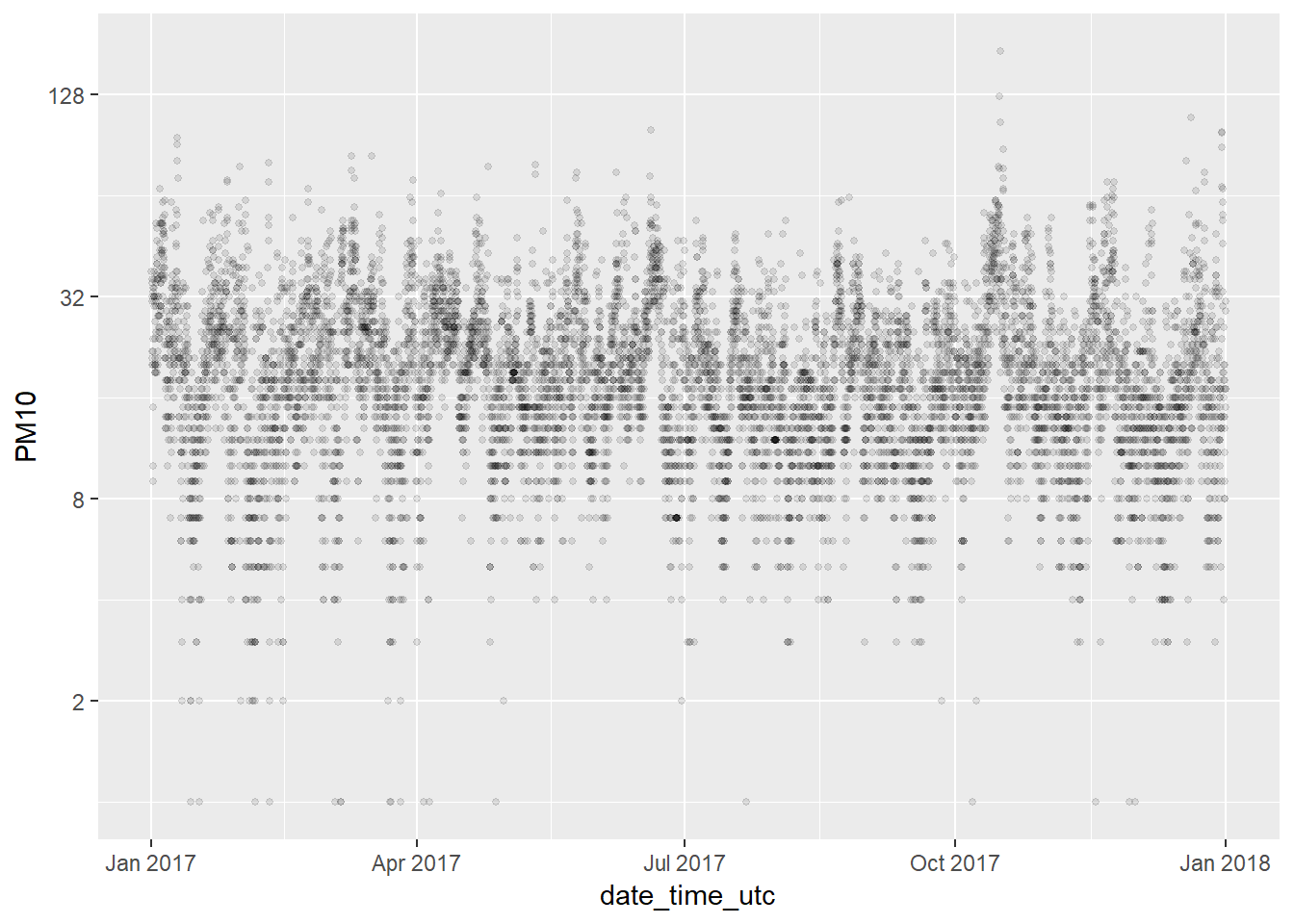
Daily averages
We create a new dataset with the PM10 daily averages and we plot them in a new graphic. We add a trend line too. There is a clear downward trend in the measurements and we have many fewer extreme values during the last decade. It seems like we have two very clear “epochs” in the data, with a clear change in the data structure starting somewhere around 2009.
pm10_day_avg <- pm10 %>% group_by(day = date(date_time_utc)) %>%
summarise(day_avg = mean(PM10, na.rm = TRUE))
ggplot(pm10_day_avg, aes(x = day, y = day_avg, , colour = day_avg)) +
geom_point(alpha = 0.5) +
geom_smooth(color = "grey", alpha = 0.2) +
scale_colour_gradientn(colours = terrain.colors(10)) +
theme(legend.position = c(0.3, 0.9),
legend.background = element_rect(colour = "transparent", fill = NA), legend.direction = "horizontal") +
labs(colour = "PM10 daily average (colour scale)", x = "Year", y = "PM10 daily average", title = "PM10 daily average - 2000-2017 evolution (Constitucion station)")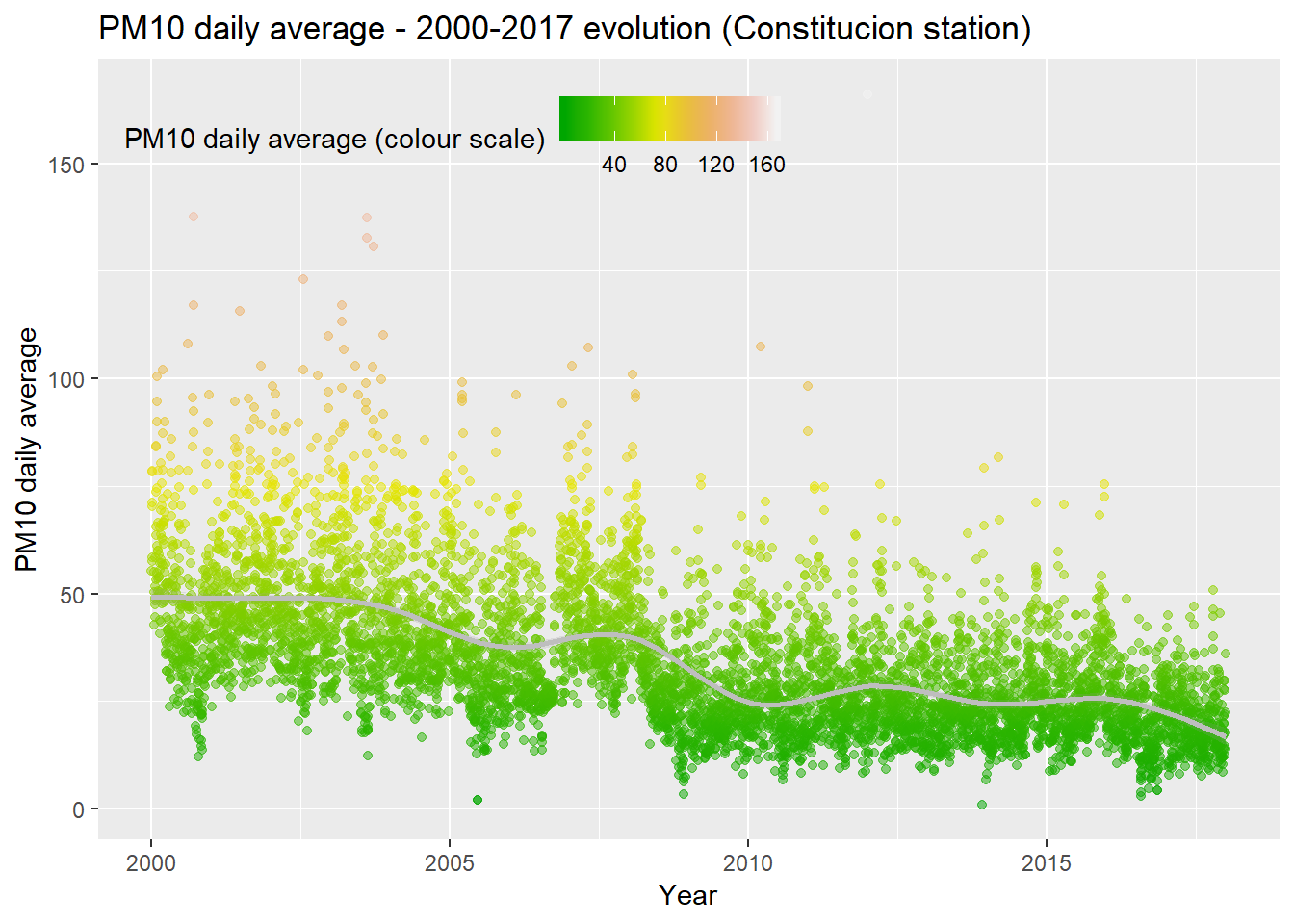 We plot the same data but through a box plot graph. Now we can see very clearly how since 2009 the behaviour of the data is sustantially different from the precedent years. The PM10 day levels are smaller, the variability is significantly less pronounced and the trend is smoother too.
We plot the same data but through a box plot graph. Now we can see very clearly how since 2009 the behaviour of the data is sustantially different from the precedent years. The PM10 day levels are smaller, the variability is significantly less pronounced and the trend is smoother too.
pm10_day_avg <- pm10 %>% group_by(day = date(date_time_utc)) %>%
summarise(day_avg = mean(PM10, na.rm = TRUE))
ggplot(pm10_day_avg, aes(y = day_avg, x = as.factor(year(day)), colour = day_avg)) +
geom_boxplot(alpha = 0.5)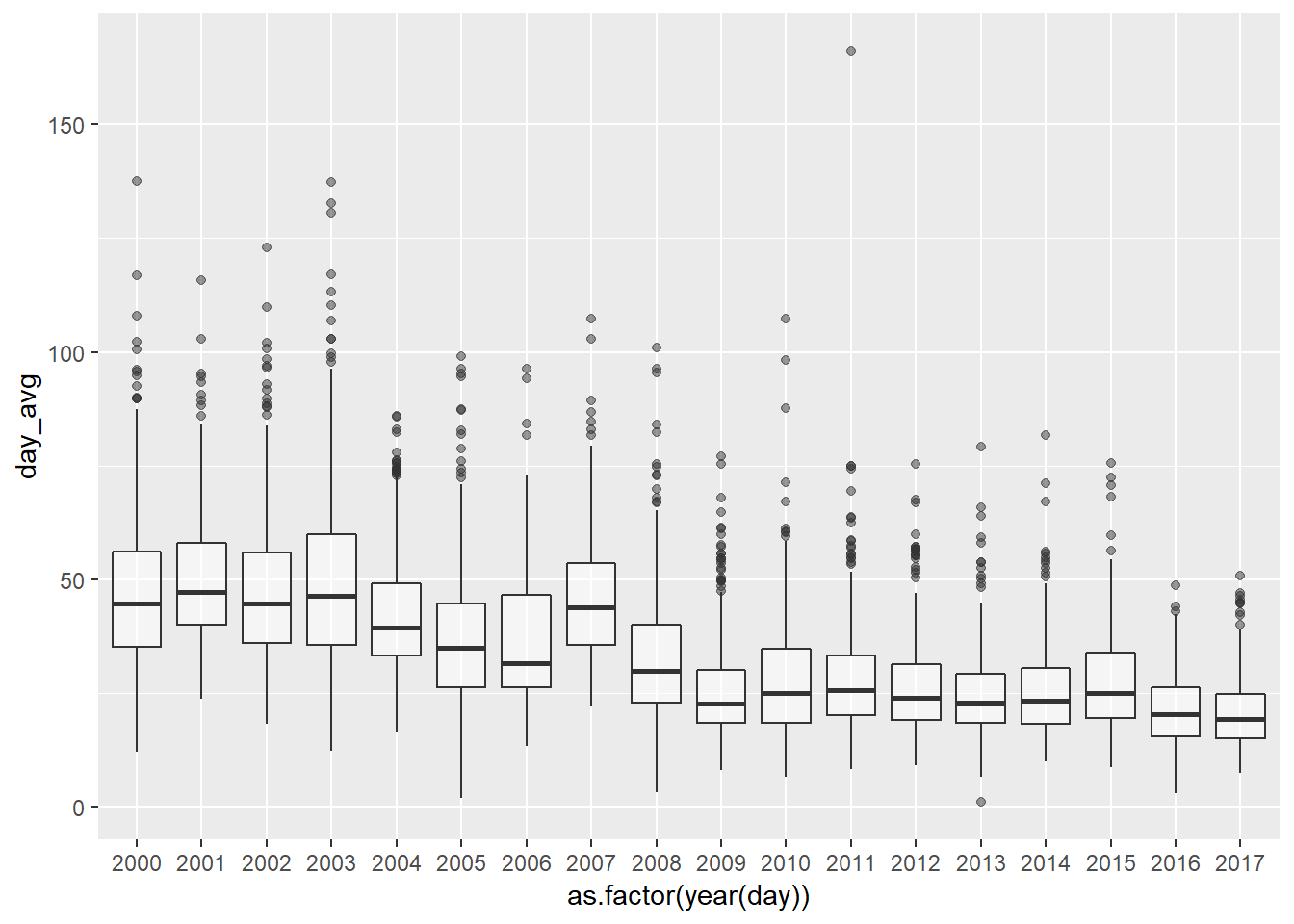
We identify a very clear trend through the years on the last graph. But, as we already saw before on the grid graphs there are other things happening at the same time.
year_const <- year_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'PM10')
plot1 <- ggplot(year_const, aes(x = year, y = value)) +
geom_line()
month_const <- month_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'PM10')
plot2 <- ggplot(month_const, aes(x = month, y = value)) +
geom_line()
week_day_const <- week_day_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'PM10')
plot3 <- ggplot(week_day_const, aes(x = week_day, y = value)) +
geom_line()
hour_const <- hour_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'PM10')
plot4 <- ggplot(hour_const, aes(x = hour, y = value)) +
geom_line()
grid.arrange(plot1, plot2, plot3, plot4, ncol = 2)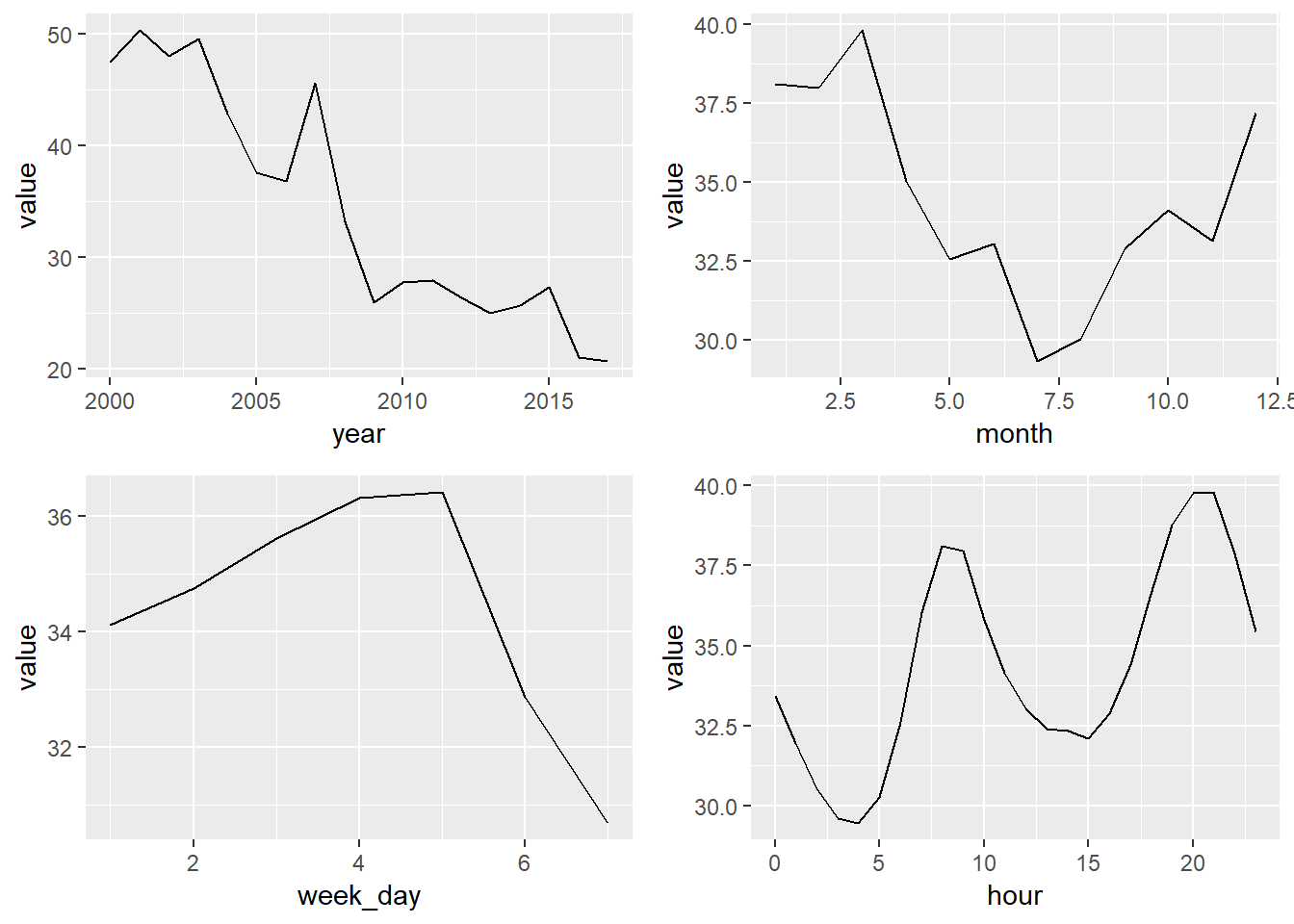
7.3 NO2 Constitucion Station
We create the dataset pm10 with PM10 values from the Constitución Station and we execute a summary
NO2 <- air_data_2 %>% filter(station == '1') %>%
select(date_time_utc, NO2)
summary(NO2)## date_time_utc NO2
## Min. :2000-01-01 00:00:00 Min. : 0.00
## 1st Qu.:2004-06-30 23:15:00 1st Qu.: 20.00
## Median :2009-01-02 00:30:00 Median : 34.00
## Mean :2008-12-31 18:50:45 Mean : 37.26
## 3rd Qu.:2013-07-02 23:45:00 3rd Qu.: 51.00
## Max. :2017-12-31 23:00:00 Max. :408.00
## NA's :2930NO2_day_avg <- NO2 %>% group_by(day = date(date_time_utc)) %>%
summarise(day_avg = mean(NO2, na.rm = TRUE))
ggplot(NO2_day_avg, aes(x = day, y = day_avg, , colour = day_avg)) +
geom_point(alpha = 0.5) +
geom_smooth(color = "grey", alpha = 0.2) +
scale_colour_gradientn(colours = terrain.colors(10)) +
theme(legend.position = c(0.3, 0.9),
legend.background = element_rect(colour = "transparent", fill = NA), legend.direction = "horizontal") +
labs(colour = "PM10 daily average (colour scale)", x = "Year", y = "PM10 daily average", title = "NO2 daily average - 2000-2017 evolution (Constitucion station)")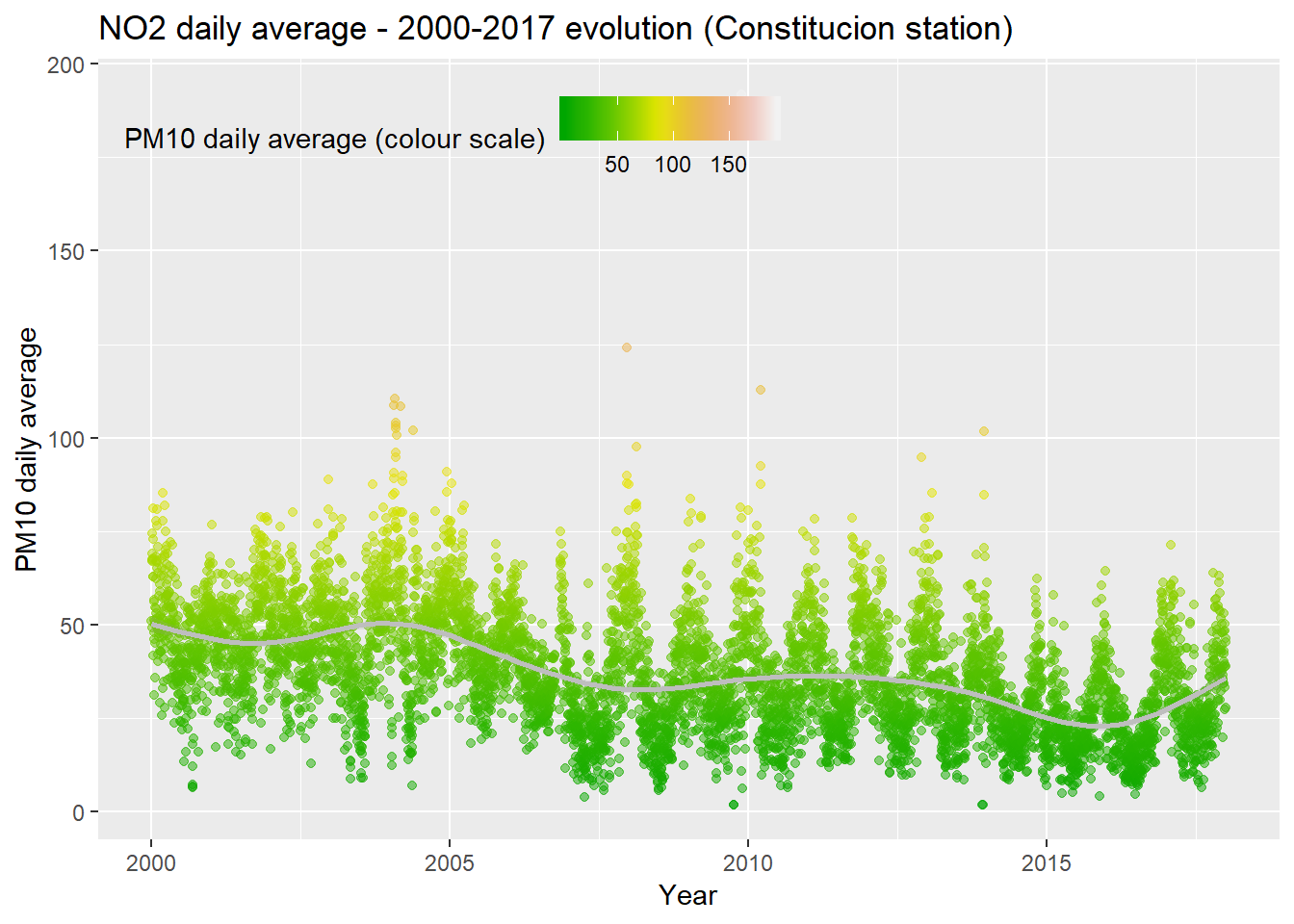
significantly less pronounced and the trend is smoother too.
NO2_day_avg <- NO2 %>% group_by(day = date(date_time_utc)) %>%
summarise(day_avg = mean(NO2, na.rm = TRUE))
ggplot(NO2_day_avg, aes(y = day_avg, x = as.factor(year(day)), colour = day_avg)) +
geom_boxplot(alpha = 0.5)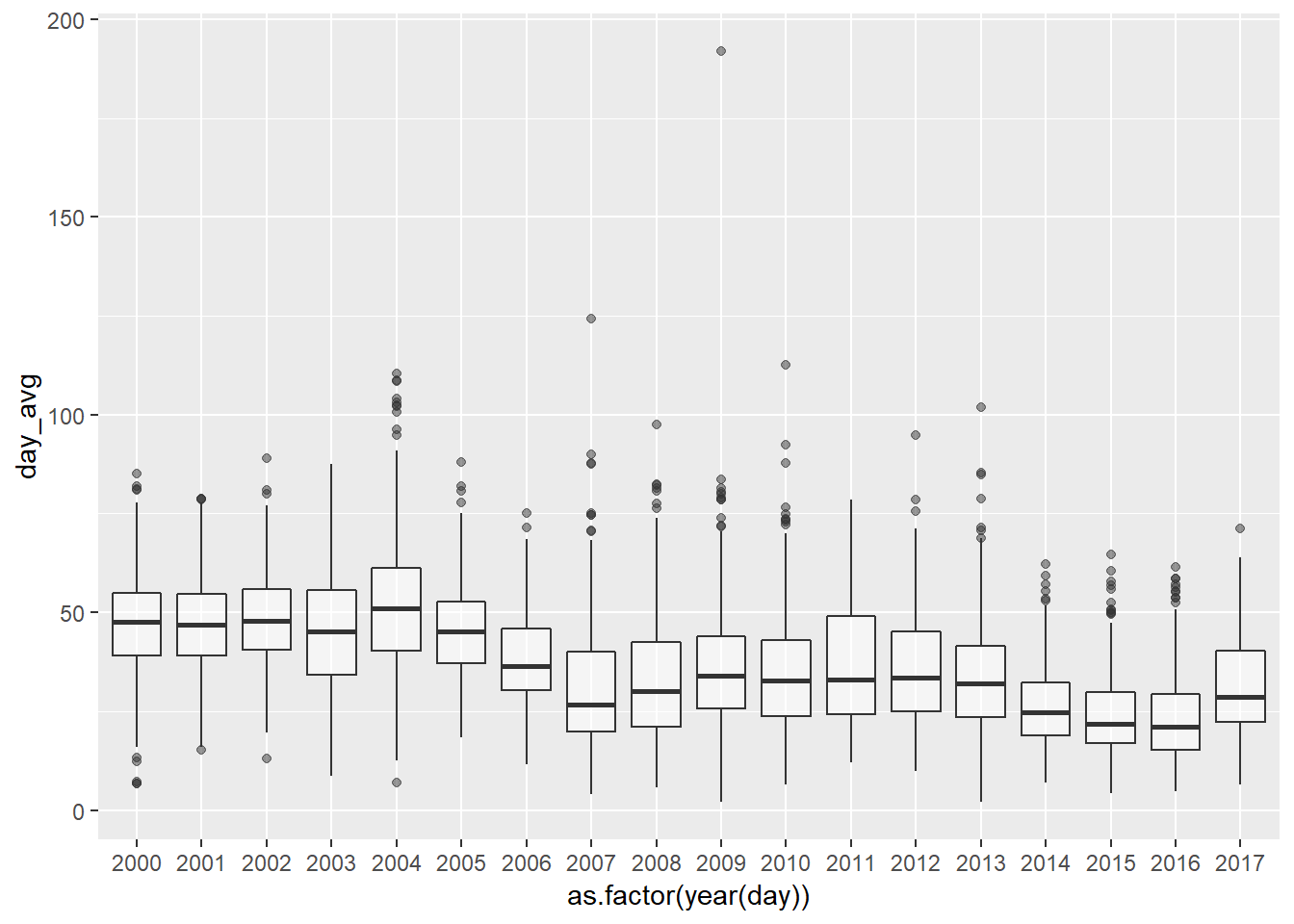
year_const <- year_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'NO2')
plot1 <- ggplot(year_const, aes(x = year, y = value)) +
geom_line()
month_const <- month_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'NO2')
plot2 <- ggplot(month_const, aes(x = month, y = value)) +
geom_line()
week_day_const <- week_day_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'NO2')
plot3 <- ggplot(week_day_const, aes(x = week_day, y = value)) +
geom_line()
hour_const <- hour_avgs_long %>% filter(grepl('Constit', station_alias), contaminante == 'NO2')
plot4 <- ggplot(hour_const, aes(x = hour, y = value)) +
geom_line()
grid.arrange(plot1, plot2, plot3, plot4, ncol = 2)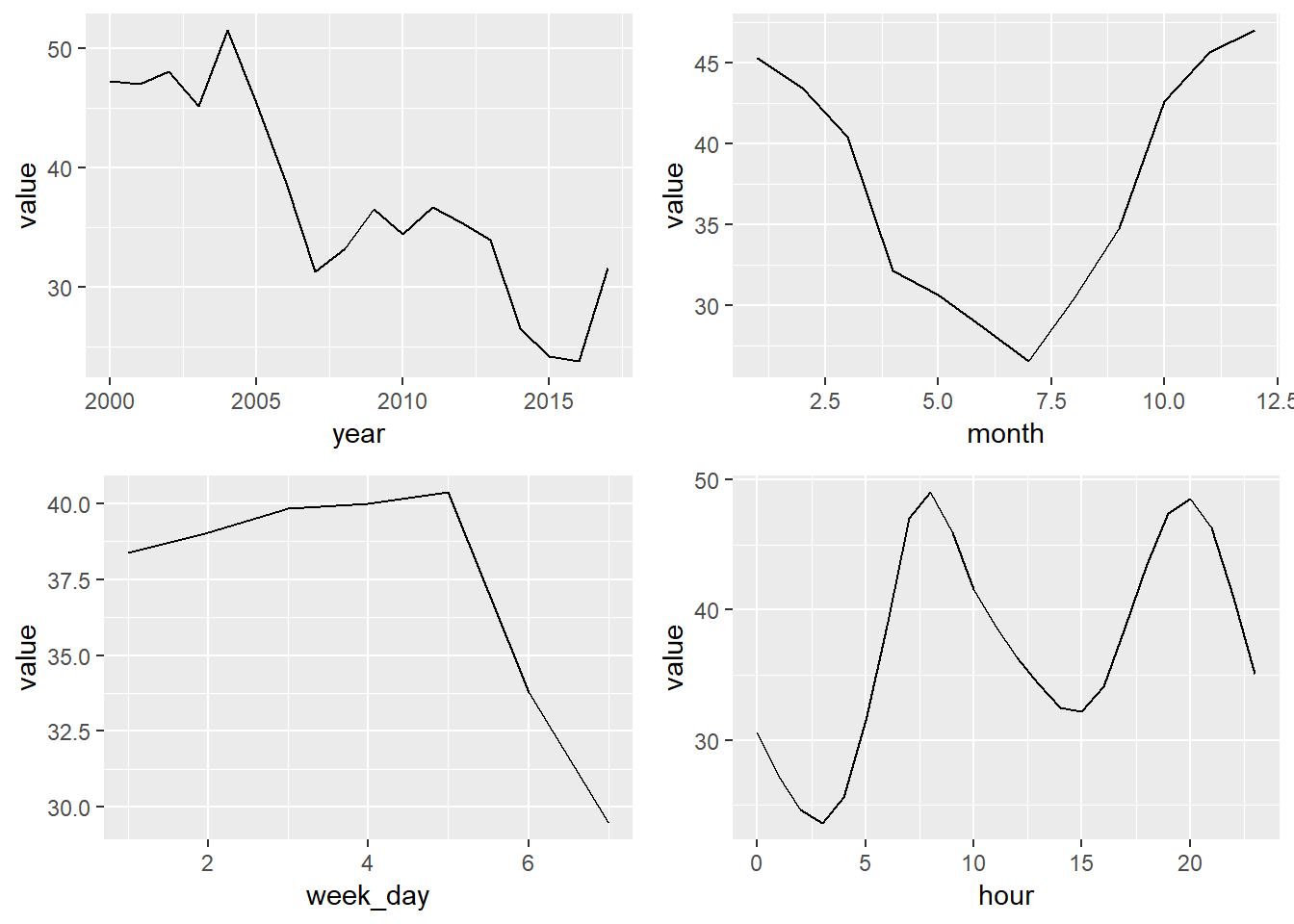
7.4 Relationships between variables
The Constitucion Station is the only station with meteorological data. So, we are going to focus our efforts of data exploration on this station.
Pollutants correlations matrix
constitucion_data <- readRDS("data_rds/constitucion_data.rds")
### Pollutants correlation matrix
# We select the pollutants variables
pollutants <- constitucion_data %>% select(PM10, PM25, NO2, NO, SO2, CO, O3, MXIL, TOL) %>%
na.omit()
# We create the pollutants correlation matrix
cor_matrix <- round(cor(pollutants), 2)
# We transform the table to long format to be handled by ggplot2
long_cor_matrix <- melt(cor_matrix)
# Get lower triangle of the correlation matrix
get_lower_tri<-function(cor_matrix){
cor_matrix[upper.tri(cor_matrix)] <- NA
return(cor_matrix)
}
# Get upper triangle of the correlation matrix
get_upper_tri <- function(cor_matrix){
cor_matrix[lower.tri(cor_matrix)]<- NA
return(cor_matrix)
}
upper_tri <- get_upper_tri(cor_matrix)
# Melt the correlation matrix
library(reshape2)
long_cor_matrix <- melt(upper_tri, na.rm = TRUE)
# Create a ggheatmap
ggheatmap <- ggplot(long_cor_matrix, aes(Var2, Var1, fill = value))+
geom_tile(color = "white")+
scale_fill_gradient2(low = "blue", high = "red", mid = "white",
midpoint = 0, limit = c(-1,1), space = "Lab",
name="Pearson\nCorrelation") +
theme_minimal()+ # minimal theme
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 12, hjust = 1))+
coord_fixed()
pollutants_heatmap <- ggheatmap + geom_text(aes(Var2, Var1, label = value), color = "black", size = 4) +
theme(
axis.title.x = element_blank(),
axis.title.y = element_blank(),
panel.grid.major = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
axis.ticks = element_blank(),
legend.justification = c(1, 0),
legend.position = c(0.6, 0.7),
legend.direction = "horizontal")+
guides(fill = guide_colorbar(barwidth = 7, barheight = 1,
title.position = "top", title.hjust = 0.5))
print(pollutants_heatmap)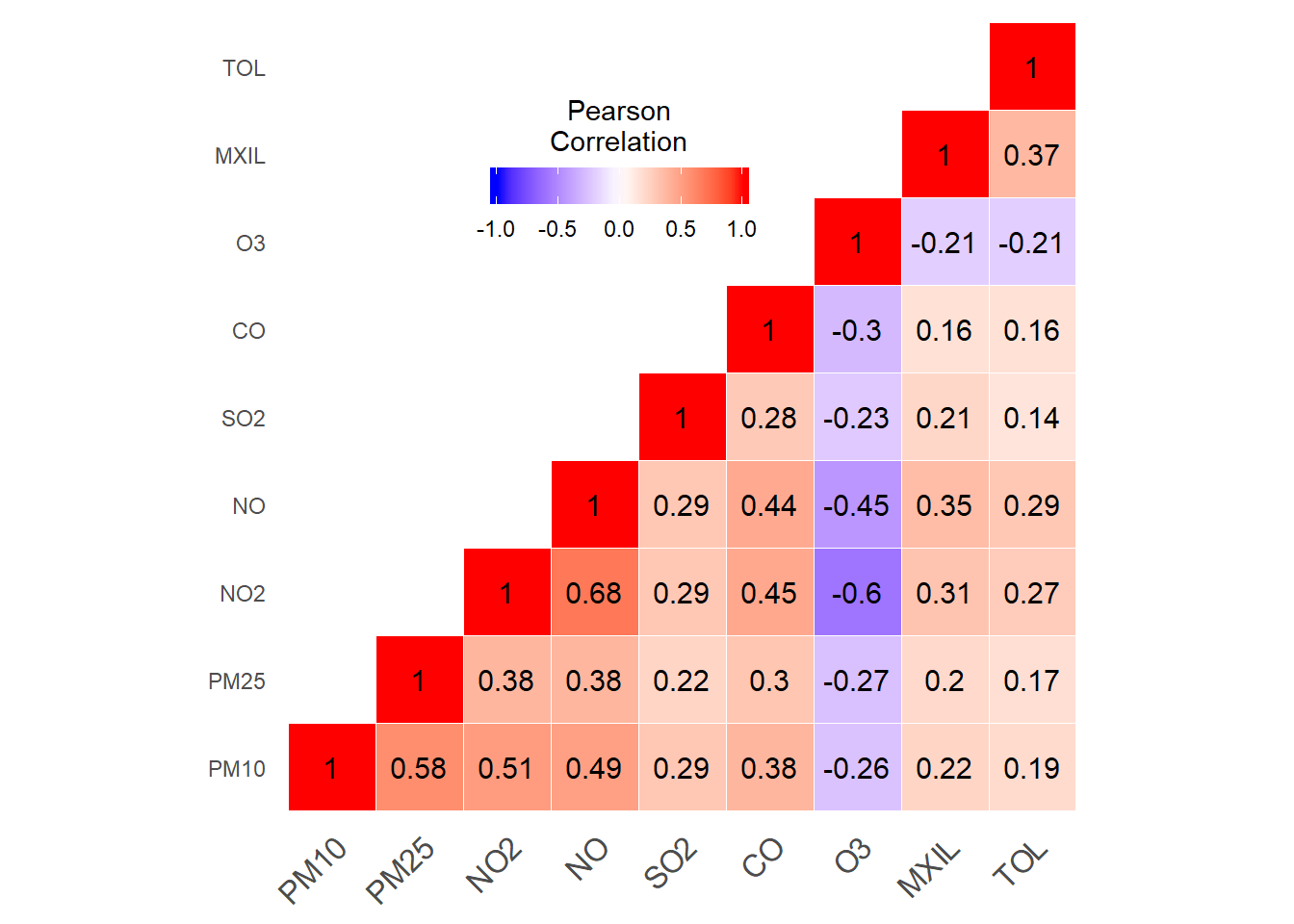
Pollutants-weather data correlations matrix
constitucion_data <- readRDS("data_rds/constitucion_data.rds")
### Pollutants correlation matrix
# We select the pollutants variables
pollutants_weather <- constitucion_data %>% select(PM10, PM25, NO2, NO, SO2, CO, O3, MXIL, TOL, dd, vv, TMP, PRB, LL, HR, RS) %>%
na.omit()
# We create the pollutants correlation matrix
cor_matrix <- round(cor(pollutants_weather), 2)
# We transform the table to long format to be handled by ggplot2
long_cor_matrix <- melt(cor_matrix)
# Get lower triangle of the correlation matrix
get_lower_tri<-function(cor_matrix){
cor_matrix[upper.tri(cor_matrix)] <- NA
return(cor_matrix)
}
# Get upper triangle of the correlation matrix
get_upper_tri <- function(cor_matrix){
cor_matrix[lower.tri(cor_matrix)]<- NA
return(cor_matrix)
}
upper_tri <- get_upper_tri(cor_matrix)
# Melt the correlation matrix
library(reshape2)
long_cor_matrix <- melt(upper_tri, na.rm = TRUE)
# Create a ggheatmap
ggheatmap <- ggplot(long_cor_matrix, aes(Var2, Var1, fill = value))+
geom_tile(color = "white")+
scale_fill_gradient2(low = "blue", high = "red", mid = "white",
midpoint = 0, limit = c(-1,1), space = "Lab",
name="Pearson\nCorrelation") +
theme_minimal()+ # minimal theme
theme(axis.text.x = element_text(angle = 45, vjust = 1,
size = 12, hjust = 1))+
coord_fixed()
pollutants_heatmap <- ggheatmap + geom_text(aes(Var2, Var1, label = value), color = "black", size = 2) +
theme(
axis.title.x = element_blank(),
axis.title.y = element_blank(),
panel.grid.major = element_blank(),
panel.border = element_blank(),
panel.background = element_blank(),
axis.ticks = element_blank(),
legend.justification = c(1, 0),
legend.position = c(0.6, 0.7),
legend.direction = "horizontal")+
guides(fill = guide_colorbar(barwidth = 7, barheight = 1,
title.position = "top", title.hjust = 0.5))
print(pollutants_heatmap)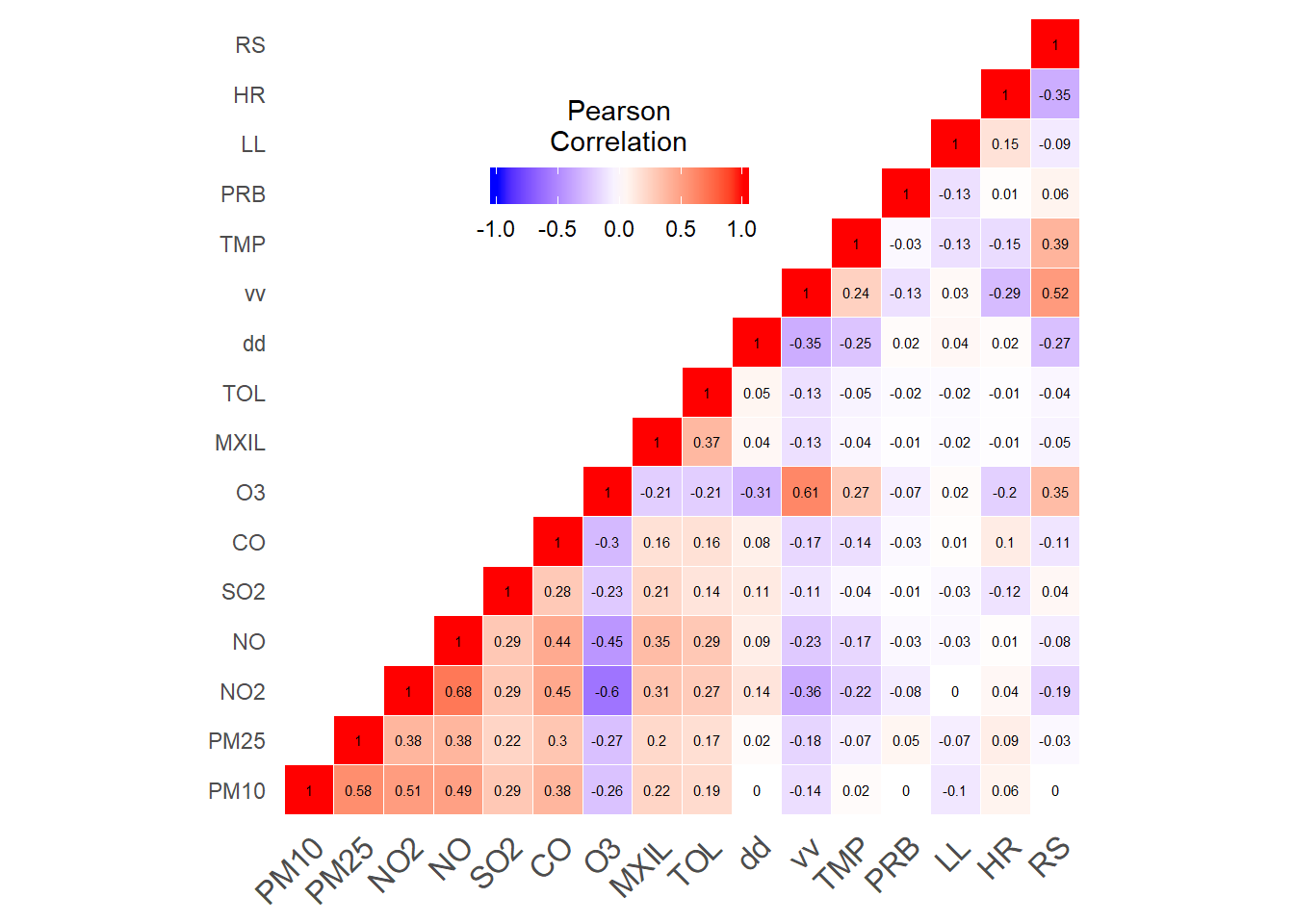
The openair package is a R package especialized in the visualizations of air quality data.
Wind visualizations (openair package)
windRose(air_data_2)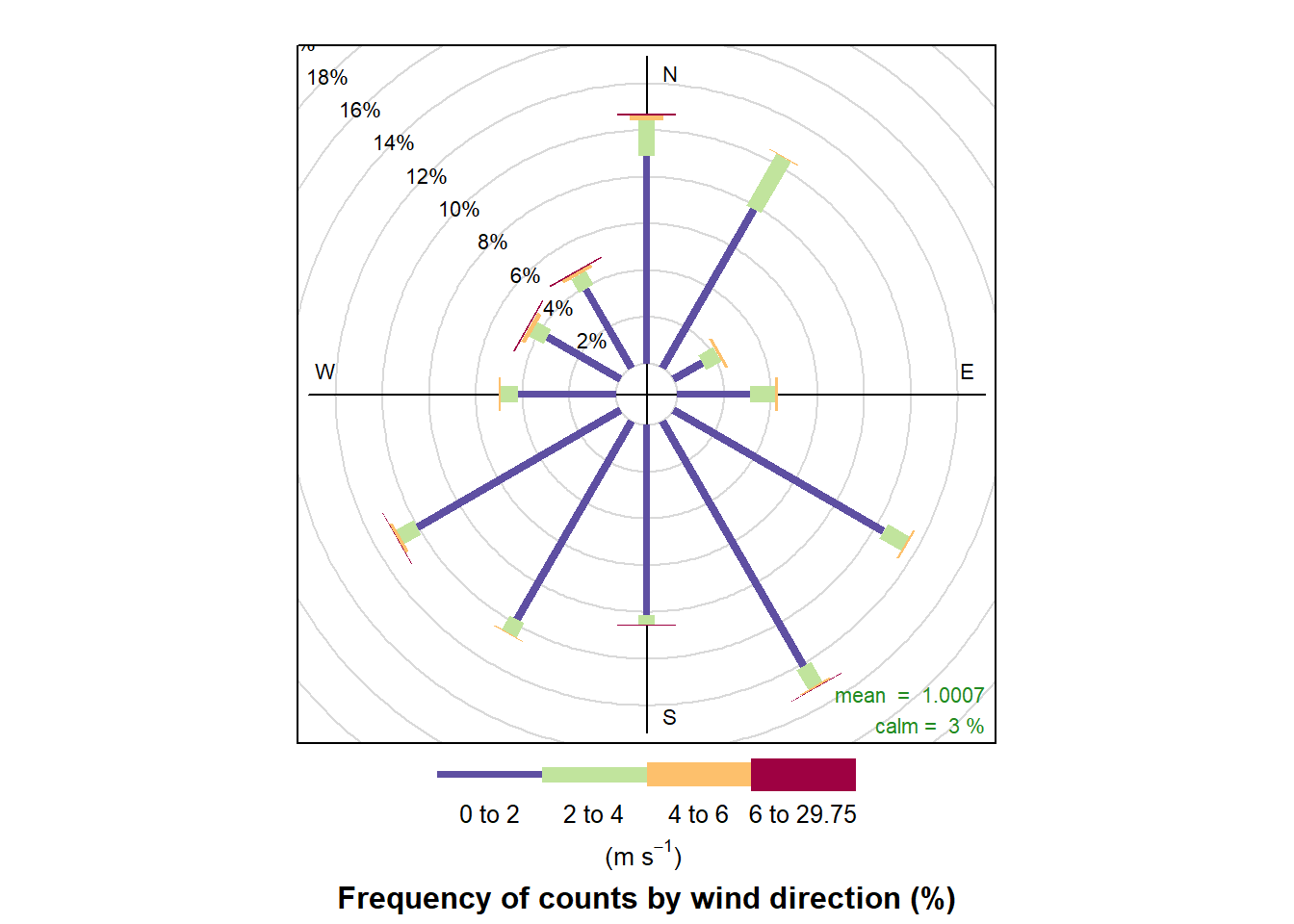
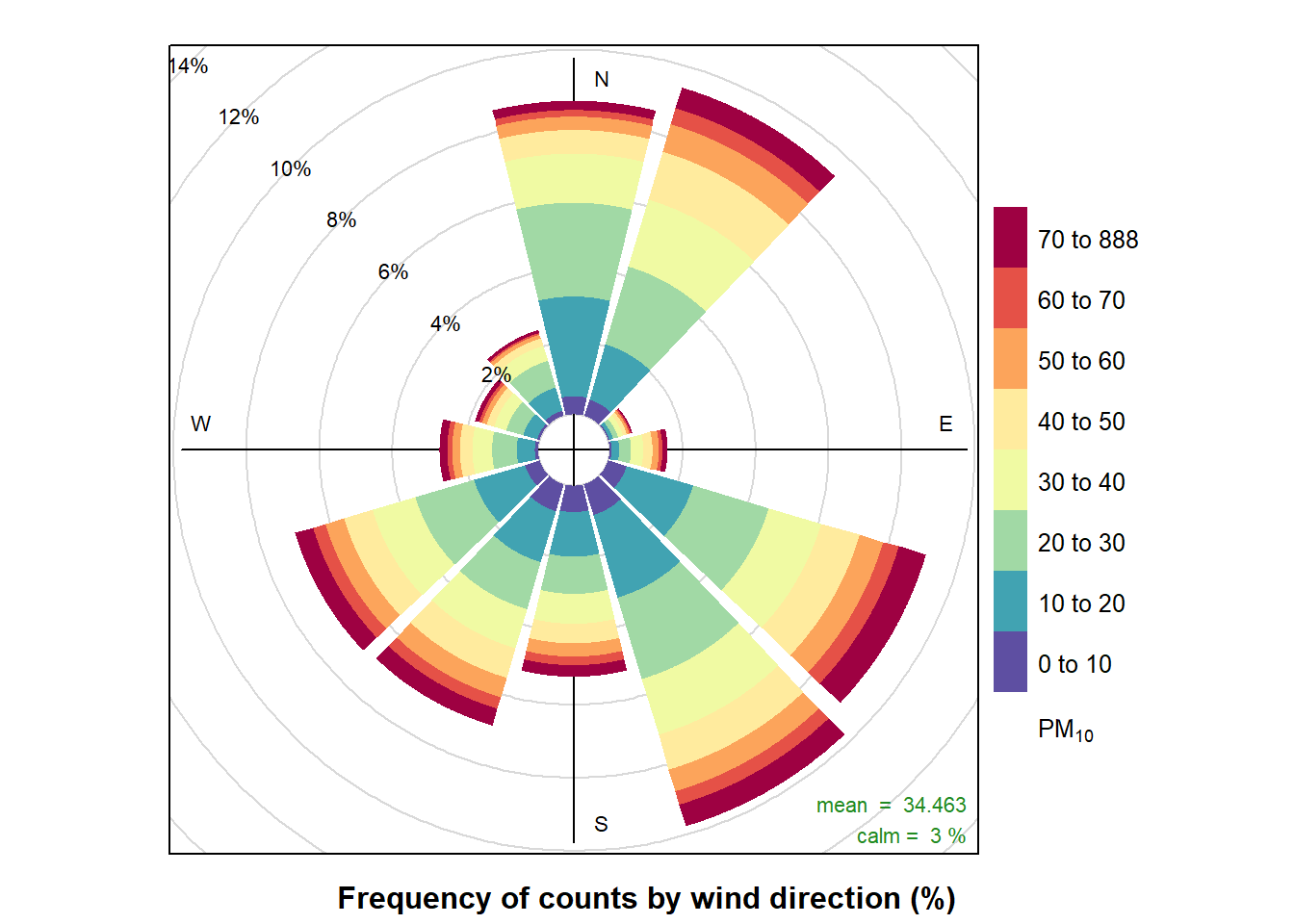
Pollution rose ((openair package))
pollutionRose(air_data_2, pollutant = "PM10")