1 Genes, Genomes and Genome Browsers
1.1 What is a gene?
There are many ways to define a gene. One definition: A gene is a segment of double-stranded DNA that directs the production of a functional product (i.e. protein). A significant number of eukaroytic genes are protein coding. Each protein-coding gene first directs the production of an RNA transcript through a process called transcription. This RNA transcript is then processed into a messenger RNA (mRNA). The most notable change that occurs during RNA processing is that intron sequences are removed (thin lines in Figure 1.1). The mRNA is then exported from the nucleus and used as a template to make a protein in a process called translation. This entire process (transcription, RNA processing and translation) is called Gene expression. The final products, often proteins, do much of the work necessary to create and maintain organismal structure and function.
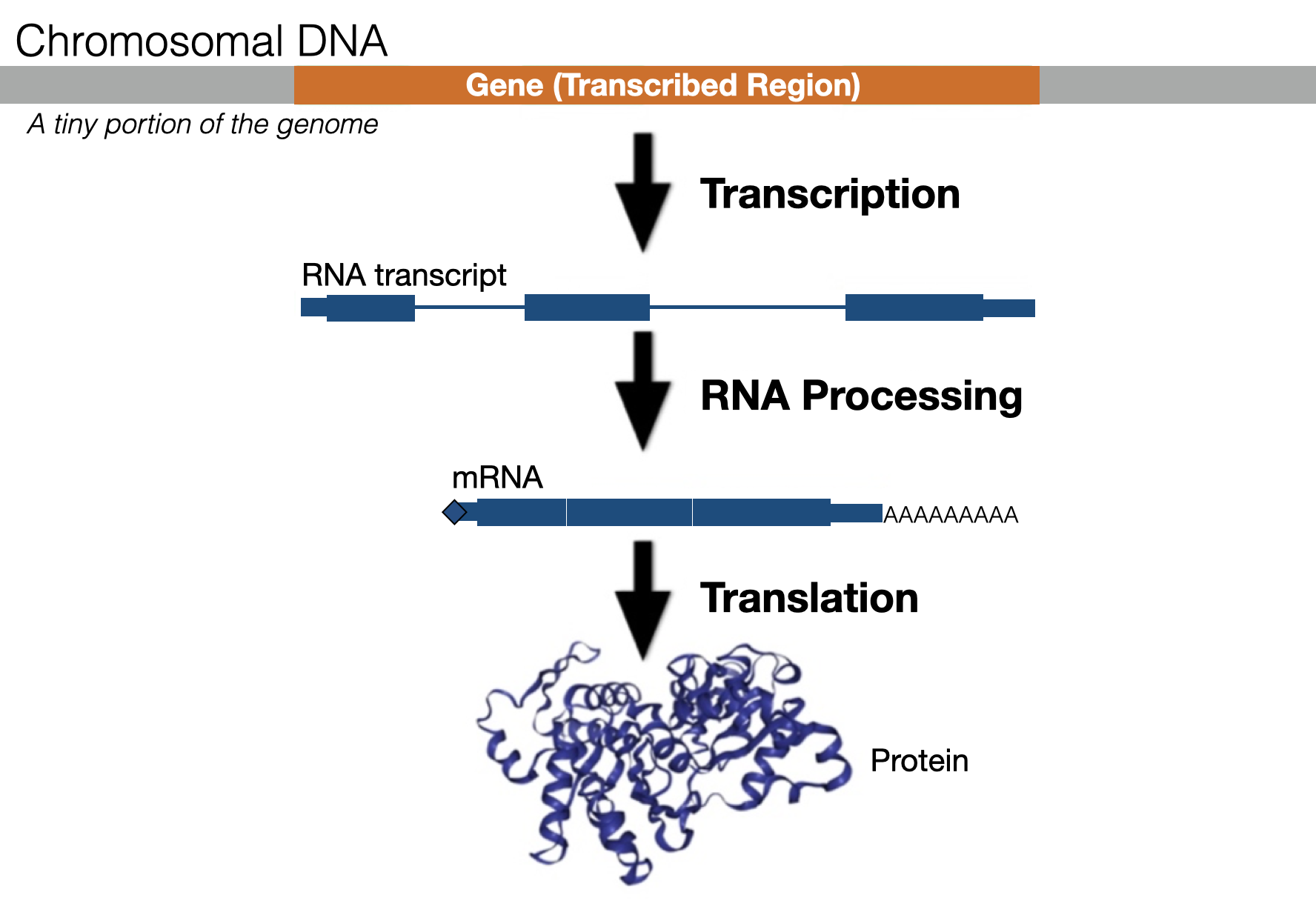
Figure 1.1: An overview of Gene Expression. A small segment of a chromosome (light grey) containing a single gene (orange) is shown. The RNA transcript is highlighted as blue rectangles (exons) and blue lines (introns). Introns are removed during RNA processing to create a mature messenger RNA (mRNA)
While the transcribed region of a gene requires adjacent sequence (i.e. the promoter, see Chapter 5) to become transcribed at the right time and place, for simplicity, the size of a gene (number of base pairs) is typically defined by where transcription begins and ends. Thus, the sites of transcription initiation and transcription termination define both gene size and RNA transcript size (but not mRNA size which is typically shorter as most eukaryotic genes have introns).
1.2 What is a genome?
Genes are distributed linearly along chromosomes2 (Figure 1.2). One complete, nonredundant set of chromosomes for a given species makes up what we call a genome (also called a “haploid genome”)3.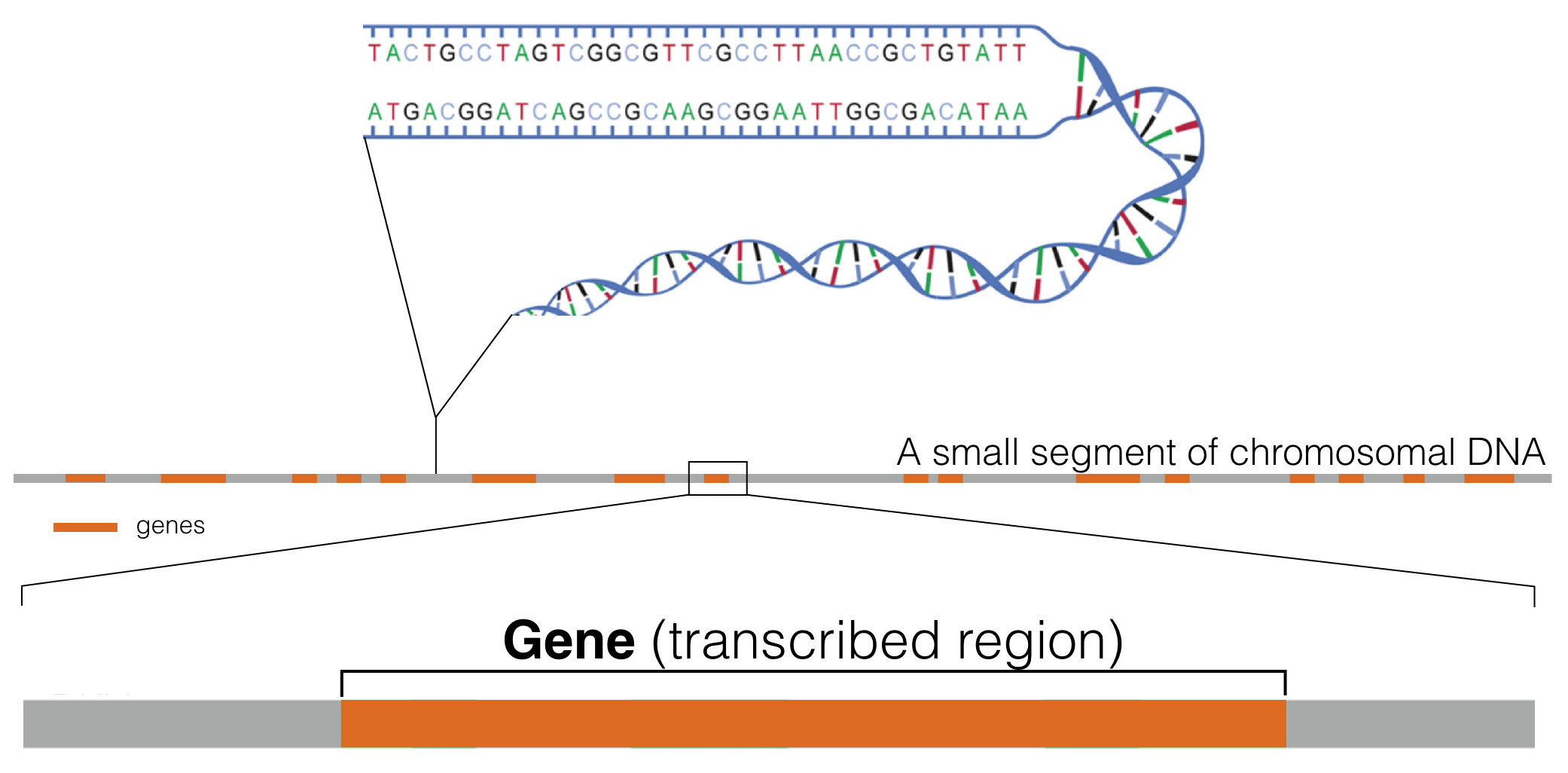
Figure 1.2: This schematic represents a hypothetical segment of a chromosome with orange rectangles representing genes distributed linearly along the DNA segment shown.
The haploid genome for humans consists of 24 linear chromosomes (Figure 1.3) and one circular mitochondrial chromosome (not shown). Each chromosome varies in length. For example, chromosome one is the longest at 248,956,422 base pairs (bp). Given that 10 bp of a double helix measures 34 angstroms in length, chromosome one is 82.7 mm (more than three inches if pulled taut)!4 (Figure 1.3).
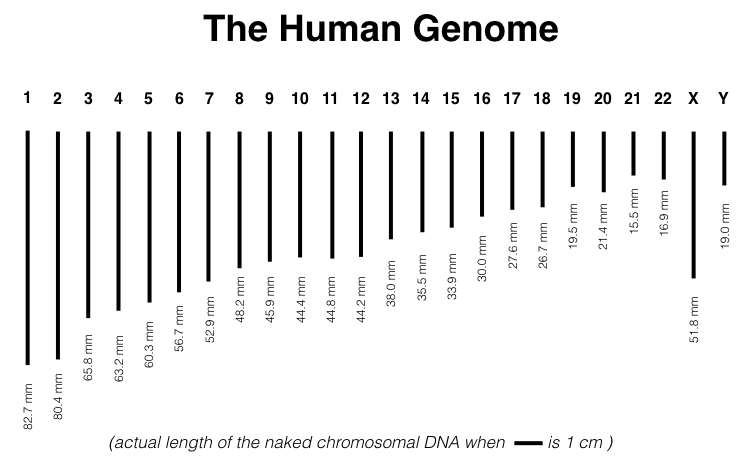
Figure 1.3: Actual length of the human genome when scale bar equals one centimeter. Naken mitochondrial DNA is too small to be shown. It is a small circular DNA molecule, with a circumference of 55 microns.
Like most animals, humans are diploid. Thus, their somatic cells5 harbor 22 pairs of autosomes6 and one pair of sex chromosomes (either XX or XY). Thus, over two meters (2000 mm) of DNA is crammed into each somatic nucleus! But the diameter of the average mammalian nucleus is only 6 microns or 0.006 of a millimeter! How does the diploid genome fit in the nucleus? And how does it prevented from becoming a tangled mess? First, the diameter of the DNA double helix is only twenty angstroms7 but also, each chromosome is carefully packaged by proteins in a systematic and stereotypical manner (Figure 1.4).
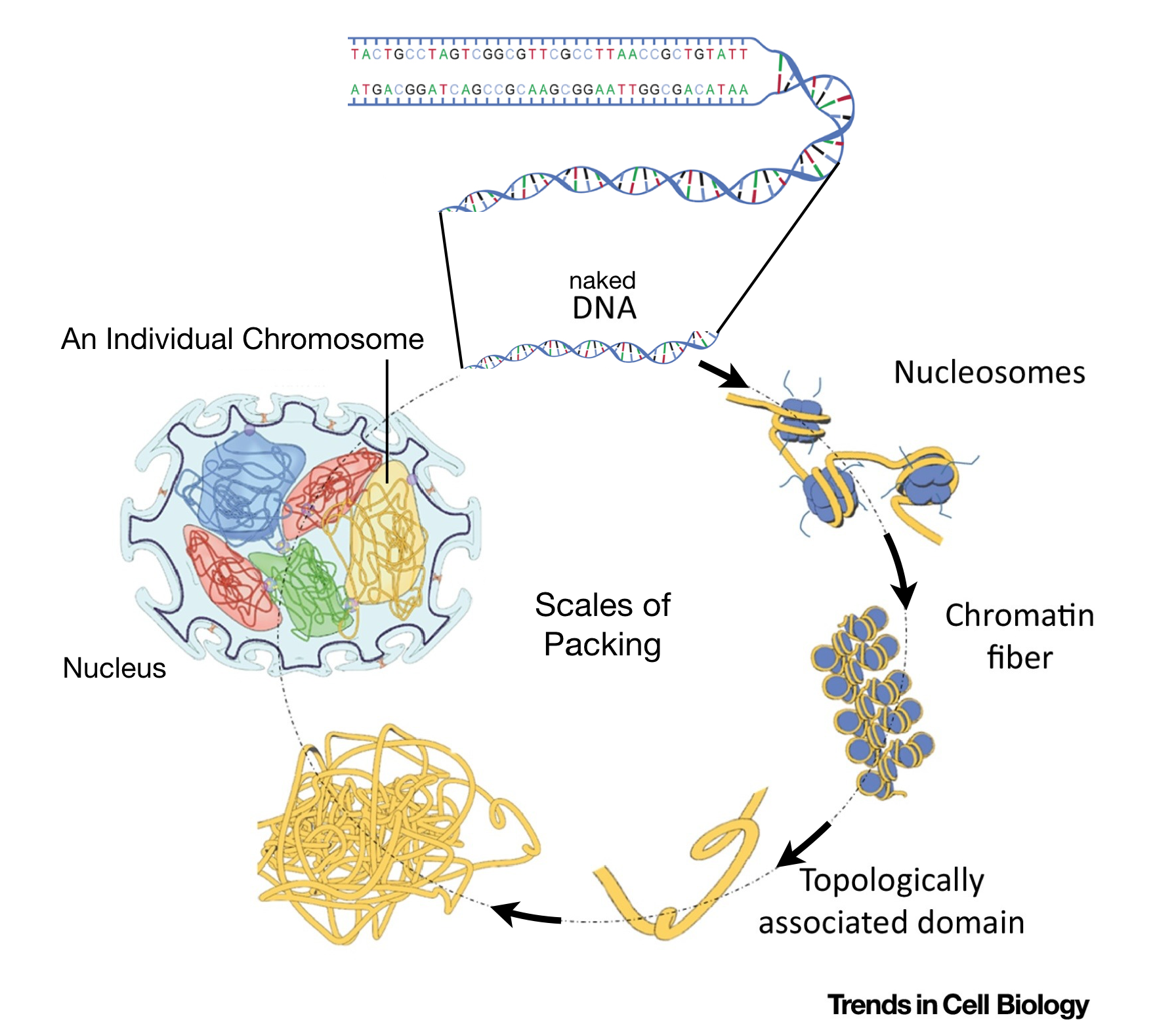
Figure 1.4: This image is modified from Uhler and Shivashankar, 2017. It shows the various levels of chromosome packing found in nuclei during interphase of the cell cycle. The first level of packing requires an octamer of histone proteins that assemble into a ball-like structure called a nucleosome. Naked DNA wraps around these octamers forming ‘beads on a string’. Additional levels of packing into chromatin fibers and topological associated domains requires additional proteins. Individual chromosomes (23 pairs in humans) are then carefully orgnanized within the cell nucleus in a cell type-dependent manner.
1.2.1 Test Your Understanding
Click the link above to see the TYU questions.
1.3 What is a Genome Browser?
According to Wikipedia, Genome browsers enable researchers to visualize genes in the genomes with annotated data including gene structure, information about gene expression, positions of disease causing mutations etc. They differ from ordinary biological databases in that they display data contributed by multiple sources in a graphical format. The top strand of the genome sequence is typically displayed (by default) along the X-axis (when the sequence is not visible, coordinates are given). The annotations and graphics that describe a given region of the genome are stacked below the genome sequence.
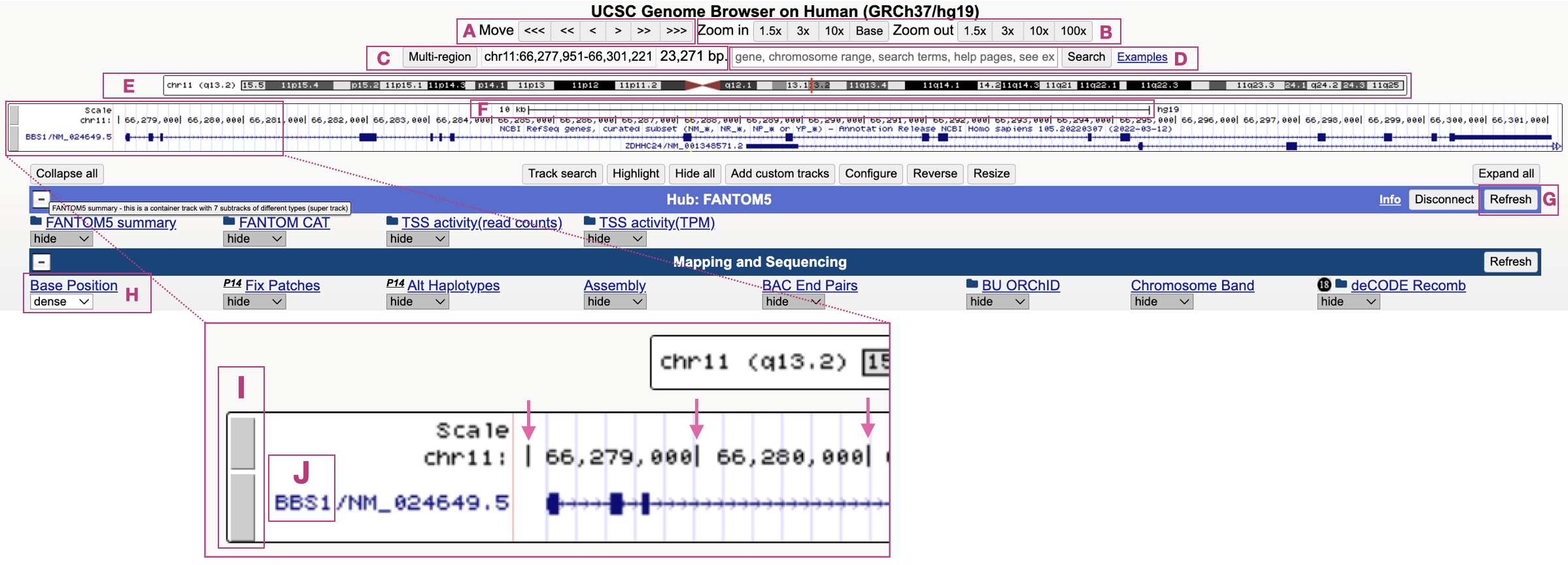
There are numerous Genome Browsers available. We will use the UCSC Genome Browser. The UCSC Genome Browser harbors the sequence of genomes representative of numerous species called “reference genomes”8. Our initial focus will be on a single gene in the human genome, BBS1. In this chapter, using BBS1 as our example, you will learn how the UCSC Genome Browser is organized, how to configure so-called “evidence tracks”9, how to navigate through Genome Browser window (scrolling left and right, zooming in and out) and how to understand the graphical data that describe specific gene features.
1.5 Addendum
Now that you have more familiarity with gene structure, the following tutorials are highly recommended. They were created by the UCSC Genome Browser team. These are also worth revisiting before you embark on your Independent Project at the end of the term.
- UCSC Genome Browser Basics. Part 1: Getting around in the Browser
- UCSC Genome Browser Basics, Part 2: Configuring the Browser
- UCSC Genome Browser Basics. Part 3: Configuration + DNA navigation
- Saving and Sharing Sessions on the UCSC Genome Browser
- Controlling the visibility of data tracks on the UCSC Genome Browser
- and there is more!
© 2025, Maria Gallegos. All rights reserved.
a chromosome is a single, long molecule of double stranded DNA↩︎
Many organisms are diploid (including humans) meaning they have two copies of every chromosome. Thus the phrase “haploid genome” is more precise, although the word “haploid” is often omitted and simply assumed↩︎
to calculate the length of a chromosome: Multiply the length of a chromosome in base pairs (bp) with 0.000000332, the length (in mm) of each bp.↩︎
Cells of a multicellular organism can be divided into two main types: germ cells and somatic cells. Germ cells are destined to become the reproductive cells like sperm and oocytes. Somatic cells are destined to become all the other cell types like skin, neurons and muscle. This distinction is made as somatic cells die with the death of the organism while germ cells have the potential to pass their DNA on to the next generation.↩︎
An autosome is one of the numbered chromosomes, as opposed to the sex chromosomes. Autosomes are numbered roughly in relation to their sizes. The largest autosome — chromosome 1 — has approximately 2,800 genes; the smallest autosome — chromosome 22 — has approximately 750 genes. This definition is found at https://www.genome.gov/genetics-glossary/Autosome↩︎
an angstrom is a unit of length equal to one ten-millionth of a millimeter!↩︎
A “reference genome” (also called a “reference assembly”) is a genome sequence created from thousands of sequence runs assembled in silico to represent the sequence of a genome of one idealized individual organism. Since it is assembled from sequence data obtained from a number of donors, reference genomes do not represent the sequence of any single individual or organism, but rather a mosaic of multiple donors↩︎
each evidence track harbors specific biological data from a single source pertaining to the sequence displayed in the browser window)↩︎
The top strand of DNA is also referred to as the “+ strand”↩︎