A
High salt DNA extraction protocol
Ingredients:
- 20mg/ml proteinase K
- DNA extraction buffer (0.4M NaCl, 10mM Tris-HCl pH 8.0, and 2mM EDTA pH 8.0)
- TE buffer (10 mM Tris, 1mM EDTA, pH 8.0)
- 20% sodium dodecyl sulfate (warm to re-dissolve)
- 5M sodium chloride (NaCl saturated dH2O: Autoclave)
- 100% isopropanol
- 70% ethanol
Sample preparation
- Select the samples to be used from the freezer and hold on ice during sample prep.
- Remove the tissue and cut a portion onto a clean kimwipe in a petri dish.
- Squash out the ethanol or scrape excess salts, weigh ~20-40mg of tissue into a 1.5ml microcentrifuge tube and return tissue to storage. Cut up the tissue. to improve digestion and prevent the tissue from sticking to side of tube.
- Clean forceps and scissors between each sub-sample by wiping with a clean paper kimwipe, dipping in 98% ethanol and sterilising by passing through a flame
Cell lysis
- Add 400\(\mu\)l of DNA extraction buffer and 80\(\mu\)l of SDS (If SDS has precipitated, dissolve it on a heatblock (>40\(^\circ\)C).
- Add 10\(\mu\)l of proteinase K.
- Place in orbital mixer for 2hrs to overnight set to 300rpm and 50\(^\circ\)C throughout digestion.
Protein precipitation and removal
- Centrifuge max speed – 5 min.
- Transfer supernatant to a new tube.
- Add 320\(\mu\)l of 5M sodium chloride to each tube and mix by inverting the tubes 60 times.
- Centrifuge max speed – 10 min.
- Transfer supernatant to a new tube.
DNA precipitation
- Add 525\(\mu\)l of chilled (-20\(^\circ\)C) 100% isopropanol to the supernatant.
- Inverted 60 times.
- Centrifuge tubes for 20 min at 13K rpm (15.7k xg) at 4\(^\circ\)C.
- Carefully remove supernatant without disturbing pellet.
- Add 1ml of chilled 70% ethanol to tubes and invert 60 times.
- Centrifuge for 10 min at 13K rpm (15.7k xg) at 4\(^\circ\)C.
- Remove the supernatant and air dry the pellet for approximately 5 min at 37\(^\circ\)C on heat block, leave tubes open and cover with kimwipe - do not over-dry.
DNA rehydration
- Add 30\(\mu\)l of TE buffer to the dried pellet (dilute further if needed) and gently agitate tube to resuspended DNA. Leave to rehydrate for ~2hrs to overnight.
- Store DNA in fridge until checking quality (no longer than 48 hrs) or store in freezer for long term storage.
DNA Quantification
- Quantify DNA on the Nanodrop. Record ng/\(\mu\)l, 260/280 ratio (>1.8 pure DNA and <1.7 indicates protein contamination) and 260/230 ratio (<1.5 indicate salt contamination, ideally >1.8).
- Wipe pedestal clean with water.
- Add 1\(\mu\)l of TE buffer and run blank.
- Wipe pedestal with kimwipe then add 1\(\mu\)l of the first sample and click analyse.
- Repeat this step for each sample, wiping the pedestal clean with a kimwipe between samples, re-blank with TE buffer after every 5 samples.
- If DNA is too concentrated, dilute then store samples in fridge for at least 2 hours before re-measuring. Gently agitate tube to resuspend DNA before re-measuring. Aiming for around 100-200ng/\(\mu\)l of DNA.
- Analyse 1\(\mu\)l of DNA on a 1% agrose gel. Compare against 1\(\mu\)l of a high molecular weight ladder such as lambda Hind 11. Run gel for 30 minutes at 90V.
- Stain gel in Ethidium bromide for 20mins, wash in H20 for 1min and visualize in the UV imaging box.
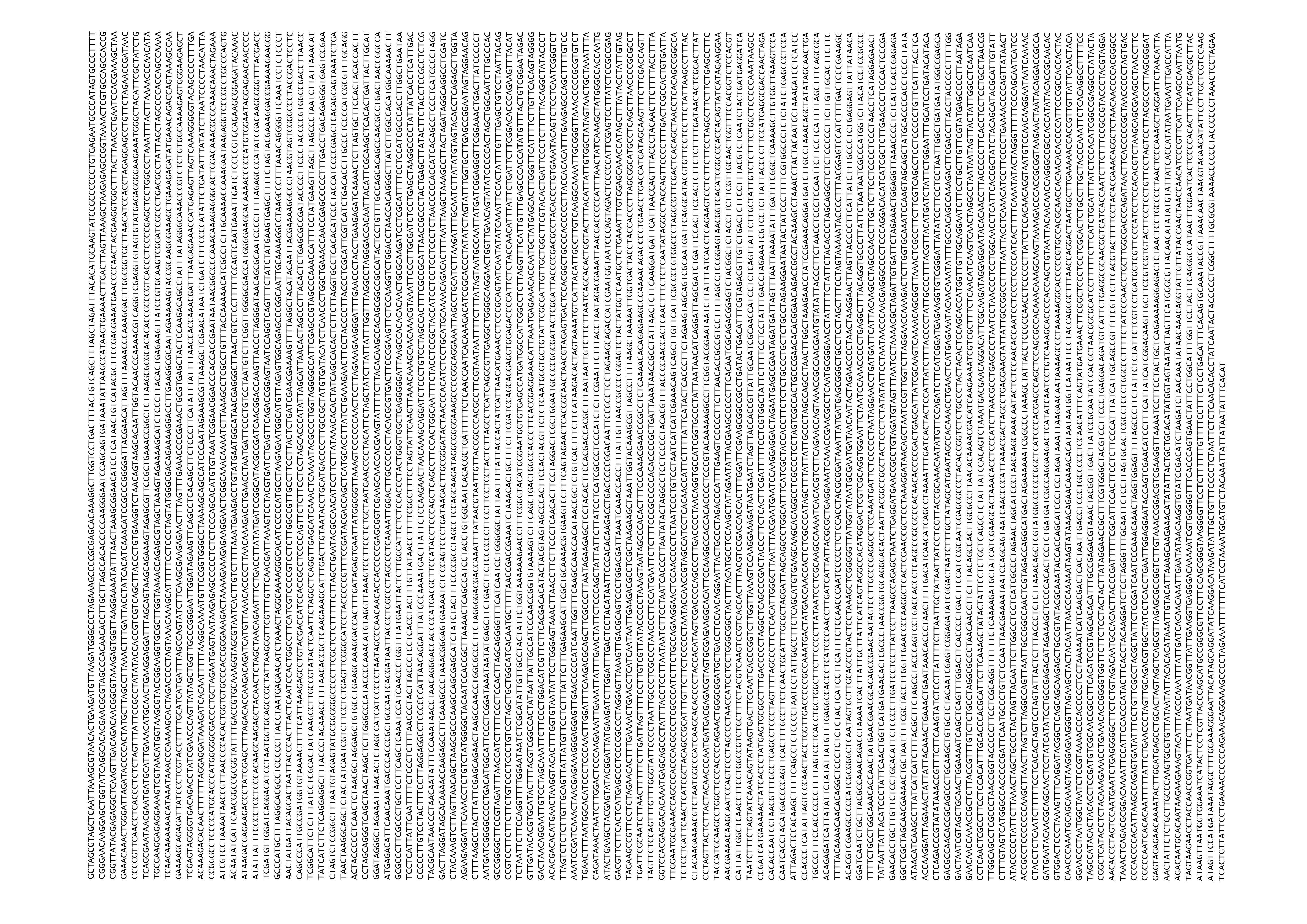
Figure A.1: Final P. georgianus mitochondrial genome sequence.
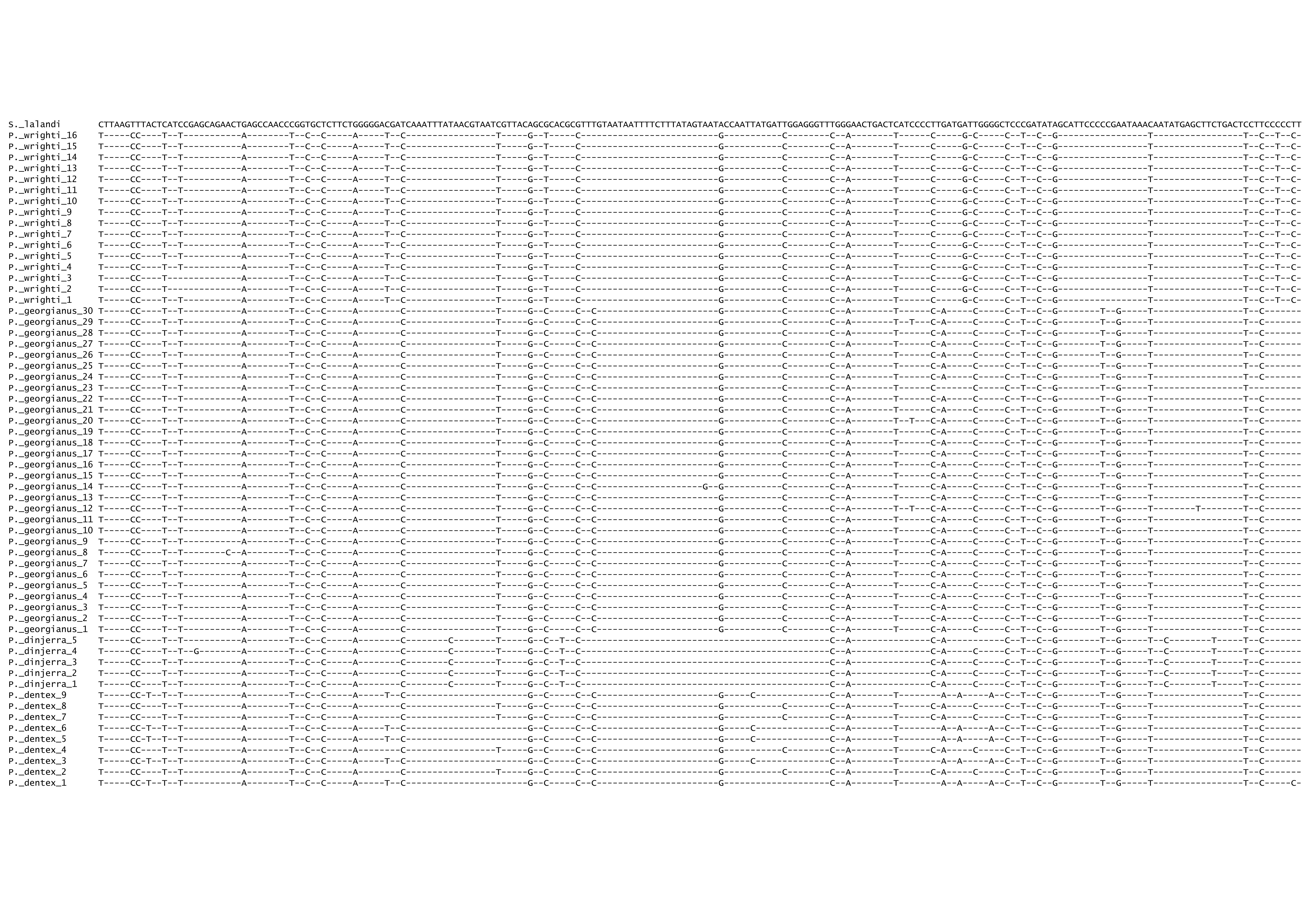
Figure A.2: Final COI alignment.

Figure A.3: Figure continued.

Figure A.4: Figure continued.
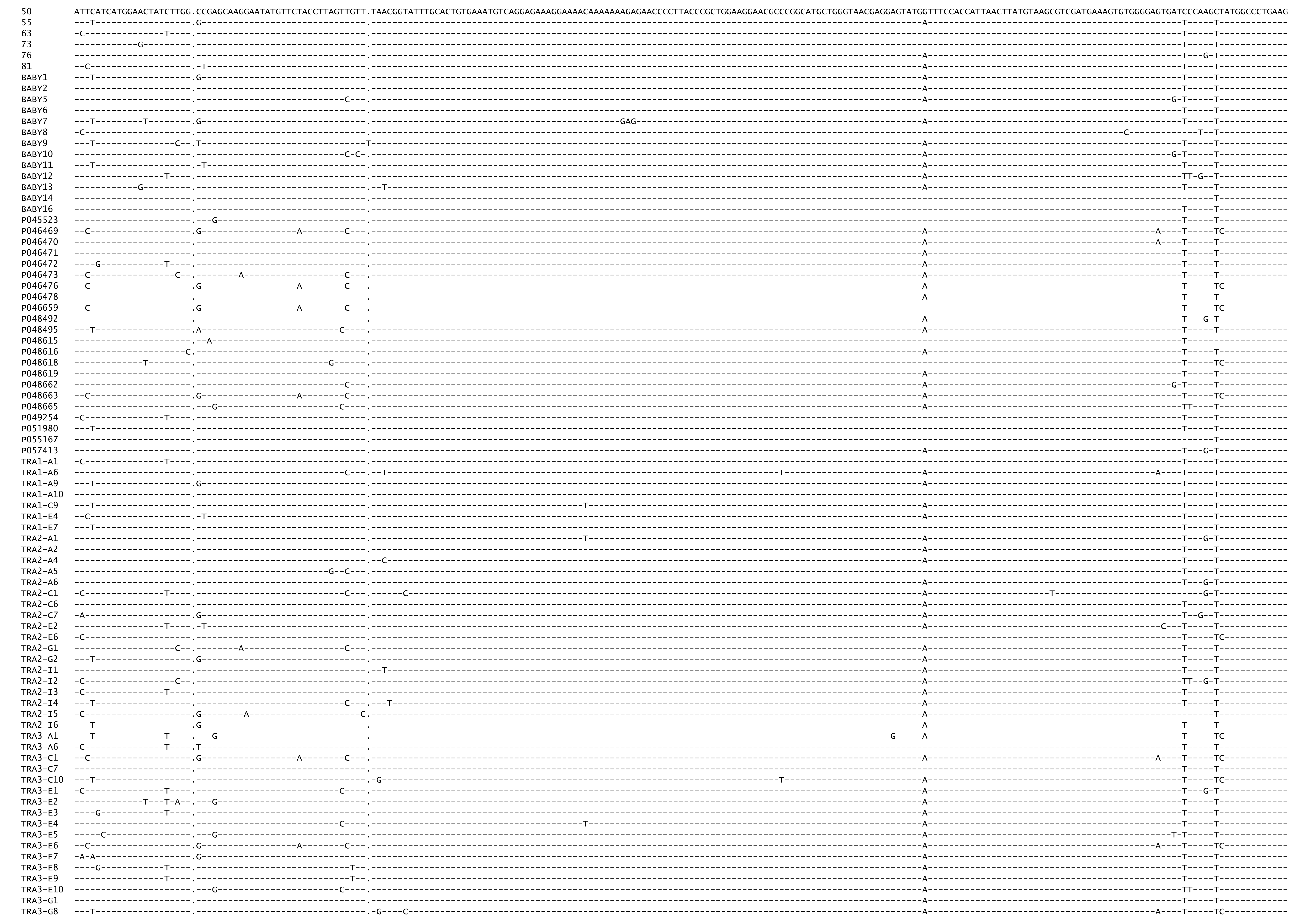
Figure A.5: Final control region alignment (83 of 304 sequences).
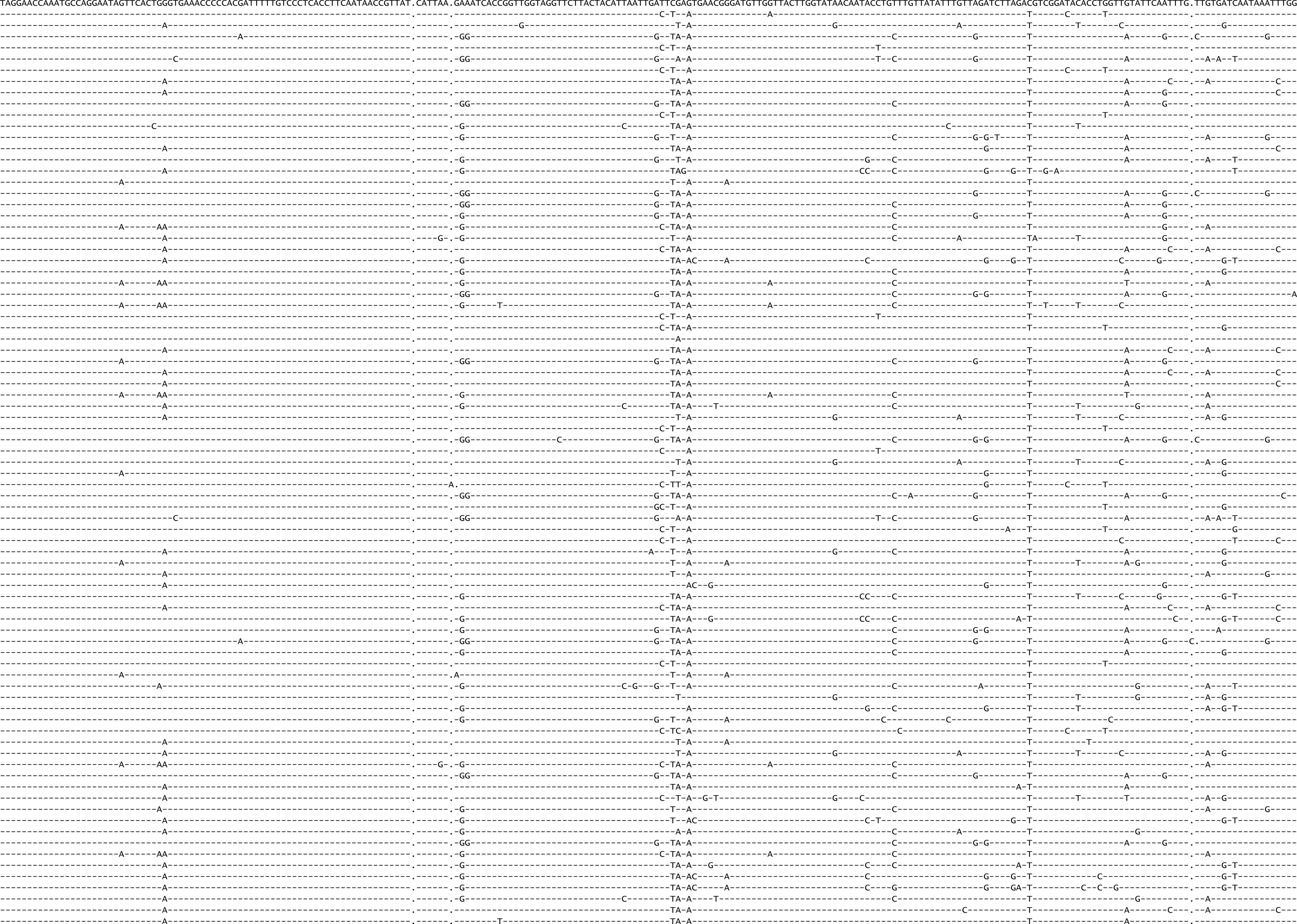
Figure A.6: Figure continued.
Figure A.7: Figure continued.

Figure A.8: Final control region alignment of five Trevally sampled from Three Kings and Kermadec Islands.
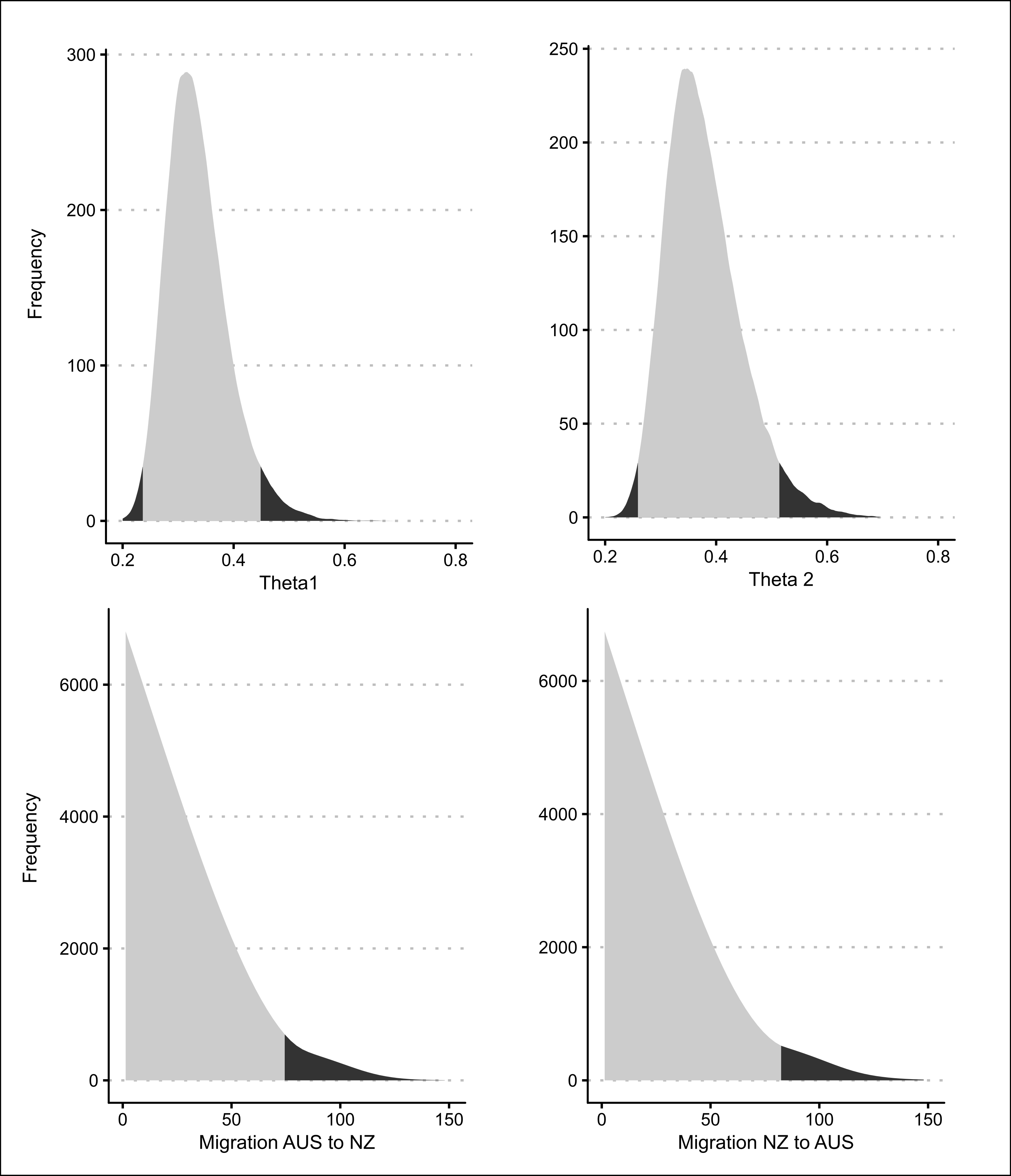
Figure A.9: Posterior probability distributions for all four Migrate model parameters.
Afonso, P., J. Fontes, K. Holland, and R. Santos. 2009. “Multi-Scale Patterns of Habitat Use in a Highly Mobile Reef Fish, the White Trevally Pseudocaranx Dentex, and Their Implications for Marine Reserve Design.” Journal Article. Marine Ecology Progress Series 381: 273–86. https://doi.org/10.3354/meps07946.
Afonso, P., J. Fontes, T. Morato, K. Holland, and R. Santos. 2008. “Reproduction and Spawning Habitat of White Trevally, Pseudocaranx Dentex, in the Azores, Central North Atlantic.” Journal Article. Scientia Marina 72 (2): 373–81.
Allaire, J., C. Gandrud, K. Russell, and C. Yetman. 2017. NetworkD3: D3 JavaScript network graphs from R. https://CRAN.R-project.org/package=networkD3.
Allaire, J., Y. Xie, J. McPherson, J. Luraschi, K. Ushey, A. Atkins, H. Wickham, J. Cheng, W. Chang, and R. Iannone. 2018. Rmarkdown: Dynamic Documents for R. https://CRAN.R-project.org/package=rmarkdown.
Allendorf, F., and N. Ryman. 2002. “The Role of Genetics in Population Viability Analysis.” Book Section. In Population Viability Analysis, edited by S. Beissinger and D. McCullough, 50–85. Chicago: University of Chicago Press. https://doi.org/10.1016/bs.adgen.2017.09.007.
Appelhans, T., F. Detsch, C. Reudenbach, and S. Woellauer. 2019. Mapview: Interactive Viewing of Spatial Data in R. https://CRAN.R-project.org/package=mapview.
Australian Fisheries Management Authority. 2010. “Norfolk Island Inshore Fishery Data Summary 2006-2009.” Report. Australian Fisheries Management Authority.
Australian Government Department of Fisheries. 2019a. “Commonwealth Trawl and Scalefish Hook Sectors.” Web Page.
———. 2019b. “Norfolk Island Fishery.” Web Page.
Avise, J., J. Arnold, R. Ball, E. Bermingham, T. Lamb, J. Neigel, C. Reeb, and N. Saunders. 1987. “Intraspecific Phylogeography - the Mitochondrial Dna Bridge Between Population Genetics and Systematics.” Journal Article. Annual Review of Ecology and Systematics 18: 489–522. https://doi.org/10.1146/annurev.ecolsys.18.1.489.
Ballard, J., and M. Whitlock. 2004. “The Incomplete Natural History of Mitochondria.” Journal Article. Molecular Ecology 13 (4): 729–44. https://doi.org/10.1046/j.1365-294X.2003.02063.x.
Bandelt, H., P. Forster, and A. Röhl. 1999. “Median-Joining Networks for Inferring Intraspecific Phylogenies.” Journal Article. Molecular Biology and Evolution 16 (1): 37–48. https://doi.org/10.1093/oxfordjournals.molbev.a026036.
Bearez, P., and C. Villaroel. 2018. “First Record of Pseudocaranx Chilensis (Carangidae) from the Continental Coast of North-Central Chile.” Journal Article. Cybium 42 (3): 303–5. https://doi.org/10.26028/cybium/2018-423-009.
Bearham, D. 2004. “A mtDNA Study of the Population Structure of Silver Trevally Pseudocaranx Dentex and the Relationship Between Silver and Sand Trevally Pseudocaranx Wrighti in Western Australian Waters.” Thesis, Murdoch University.
Beerli, P. 1998. “Estimation of Migration Rates and Population Sizes in Geographically Structured Populations.” Journal Article. Advances in Molecular Ecology 306: 39–53.
Beerli, P., and J. Felsenstein. 1999. “Maximum-Likelihood Estimation of Migration Rates and Effective Population Numbers in Two Populations Using a Coalescent Approach.” Journal Article. Genetics 152 (2): 763–73. https://doi.org/10.1073/pnas.081068098.
Bernatchez, L., M. Wellenreuther, C. Araneda, D. Ashton, J. Barth, T. Beacham, G. Maes, et al. 2017. “Harnessing the Power of Genomics to Secure the Future of Seafood.” Journal Article. Trends in Ecology and Evolution 32 (9): 665–80. https://doi.org/10.1016/j.tree.2017.06.010.
Bhaska, R., and S. Barret. 2018. Leaflet.extras: Extra Functionality for ’Leaflet’ Package. https://CRAN.R-project.org/package=leaflet.extras.
Biomatters. 2018. “Geneious 11.1.” Report. Geneious.
Brooks, A. 2019. “Using Ecological Niche Modelling to Predict Climate Change Responses of Ten Key Fishery Species in Aotearoa New Zealand.” Thesis, Victoria University of Wellington.
Burzynski, A., M. Zbawicka, D. Skibinski, and R. Wenne. 2003. “Evidence for Recombination of mtDNA in the Marine Mussel Mytilus Trossulus from the Baltic.” Journal Article. Molecular Biology and Evolution 20 (3): 388–92. https://doi.org/10.1093/molbev/msg058.
Campos, P., M. de Castro, and A. Bonecker. 2010. “Occurrence and Distribution of Carangidae Larvae (Teleostei, Perciformes) from the Southwest Atlantic Ocean, Brazil (12-23 Degrees S).” Journal Article. Journal of Applied Ichthyology 26 (6): 920–24. https://doi.org/10.1111/j.1439-0426.2010.01511.x.
Casillas, S., and A. Barbadilla. 2017. “Molecular Population Genetics.” Journal Article. Genetics 205 (3): 1003–35. https://doi.org/10.1534/genetics.116.196493.
Castro, J., J. Santiago, and V. Hernandez-Garcia. 1999. “Fish Associated with Fish Aggregation Devices Off the Canary Islands (Central-East Atlantic).” Journal Article. Scientia Marina 63 (3-4): 191–98. https://doi.org/10.3989/scimar.1999.63n3-4191.
Charlop-Powers, Z. 2017. Phylocanvas: Interactive Phylogenetic Trees Using the ’Phylocanvas’ JavaScript Library. https://CRAN.R-project.org/package=phylocanvas.
Cheng, J., B. Karambelkar, and Y. Xie. 2018. Leaflet: Create Interactive Web Maps with the JavaScript ’Leaflet’ Library. https://CRAN.R-project.org/package=leaflet.
Cryer, M., P. Mace, and K. Sullivan. 2016. “New Zealand’s Ecosystem Approach to Fisheries Management.” Journal Article. Fisheries Oceanography 25: 57–70. https://doi.org/10.1111/fog.12088.
Damerau, M., M. Freese, and R. Hanel. 2018. “Multi-Gene Phylogeny of Jacks and Pompanos (Carangidae), Including Placement of Monotypic Vadigo Campogramma Glaycos.” Journal Article. Journal of Fish Biology 92: 190–202. https://doi.org/10.1111/jfb.13509.
Darling, A., B. Mau, and N. Perna. 2010. “progressiveMauve: Multiple Genome Alignment with Gene Gain, Loss and Rearrangement.” Journal Article. Plos One 5 (6). https://doi.org/10.1371/journal.pone.0011147.
Darriba, D., G. Taboada, R. Doallo, and D. Posada. 2012. “JModelTest 2: More Models, New Heuristics and Parallel Computing.” Journal Article. Nature Methods 9 (8): 772–72. https://doi.org/10.1038/nmeth.2109.
Davila, J., M. Arrieta-Montiel, Y. Wamboldt, J. Cao, J. Hagmann, V. Shedge, Y. Xu, D. Weigel, S. Mackenzie, and Sally A. 2011. “Double-Strand Break Repair Processes Drive Evolution of the Mitochondrial Genome in Arabidopsis.” Journal Article. BMC Biology 9 (64): 1–14. https://doi.org/10.1186/1741-7007-9-64.
Department of Agriculture and Water Resources. 2018. “Fishery Status Reports 2018.” Report. Australian Government.
Dudley, B., N. Tolimieri, and J. Montgomery. 2000. “Swimming Ability of the Larvae of Some Reef Fishes from New Zealand Waters.” Journal Article. Marine and Freshwater Research 51 (8): 783–87. https://doi.org/10.1071/mf00062.
Duffy, C., and S. Ahyong. 2015. “Annotated Checklist of the Marine Flora and Fauna of the Kermadec Islands Marine Reserve and Northern Kermadec Ridge.” Report. Auckland War Memorial Museum.
Dyer, B., and M. Westneat. 2010. “Taxonomy and Biogeography of the Coastal Fishes of Juan Fernandez Archipelago and Desventuradas Islands, Chile.” Journal Article. Revista de Biologia Marina Y Oceanografia 45: 589–617. https://doi.org/10.4067/s0718-19572010000400007.
Excoffier, L., and H. Lischer. 2010. “Arlequin Suite Ver 3.5: A New Series of Programs to Perform Population Genetics Analyses Under Linux and Windows.” Journal Article. Molecular Ecology Resources 10 (3): 564–67. https://doi.org/10.1111/j.1755-0998.2010.02847.x.
Excoffier, L., P. Smouse, and J. Quattro. 1992. “Anaysis of Molecular Variance Inferred from Metric Distances Among DNA Haplotypes - Application to Human Mitochondrial DNA Restriction Data.” Journal Article. Genetics 131 (2): 479–91.
Fairclough, D., I. Potter, E. Lek, A. Bivoltsis, and R. Babcock. 2011. “The Fish Communities and Main Fish Populations of the Jurien Bay Marine Park.” Report. Centre for Fish; Fisheries Research.
Farmer, B., D. French, I. Potter, S. Hesp, and N. Hall. 2005. “Determination of Biological Parameters for Managing the Fisheries for Mulloway and Silver Trevally in Western Australia.” Report. Murdoch University.
Farris, J. 1970. “Methods for Computing Wagner Trees.” Journal Article. Systematic Zoology 19 (1): 83–92. https://doi.org/10.2307/2412028.
Fisheries Science Group. 2018. “Fisheries Assessment Plenary, May 2018: Stock Assessments and Stock Status.” Report. Fisheries New Zealand.
Fowler, A., R. Chick, and J. Stewart. 2018. “Patterns and Drivers of Movement for a Coastal Benthopelagic Fish, Pseudocaranx Georgianus, on Australia’s Southeast Coast.” Journal Article. Scientific Reports 8 (1): 16738–8. https://doi.org/10.1038/s41598-018-34922-6.
Francis, M., M. Morrison, J. Leathwick, C. Walsh, and C. Middleton. 2005. “Predictive Models of Small Fish Presence and Abundance in Northern New Zealand Harbours.” Journal Article. Estuarine Coastal and Shelf Science 64 (2-3): 419–35. https://doi.org/10.1016/j.ecss.2005.03.007.
Froese, R., and D. Pauly. 2019. “FishBase.”
Fu, Y. 1996. “New Statistical Tests of Neutrality for DNA Samples from a Population.” Journal Article. Genetics 143 (1): 557–70.
Gebbies, C. 2014. “Population Genetic Structure of New Zealand Blue Cod (Parapercis Colias) Based on Mitochondrial and Microsatellite DNA Markers.” Thesis, Victoria University of Wellington.
Guindon, S., J. Dufayard, V. Lefort, M. Anisimova, W. Hordijk, and O. Gascuel. 2010. “New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0.” Journal Article. Systematic Biology 59 (3): 307–21. https://doi.org/10.1093/sysbio/syq010.
Guindon, S., and O. Gascuel. 2003. “A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood.” Journal Article. Systematic Biology 52 (5): 696–704. https://doi.org/10.1080/10635150390235520.
Hahn, C., L. Bachmann, and B. Chevreux. 2013. “Reconstructing Mitochondrial Genomes Directly from Genomic Next-Generation Sequencing Reads-a Baiting and Iterative Mapping Approach.” Journal Article. Nucleic Acids Research 41 (13). https://doi.org/10.1093/nar/gkt371.
Hellberg, M. 2009. “Gene Flow and Isolation Among Populations of Marine Animals.” Journal Article. Annual Review of Ecology Evolution and Systematics 40: 291–310. https://doi.org/10.1146/annurev.ecolsys.110308.120223.
Hodgson, B. 2011. “Kahawai Phylogny and Phylogenetics: A Genetic Investigation into Commercial and Recreational Fisheries Management and Practice.” Thesis, Victoria University of Wellington.
Holland, B., K. Huber, V. Moulton, and P. Lockhart. 2004. “Using Consensus Networks to Visualize Contradictory Evidence for Species Phylogeny.” Journal Article. Molecular Biology and Evolution 21 (7). https://doi.org/10.1093/molbev/msh145.
Horn, P. 1986. “Distribution and Growth of Snapper Chrysophrys Auratus in the North Taranaki Bight, and Managment Implications of These Data.” Journal Article. New Zealand Journal of Marine and Freshwater Research 20 (3): 419–30. https://doi.org/10.1080/00288330.1986.9516161.
Hudson, R. 1990. “Gene Genealogies and the Coalescent Process.” Journal Article. Oxford Surveys in Evolutionary Biology, 1–44.
Huelsenbeck, J., and B. Rannala. 2004. “Frequentist Properties of Bayesian Posterior Probabilities of Phylogenetic Trees Under Simple and Complex Substitution Models.” Journal Article. Systematic Biology 53: 904–13. https://doi.org/10.1080/10635150490522629.
Huelsenbeck, J., and F. Ronquist. 2001. “MRBAYES: Bayesian Inference of Phylogenetic Trees.” Journal Article. Bioinformatics 17 (8): 754–55. https://doi.org/10.1093/bioinformatics/17.8.754.
Jaafar, T., M. Taylor, S. Nor, M. de Bruyn, and G. Carvalho. 2012. “DNA Barcoding Reveals Cryptic Diversity Within Commercially Exploited Indo-Malay Carangidae (Teleosteii: Perciformes).” Journal Article. Plos One 7 (11). https://doi.org/10.1371/journal.pone.0049623.
James, G. 1976. “Eggs and Larvae of the Trevally Caranx Georgianus (Teleostei: Carangidae).” Journal Article. New Zealand Journal of Marine and Freshwater Research 10 (2): 301–10. https://doi.org/10.1080/00288330.1976.9515614.
———. 1978. “Trevally and Koheru - Biology and Fisheries.” Journal Article. Fisheries Research Division Occasional Publication (New Zealand) 15: 50–54.
———. 1980. “Tagging Experiments on Trawl-Caught Trevally, Caranx Georgianus, Off North-East New Zealand, 1973-79.” Journal Article. New Zealand Journal of Marine and Freshwater Research 14 (3): 249–54. https://doi.org/10.1080/00288330.1980.9515867.
James, G., and A. Stephenson. 1974. “Caranx Georgianus Cuvier, 1833 (Pisces: Carangidae) in Temperate Australasian Waters.” Journal Article. Journal of the Royal Society of New Zealand 4 (4): 401–10. https://doi.org/10.1080/03036758.1974.10419384.
Kearse, M., R. Moir, A. Wilson, S. Stones-Havas, M. Cheung, S. Sturrock, S. Buxton, et al. 2012. “Geneious Basic: An Integrated and Extendable Desktop Software Platform for the Organization and Analysis of Sequence Data.” Journal Article. Bioinformatics 28 (12): 1647–9. https://doi.org/10.1093/bioinformatics/bts199.
Kingman, J. 1982a. “On the Genealogy of Large Populations.” Journal Article. Journal of Applied Probability 19: 22–43.
———. 1982b. “The Coalescent.” Journal Article. Stochastic Processes and Their Applications 14: 235–48.
———. 2000. “Origins of the Coalescent: 1974-1982.” Journal Article. Genetics 156 (4): 1461–3.
Kress, W., C. Garcia-Robledo, M. Uriarte, and D. Erickson. 2015. “DNA Barcodes for Ecology, Evolution, and Conservation.” Journal Article. Trends in Ecology and Evolution 30 (1): 25–35. https://doi.org/10.1016/j.tree.2014.10.008.
Kruskal, J. 1956. “On the Shortest Spanning Subtree of a Graph and the Traveling Salesman Problem.” Journal Article. Proceedings of the American Mathematical Society 7: 48–50. https://doi.org/10.2307/2033241.
Laikre, L., S. Palm, and N. Ryman. 2005. “Genetic Population Structure of Fishes: Implications for Coastal Zone Management.” Journal Article. Ambio 34 (2): 111–19. https://doi.org/10.1639/0044-7447(2005)034[0111:Gpsofi]2.0.Co;2.
Langley, A. 2004. “Length and Age Composition of Trevally (Pseudocaranx Dentex) in Commercial Landings from TRE1 Purse-Seine Fishery, 2002-03.” Journal Article. New Zealand Fisheries Assessment Research Document Series.
Langley, A., T. Kendrick, and N. Bentley. 2015. “Stock Assessment of Trevally in TRE7.” Report. New Zealand Fisheries Assessment Report.
Leducq, J., M. Henault, G. Charron, L. Nielly-Thibault, Y. Terrat, H. Fiumera, B. Shapiro, and C. Landry. 2017. “Mitochondrial Recombination and Introgression During Speciation by Hybridization.” Journal Article. Molecular Biology and Evolution 34 (8): 1947–59. https://doi.org/10.1093/molbev/msx139.
Leigh, J., and D. Bryant. 2015. “POPART: full-feature software for haplotype network construction.” Journal Article. Methods in Ecology and Evolution 6 (9): 1110–6. https://doi.org/10.1111/2041-210x.12410.
Masuda, R., T. Kamaishi, T. Kobayashi, K. Tsukamoto, and K. Numachi. 1995. “Mitochondrial DNA Differentiation Between Two Sympatric Morphs of Striped Jack Near Japan.” Journal Article. Journal of Fish Biology 46 (6): 1003–10. https://doi.org/10.1111/j.1095-8649.1995.tb01405.x.
Meirmans, S., P. Meirmans, and L. Kirkendall. 2012. “The Costs of Sex: Facing Real-World Complexities.” Journal Article. Quarterly Review of Biology 87 (1): 19–40. https://doi.org/10.1086/663945.
Miya, M., Y. Sato, T. Fukunaga, T. Sado, J. Y. Poulsen, K. Sato, T. Minamoto, et al. 2015. “MiFish, a Set of Universal PCR Primers for Metabarcoding Environmental DNA from Fishes: Detection of More Than 230 Subtropical Marine Species.” Journal Article. Royal Society Open Science 2 (7). https://doi.org/10.1098/rsos.150088.
Nath, H., and R. Griffiths. 1993. “The Coalescent in 2 Colonies with Symmetrical Migration.” Journal Article. Journal of Mathematical Biology 31 (8): 841–51. https://doi.org/10.1007/bf00168049.
National Institute of Water and Atmospheric Research. 2019. “Feature Layers by MPI Geospatial Management.” Web Page.
Neira, F., R. Perry, C. Burridge, J. Lyle, and J. Keane. 2015. “Molecular Discrimination of Shelf-Spawned Eggs of Two Co-Occurring Trachurus Spp. (Carangidae) in Southeastern Australia: A Key Step to Future Egg-Based Biomass Estimates.” Journal Article. Ices Journal of Marine Science 72 (2): 614–24. https://doi.org/10.1093/icesjms/fsu151.
Nogueira, N., M. Ferreira, N. Cordeiro, and P. Canada. 1990. “The Coalescent and the Genealogical Proccess in Geographically Structured Population.” Journal Article. Journal of Mathematical Biology 29 (1): 59–75. https://doi.org/10.1007/bf00173909.
Ojala, D., J. Montoya, and G. Attardi. 1981. “Transfer-RNA Punctuation Model of RNA Processing in Human Mitochondria.” Journal Article. Nature 290 (5806): 470–74. https://doi.org/10.1038/290470a0.
Paradis, E. 2018. “Analysis of Haplotype Networks: The Randomized Minimum Spanning Tree Method.” Journal Article. Methods in Ecology and Evolution 9 (5): 1308–17. https://doi.org/10.1111/2041-210x.12969.
Pebesma, E. 2018. Sf: Simple Features for R. https://CRAN.R-project.org/package=sf.
Pebesma, E., and R. Bivand. 2018. Sp: Classes and Methods for Spatial Data. https://CRAN.R-project.org/package=sp.
Philippe, H., H. Brinkmann, D. Lavrov, D. Timothy J. Littlewood, M. Manuel, G. Woerheide, and D. Baurain. 2011. “Resolving Difficult Phylogenetic Questions: Why More Sequences Are Not Enough.” Journal Article. Plos Biology 9 (3). https://doi.org/10.1371/journal.pbio.1000602.
Philippe, H., and B. Roure. 2011. “Difficult Phylogenetic Questions: More Data, Maybe; Better Methods, Certainly.” Journal Article. Bmc Biology 9. https://doi.org/10.1186/1741-7007-9-91.
Pyron, A. 2015. “Post-Molecular Systematics and the Future of Phylogenetics.” Journal Article. Trends in Ecology and Evolution 30 (7): 384–89. https://doi.org/10.1016/j.tree.2015.04.016.
Ray, N., M. Currat, and L. Excoffier. 2003. “Intra-Deme Molecular Diversity in Spatially Expanding Populations.” Journal Article. Molecular Biology and Evolution 20 (1): 76–86. https://doi.org/10.1093/molbev/msg009.
Rogers, A., A. Fraley, M. Bamshad, S. Watkins, and L. Jorde. 1996. “Mitochondrial Mismatch Analysis Is Insensitive to the Mutational Process.” Journal Article. Molecular Biology and Evolution 13 (7): 895–902. https://doi.org/10.1093/molbev/13.7.895.
Rogers, A., and H. Harpending. 1992. “Population Growth Makes Waves in the Distribution of Pairwise Genetic Differences.” Journal Article. Molecular Biology and Evolution 9 (3): 552–69. https://doi.org/10.1093/oxfordjournals.molbev.a040727.
Rosenberg, N., and M. Nordborg. 2002. “Genealogical Trees, Coalescent Theory and the Analysis of Genetic Polymorphisms.” Journal Article. Nature Reviews Genetics 3 (5): 380–90. https://doi.org/10.1038/nrg795.
Rowling, K., and L. Raines. 2000. “Description of the Biology and an Assessment of the Fishery for Silver Trevally Pseudocaranx Dentex Off New South Wales.” Report. NSW Fisheries.
Rozas, J., A. Ferrer-Mata, J. Sanchez-DelBarrio, S. Guirao-Rico, P. Librado, S. Ramos-Onsins, and A. Sanchez-Gracia. 2017. “DnaSP 6: DNA Sequence Polymorphism Analysis of Large Data Sets.” Journal Article. Molecular Biology and Evolution 34 (12): 3299–3302. https://doi.org/10.1093/molbev/msx248.
Santini, F., and G. Carnevale. 2015. “First Multilocus and Densely Sampled Timetree of Trevallies, Pompanos and Allies (Carangoidei, Percomorpha) Suggests a Cretaceous Origin and Eocene Radiation of a Major Clade of Piscivores.” Journal Article. Molecular Phylogenetics and Evolution 83: 33–39. https://doi.org/10.1016/j.ympev.2014.10.018.
Satoh, T., M. Miya, K. Mabuchi, and M. Nishida. 2016. “Structure and Variation of the Mitochondrial Genome of Fishes.” Journal Article. BMC Genomics 17. https://doi.org/10.1186/s12864-016-3054-y.
Scott, S. 2010. “Phylogeography of the Common New Zealand Wrasse Species, Notolabrus Celidotus, and the Phylogenetics of the Pseudolabrine Tribe).” Thesis, Victoria University of Wellington.
Shen, X., C. Hittinger, and A. Rokas. 2017. “Contentious Relationships in Phylogenomic Studies Can Be Driven by a Handful of Genes.” Journal Article. Nature Ecology and Evolution 1 (5). https://doi.org/10.1038/s41559-017-0126.
Sievert, C., C. Parmer, S. Hocking T. Chamberlain, K. Ram, M. Corvellec, and P. Despouy. 2018. Plotly: Create Interactive Web Graphics via ’Plotly.js’. https://CRAN.R-project.org/package=plotly.
Slatkin, M. 1987. “Gene Flow and the Geographic Structure of Natural Populations.” Journal Article. Science 236 (4803): 787–92. https://doi.org/10.1126/science.3576198.
Smith, H. 2012. “Characterisation of the Mitochondrial Genome and the Phylogeographic Structure of Blue Cod (Parapercis Colias).” Thesis, Victoria University of Wellington.
Smith-Vaniz, W., and H. Jelks. 2006. “Australian Trevallies of the Genus Pseudocaranx (Teleostei: Carangidae), with Description of a New Species from Western Australia.” Journal Article. Memoirs of Museum Victoria 63 (1): 97–106. https://doi.org/10.24199/j.mmv.2006.63.12.
Springer, M., and J. Gatesy. 2018. “On the Importance of Homology in the Age of Phylogenomics.” Journal Article. Systematics and Biodiversity 16 (3): 210–28. https://doi.org/10.1080/14772000.2017.1401016.
Tajima, F. 1989. “Statistical Method for Testing the Neutral Mutation Hypothesis by DNA Polymorphism.” Journal Article. Genetics 123 (3): 585–95.
Teske, P., T. Golla, J. Sandoval-Castillo, A. Emami-Khoyi, C. van der Lingen, S. von der Heyden, B. Chiazzari, B. van Vuuren, and L. Beheregaray. 2018. “Mitochondrial DNA Is Unsuitable to Test for Isolation by Distance.” Journal Article. Scientific Reports 8. https://doi.org/10.1038/s41598-018-25138-9.
Untergasser, A., I. Cutcutache, T. Koressaar, J. Ye, B. Faircloth, M. Remm, and S. Rozen. 2012. “Primer3-New Capabilities and Interfaces.” Journal Article. Nucleic Acids Research 40 (15). https://doi.org/10.1093/nar/gks596.
Waples, R., A. Punt, and J. Cope. 2008. “Integrating Genetic Data into Management of Marine Resources: How Can We Do It Better?” Journal Article. Fish and Fisheries 9 (4): 423–49. https://doi.org/10.1111/j.1467-2979.2008.00303.x.
Ward, R., R. Hanner, and P. Hebert. 2009. “The Campaign to DNA Barcode All Fishes.” Journal Article. Journal of Fish Biology 74 (2): 329–56. https://doi.org/10.1111/j.1095-8649.2008.02080.x.
Wickham, H., W. Chang, L. Henry, T. Pedersen, K. Takahashi, C. Wilke, and K. Woo. 2018. Ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. https://CRAN.R-project.org/package=ggplot2.
Wickham, H., R. François, L. Henry, and K. Müller. 2019. Dplyr: A Grammar of Data Manipulation. https://CRAN.R-project.org/package=dplyr.
Wilcox, R. 2015. “A Population Genetic Analysis of the New Zealand Spotty (Notolabrus Celidotus) Using Mitochondrial DNA and Microsatellite DNA Markers.” Thesis, Victoria University of Wellington.
Wright, S. 1943. “Isolation by Distance.” Journal Article. Genetics 28 (2): 114–38.
Xie, Y. 2018a. Bookdown: Authoring Books and Technical Documents with R Markdown. https://CRAN.R-project.org/package=bookdown.
———. 2018b. Knitr: A General-Purpose Package for Dynamic Report Generation in R. https://CRAN.R-project.org/package=knitr.
———. 2019. Tinytex: Helper Functions to Install and Maintain ’Tex Live’, and Compile ’Latex’ Documents. https://CRAN.R-project.org/package=tinytex.
Yamaoka, K., H. Han, and N. Taniguchi. 1992. “Genetic Dimorphism in Pseudocaranx Dentex from Tosa Bay, Japan.” Journal Article. Bulletin of the Japanese Society of Scientific Fisheries 58 (1): 39–44. https://doi.org/10.2331/suisan.58.39.
Yang, Z., and B. Rannala. 2012. “Molecular Phylogenetics: Principles and Practice.” Journal Article. Nature Reviews Genetics 13 (5): 303–14. https://doi.org/10.1038/nrg3186.
Ye, F., D. Samuels, T. Clark, and Y. Guo. 2014. “High-Throughput Sequencing in Mitochondrial DNA Research.” Journal Article. Mitochondrion 17: 157–63. https://doi.org/10.1016/j.mito.2014.05.004.
Yu, Y., Y. Ouyang, and W. Yao. 2018. “ShinyCircos: An R/Shiny Application for Interactive Creation of Circos Plot.” Journal Article. Bioinformatics 34 (7): 1229–31. https://doi.org/10.1093/bioinformatics/btx763.
Zhu, H. 2019. KableExtra: Construct Complex Table with ’Kable’ and Pipe Syntax. https://CRAN.R-project.org/package=kableExtra.
Zou, K., and M. Li. 2016. “Characterization of the Mitochondrial Genome of the Whitefin Trevally Carangoides Equula (Perciformes: Carangidae): A Novel Initiation Codon for ATP6 Gene.” Journal Article. Mitochondrial DNA Part A 27 (3): 1779–80. https://doi.org/10.3109/19401736.2014.963809.