Chapter 4 Logistic regression models
Machine learning assumptions: (i) a dataset is available with observations for a number of explanatory X-variables and the corresponding value of a response Y-variable and (ii) there is a function f that describes the relationship between the X variables and the Y variable: Y = f(X1, X2,…, X3) + \(\epsilon\).
Machine Learning objective: to find the function f.
If the Y variable is categorical, we have a classification problem. The aim of constructing a model is to predict, given X values of new observations, the corresponding Y value. In other words to predict to which Y category the observation belongs. Models are usually constructed in such a way that the f-value per category gives a probability that the observation belongs to that category. The user of the model can then classify the observations himself based on these probabilities. This does not necessarily mean that the category with the highest probability is chosen. For example, if a doctor decides on the basis of observations that a patient has a 30% probability of having a certain disease, he may decide to prescribe drugs to combat this disease, although there is a 70% probability that this is unnecessary.
4.1 Binary Y-variable
If the Y-variable is dichotomous, i.e. has two categories, these are often labeled POS and NEG or 1 and 0 and sometimes -1 and +1. The latter is especially the case when a perceptron model is used to distinguish between the two Y-categories.
4.1.1 Linear regression with a binary Y-variable
If the Y-categories are labeled with the number 0 and 1, theoretical a regression model can be used, after all the Y-variable is numeric.
However, this method has some disadvantages:
- assumptions of a linear regression model are violated
- interpretation of predicted values; in a linear regression model, the model Y-value can be interpreted as the average Y-value, givena certain X-value; because the Y-variable is binary, an average Y-value given an X-value should be a value in the interval [0, 1] and can be interpreted as a probability; predicted values based on a linear regression model however, can be far outside this interval.
A model that can be used in case of a dichotomous Y variable, models the probabilities of Y=1 as a function of X: P (Y=1 | X) = f(X), where f is a function that only takes on values in the interval [0, 1]. An example of such a model is the widely used logistic regression model.
4.1.2 Logistic Regression Model
The simple logistic regression model:
P(Y=1 | X) = \(\frac{e^{\beta_0+\beta_1X}}{1+e^{\beta_0+\beta_1X}}\) or in short: P(X) = \(\frac{e^{\beta_0+\beta_1X}}{1+e^{\beta_0+\beta_1X}}\)
with e Euler’s number (2.7182…), known from mathematics.
The formula can be rewritten into: \(log(\frac{P(X)}{1-P(X)}) = \beta_0 + \beta_1X\)
The expression between the brackets has the form \(\frac{probability}{1-probability}\) which is known as an odds ratio.2 A logistic regression model is also known as a log-odds-model or logit-model.
If this model is applied, the parameters of the model have to be estimated based on the observations in the training data. For this the so called maximum likelihood criterion is used.
Maximum Likelihood Criterion
Based on n observations for the X-variables, a logit model estimates the probabilities Y belongs to class 1, i.e. P(Y=1 | X).
Assume we have 5 observations, (X1, 1), (X2, 1), (X3, 1), (X4, 0), (X5, 0).
Model probabilities are p1, p2, p3, p4, p5.
For a model to be a good model, p1, p2 and p3 should be close to 1 and p4 and p5 close to 0.
The maximum likelihood criterion for choosing the best model is:
choose the model which maximizes p1 x p2 x p3 x (1-p4) x (1-p5).
This criterion can be easily generalized.
4.1.2.1 Example Logistic Regression Model
The example described in this section makes use of the Breast Cancer Wisconson Data Set, which can be found here.
library(tidyverse)
library(caret)
library(GGally) #vanwege ggpairs() plot
library(flextable)
df_read <- read_csv("Data/wbcd.csv")
df_read$id <- as.character(df_read$id)Table
First Six Rows and Six Columns of the WBCD Data Set
id | diagnosis | radius_mean | taxture_mean | perimeter_mean | area_mean |
842302 | M | 18 | 10 | 123 | 1,001 |
842517 | M | 21 | 18 | 133 | 1,326 |
84300903 | M | 20 | 21 | 130 | 1,203 |
84348301 | M | 11 | 20 | 78 | 386 |
84358402 | M | 20 | 14 | 135 | 1,297 |
843786 | M | 12 | 16 | 83 | 477 |
The first column is a unique identifier, the second column - diagnosis - is the target variable. Thw data set contains 30 X-variables. In this example only the first six X-variables are taken into account.
Figure
Summary of the WBCD Data Set
diagnosis radius_mean taxture_mean perimeter_mean
Length:569 Min. : 6.981 Min. : 9.71 Min. : 43.79
Class :character 1st Qu.:11.700 1st Qu.:16.17 1st Qu.: 75.17
Mode :character Median :13.370 Median :18.84 Median : 86.24
Mean :14.127 Mean :19.29 Mean : 91.97
3rd Qu.:15.780 3rd Qu.:21.80 3rd Qu.:104.10
Max. :28.110 Max. :39.28 Max. :188.50
area_mean smoothness_mean compactness_mean
Min. : 143.5 Min. :0.05263 Min. :0.01938
1st Qu.: 420.3 1st Qu.:0.08637 1st Qu.:0.06492
Median : 551.1 Median :0.09587 Median :0.09263
Mean : 654.9 Mean :0.09636 Mean :0.10434
3rd Qu.: 782.7 3rd Qu.:0.10530 3rd Qu.:0.13040
Max. :2501.0 Max. :0.16340 Max. :0.34540 Note. Only 6 of the 30 available X-variables are used in the example and summarized in this overview.
The diagnosis-variable is transformed into a factor variable. The GGally::ggpairs() function is used to examine the correlation between the different variables.
Figure
Correlation Plot Matrix
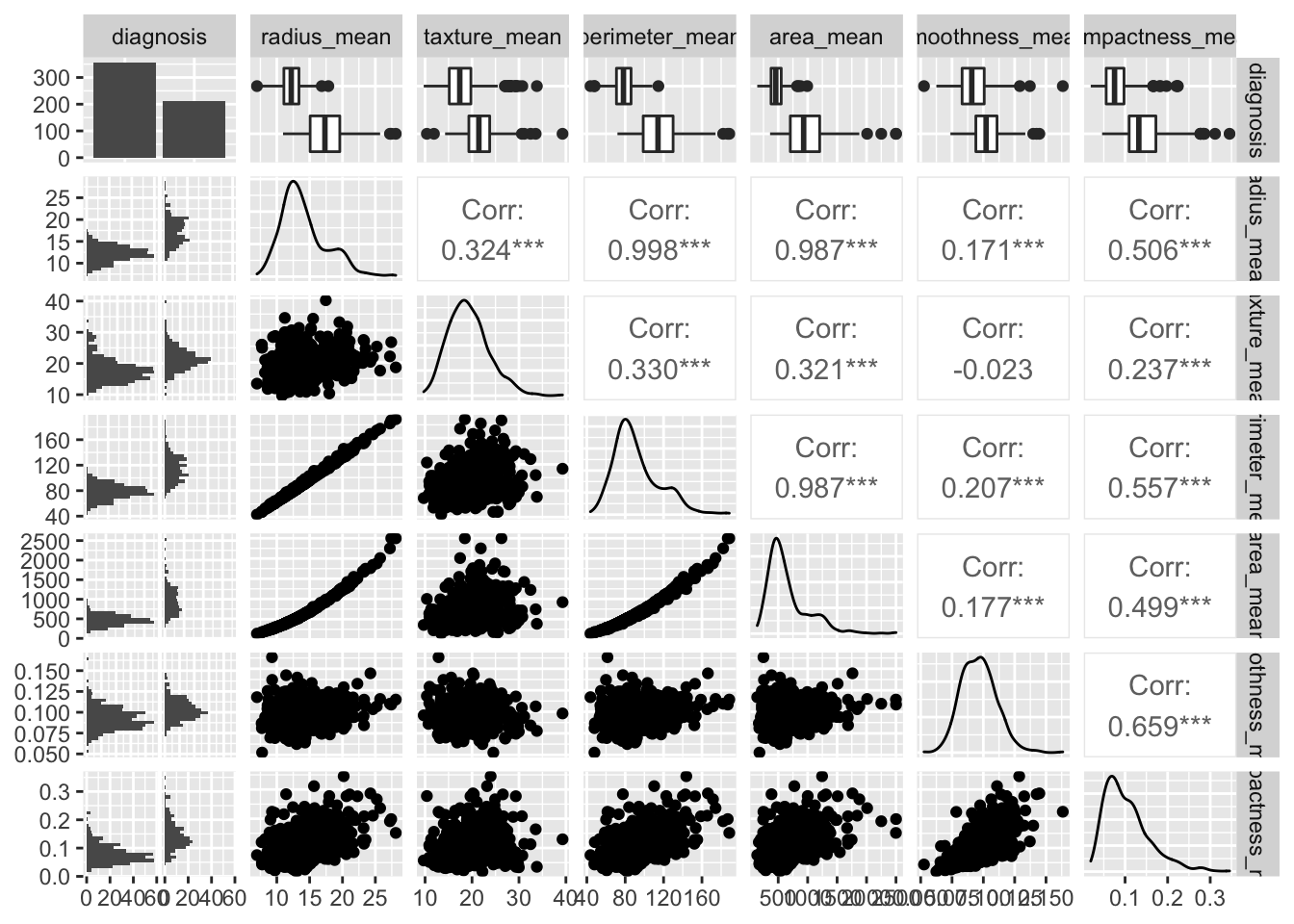 The area_mean variable seems to make a good candidate to distinguish diagnosis M from diagnosis B.
The area_mean variable seems to make a good candidate to distinguish diagnosis M from diagnosis B.
The first logistic regression model usus this variable as the only predictor.
logreg_model1 <- glm(diagnosis~area_mean, data = df,
family = "binomial")
preds_logreg1 <- predict(logreg_model1, type = 'response')
threshold <- .5
preds_logreg1_diag <- ifelse(preds_logreg1>.5, "POS", "NEG") %>%
factor()
confusionMatrix(preds_logreg1_diag, df$diagnosis,
positive = "POS")Confusion Matrix and Statistics
Reference
Prediction NEG POS
NEG 337 46
POS 20 166
Accuracy : 0.884
95% CI : (0.8548, 0.9091)
No Information Rate : 0.6274
P-Value [Acc > NIR] : < 2.2e-16
Kappa : 0.7456
Mcnemar's Test P-Value : 0.002089
Sensitivity : 0.7830
Specificity : 0.9440
Pos Pred Value : 0.8925
Neg Pred Value : 0.8799
Prevalence : 0.3726
Detection Rate : 0.2917
Detection Prevalence : 0.3269
Balanced Accuracy : 0.8635
'Positive' Class : POS
## ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.0 ──## ✓ ggplot2 3.3.3 ✓ purrr 0.3.4
## ✓ tibble 3.0.4 ✓ dplyr 1.0.2
## ✓ tidyr 1.1.2 ✓ stringr 1.4.0
## ✓ readr 1.4.0 ✓ forcats 0.5.0## ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
## x dplyr::filter() masks stats::filter()
## x dplyr::lag() masks stats::lag()## Loading required package: lattice##
## Attaching package: 'caret'## The following object is masked from 'package:purrr':
##
## lift##
## Attaching package: 'flextable'## The following object is masked from 'package:purrr':
##
## composeif the probability of a particular event equals 1/6, then the odds ratio equals 1/5 or 1 against 5; the probability that the event will not occur is 5 times higher than that it will occur. If for a particular event the odd ratio equals 4 (4 against 1), then the probability the event happens is 4/5 (80%).↩︎