Chapter 6 KNN Algorithm
The KNN, K Nearest Neighbours, algorithm is an algorithm that can be used for both unsupervised and supervised learning. It can be used in a regression and in a classification context.
6.1 KNN in Classification
The idea behind the KNN algorithm is simple. Suppose a binary classification problem, i.e. the dataset provides a couple of features and a binary target Y-variable. The dataset contains hostorical data.
To predict the value of the target variable for a new observation, based on the values of the features, the distance of the new observation to the observations in the data set is calculated. The K observatioons wtih the lowest distance are filtered. The majority Y-value for these K observations is used as the predicted Y-value for the new observation.
Before this algorithm can be used two important choices must be made: (1) which value of K to use and (2) which distance metric to use.
Besides the choice of a distance metric two important preprocessing steps have to be performed, (1) categorical variables must be transformed into dummy variables and (2) numeric variables must be standardized/ normalized. The first step is necessary, because calculating a distance requires numerical values. The second step is performed, because otherwise the units chosen largely determine the distances.
Different techniques can be used for normalizing the values of a numeric variable. Standardizing the values, i.e. transforming the values in the z-score is an option, z-score = \(\frac{x-mean}{SD}\). Another option is standardizing the values using the formula below, xnorm = \(\frac{max-x}{max-min}\).
As in all classification problems, a choice about the metric to use to assess the model must be made.
6.1.1 Choosing appropriate K value
There is no general rule for the choice of the best K-value. It is most common to divide the data in a training an a test set, test a number of choices for K and check which choice leads to the best result.
6.1.2 Distance metrics
Commonly used distance metrics are (1) Eucledian distance and (2) Manhattan distance. Be aware that there are lots of other distance metrics as well.
Example 1
Predicting diabetes
Data set: example from UCI website
The data set consists of 520 observations on 17 features. The target variable is the Class variable which can take on two values, Positive and Negative. All but one of the features are binary. The non binary feature is the Age feature.
Table 1
First Six Observations of the Diabetes Data Set
Age | Gender | Polyuria | Polydipsia | sudden weight loss | weakness | Polyphagia | Genital thrush | visual blurring | Itching | Irritability | delayed healing | partial paresis | muscle stiffness | Alopecia | Obesity | class |
40 | Male | No | Yes | No | Yes | No | No | No | Yes | No | Yes | No | Yes | Yes | Yes | Positive |
58 | Male | No | No | No | Yes | No | No | Yes | No | No | No | Yes | No | Yes | No | Positive |
41 | Male | Yes | No | No | Yes | Yes | No | No | Yes | No | Yes | No | Yes | Yes | No | Positive |
45 | Male | No | No | Yes | Yes | Yes | Yes | No | Yes | No | Yes | No | No | No | No | Positive |
60 | Male | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Positive |
55 | Male | Yes | Yes | No | Yes | Yes | No | Yes | Yes | No | Yes | No | Yes | Yes | Yes | Positive |
Table 2
Summary of the data
summary(df)
## Age Gender Polyuria Polydipsia
## Min. :16.00 Length:520 Length:520 Length:520
## 1st Qu.:39.00 Class :character Class :character Class :character
## Median :47.50 Mode :character Mode :character Mode :character
## Mean :48.03
## 3rd Qu.:57.00
## Max. :90.00
## sudden weight loss weakness Polyphagia Genital thrush
## Length:520 Length:520 Length:520 Length:520
## Class :character Class :character Class :character Class :character
## Mode :character Mode :character Mode :character Mode :character
##
##
##
## visual blurring Itching Irritability delayed healing
## Length:520 Length:520 Length:520 Length:520
## Class :character Class :character Class :character Class :character
## Mode :character Mode :character Mode :character Mode :character
##
##
##
## partial paresis muscle stiffness Alopecia Obesity
## Length:520 Length:520 Length:520 Length:520
## Class :character Class :character Class :character Class :character
## Mode :character Mode :character Mode :character Mode :character
##
##
##
## class
## Length:520
## Class :character
## Mode :character
##
##
## Data cleaning and Preprocessing
Correct variable names (no spaces)
Preprocessing: normalize Age variable
Transform other variables into dummies
Transform class variable into a factor variable, this is a requirement for the caret::knn3() function
Split data set in training and test set; 70% in training, 30% in test set
names(df) <- make.names(names(df)) %>%
toupper() %>% str_replace_all("[\\.]", "_")
df_prep <- df %>%
mutate(AGE_NORM = (AGE - min(AGE))/(max(AGE)-min(AGE))) %>%
select(AGE_NORM, everything(), -AGE) %>%
mutate(GENDER = ifelse(GENDER=="Male", 1, 0))
#variables in column 3-16 are all variables which only take on "Yes" or "No" values; these can be transformed into 0/1 variable with one line of code
df_prep[,3:16] <- apply(df_prep[,3:16], 2, function(x) ifelse(x=="Yes", 1, 0))
#transform CLASS variable into type factor variable
df_prep$CLASS <- factor(df_prep$CLASS, levels = c("Positive", "Negative"),
labels = c("POS", "NEG"))
#split in training end test set
set.seed(20210309)
train <- sample(1:nrow(df), size = .7*nrow(df))
df_train <- df_prep[train,]
df_test <- df_prep[-train,]Generate a knn-model. Choice to make beforehand: value of k; first attempt k = 5
#generate model using class::knn3() function
#forst attempt with k=5 as an arbitrary choice
knn_model_1 <- knn3(CLASS~., data = df_train, k = 5)
knn_model_15-nearest neighbor model
Training set outcome distribution:
POS NEG
234 130 Assess model
Choose metric to assess model: Accuracy
Use model to make predictions for test data
Calculate Accuracy
#make predcitions with the generated model
knn_model_1_preds <- predict(knn_model_1, newdata = df_test, type = 'class')
#construct a confusion matrix
cf_1 <- table(knn_model_1_preds, df_test$CLASS)
#calculate Accuracy
acc_1 <- (cf_1[1,1] + cf_1[2,2]) / sum(cf_1)Table 3
Confusion Matrix knn_1 model on Test Data Set
knn_model_1_preds POS NEG
POS 77 5
NEG 9 65
Accuracy = (77 + 65) / (77 + 5 +9 + 65) = 0.91.
confusionMatrix(knn_model_1_preds, df_test$CLASS)
#knn_model_1 <- train(class~., data = df_prep, method = ‘knn’) #knn_model_1
Example 1
Predicting credibility bank customers
Data set: example from UCI website
The data set contains the following 23 variables as explanatory variables:
- X1: Amount of the given credit (NT dollar): it includes both the individual consumer credit and his/her family (supplementary) credit.
- X2: Gender (1 = male; 2 = female).
- X3: Education (1 = graduate school; 2 = university; 3 = high school; 4 = others).
- X4: Marital status (1 = married; 2 = single; 3 = others).
- X5: Age (year).
- X6 - X11: History of past payment. We tracked the past monthly payment records (from April to September, 2005) as follows: X6 = the repayment status in September, 2005; X7 = the repayment status in August, 2005; . . .;X11 = the repayment status in April, 2005. The measurement scale for the repayment status is: -1 = pay duly; 1 = payment delay for one month; 2 = payment delay for two months; . . .; 8 = payment delay for eight months; 9 = payment delay for nine months and above.
- X12-X17: Amount of bill statement (NT dollar). X12 = amount of bill statement in September, 2005; X13 = amount of bill statement in August, 2005; . . .; X17 = amount of bill statement in April, 2005.
- X18-X23: Amount of previous payment (NT dollar). X18 = amount paid in September, 2005; X19 = amount paid in August, 2005; . . .;X23 = amount paid in April, 2005.
There is one binary variable, default payment (Yes = 1, No = 0), as the response variable.
6.1.2.1 Example 1: Predicting stockmarket going up or down
Dataset: ISLR:: Smarket.
See ?ISLR::Smarket for more information.
Responsive variable: Direction; two possible values: “Up”, “Down”.
Predictors: Lag1, Lag2, Lag3, Lag4, Lag5.
Choices made:
- distance metric: eucledian distance
- k: comparing different values to find the optimal value
- model assessment metric: accuracy
df <- ISLR::Smarket
#data from 2001 to 2004 as training data
#data from 2005 as test data
df_train <- filter(df, Year < 2005) %>%
select(Lag1, Lag2, Lag3, Lag4, Lag5, Direction)
df_test <- filter(df, Year == 2005) %>%
select(Lag1, Lag2, Lag3, Lag4, Lag5, Direction)
# model 1: k = 1
model_k1 <- knn(train = df_train[, 1:5],
test = df_test[, 1:5],
cl = df_train[, 6],
k = 1)
table(model_k1, df_test$Direction)
model_k1 Down Up
Down 55 66
Up 56 75acc_k1 <- round(sum(model_k1 == df_test$Direction)/nrow(df_test), 1)
# accuracy for various k values
accuracies <- data.frame(K = 1:150, ACCURACY = NA)
for (k in 1:150) {
model_knn <- knn(train = df_train[, 1:5],
test = df_test[, 1:5],
cl = df_train$Direction,
k = k)
accuracies[k, 2] <- round(sum(model_knn == df_test$Direction)/nrow(df_test), 3)
}
ggplot(accuracies, aes(x = K, y = ACCURACY)) +
geom_line() +
ylim(0.4, 0.65) +
theme_minimal()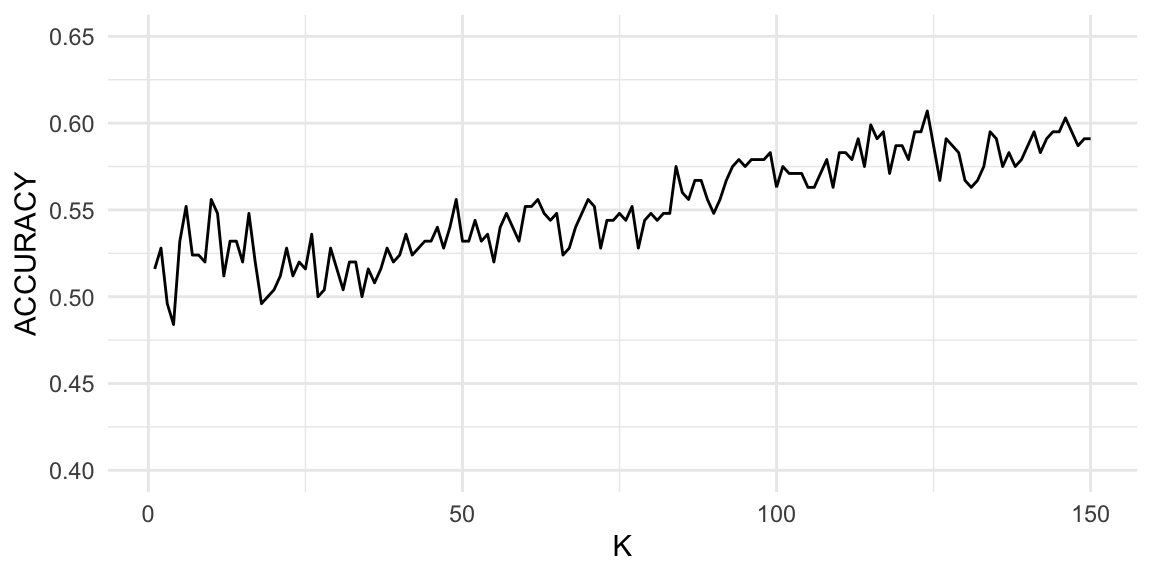
Same example using caret.
knn_model_caret <- train(Direction~.,
data = df[, c("Lag1", "Lag2", "Lag3", "Lag4", "Lag5", "Direction")],
method = "knn",
tuneGrid = data.frame(k = seq(1, 40, 2)))
preds_caret <- predict(knn_model_caret, newdata = df_test)
confusionMatrix(preds_caret, df_test$Direction)Confusion Matrix and Statistics
Reference
Prediction Down Up
Down 39 37
Up 72 104
Accuracy : 0.5675
95% CI : (0.5038, 0.6295)
No Information Rate : 0.5595
P-Value [Acc > NIR] : 0.425500
Kappa : 0.092
Mcnemar's Test P-Value : 0.001128
Sensitivity : 0.3514
Specificity : 0.7376
Pos Pred Value : 0.5132
Neg Pred Value : 0.5909
Prevalence : 0.4405
Detection Rate : 0.1548
Detection Prevalence : 0.3016
Balanced Accuracy : 0.5445
'Positive' Class : Down
library(tidyverse)
library(openxlsx)
library(flextable)
library(lubridate)
library(caret)
library(tm)
library(e1071)
#options(scipen = 999)
df_train <- tibble(Y = c("POS", "POS","POS" ,"POS" ,"POS",
"POS", "NEG","NEG", "NEG", "NEG"),
X1 = c(1,1,1,1,0,0,1,0,0,0))
df_train_2 <- tibble(Y = c("POS", "POS","POS" ,"POS" ,"POS",
"POS", "NEG","NEG", "NEG", "NEG"),
X1 = c(1,1,1,1,0,0,1,0,0,0),
X2 = c(1,0,1,0,1,0,1,0,1,0))