pop <- read.csv("./_dat/data_mal.csv") %>%
filter(ESTADO == "Sucre") %>%
dplyr::select(4:9,11,13,15) %>%
gather(t, pob, pob_2014:c2017) %>%
mutate(year = as.numeric(str_sub(t,-4,-1)),
t = str_sub(t,1,1),
CODIGO_INE = as.character(CODIGO_INE)) %>%
spread(t,pob) %>%
dplyr::rename(cases = c, pob = p)
vzla.sf <- st_read("./_dat/vzla_mun_mal2014_2/vzla_mun_mal2014_2.shp") %>%
mutate(state = str_sub(CODIGO_INE,1,2)) %>%
filter(state ==19)
vzla <- vzla.sf %>%
st_set_geometry(NULL) %>%
dplyr:: rename(X2014 = Malaria__3,
X2015 = Malaria__5,
X2016 = Malaria__7,
X2017 = Malaria__9) %>%
gather(year, cases, X2014:X2017) %>%
mutate(year = as.numeric(str_sub(year,2,5)))
Assemble
nl_ven_north <- readRDS("./_dat/nl_ven_north.rds") %>%
st_set_geometry(NULL) %>%
dplyr::select(-c(Malaria__3:Malaria__9)) %>%
gather(date, viirs, X20140101:X20171201) %>%
mutate(year = as.numeric(str_sub(date,2,5)),
month = as.numeric(str_sub(date,6,7))) %>%
group_by(CODIGO_INE, MUNICIPI0, state, year) %>%
summarise(viirs = mean(viirs, na.rm = T))
vzla <- vzla %>%
dplyr::select(-cases) %>%
inner_join(nl_ven_north, by = c("CODIGO_INE", "MUNICIPI0", "state", "year")) %>%
inner_join(pop, by = c("CODIGO_INE", "year"))
source("./_dat/nl_functions.R")
vzla %>%
ungroup() %>%
group_by(year) %>%
summarise(cases = sum(cases, na.rm = T),
viirs = mean(viirs, na.rm = T)) %>%
dual_plot(x = year, y_left = cases, y_right = viirs,
col_right = "#69b3a2", col_left = "#CD3700") +
labs(y= "Malaria cases")
vzla %>%
dual_plot(x = year, y_left = cases, y_right = viirs, group="MUNICIPI0",
col_right = "#69b3a2", col_left = "#CD3700") +
labs(y= "Malaria cases")
Functions
library(INLA)
library(spdep)
library(purrr)
devtools::source_gist("https://gist.github.com/gcarrascoe/89e018d99bad7d3365ec4ac18e3817bd")
inla.batch <- function(formula, dat1 = dt_final2) {
result = inla(formula, data=dat1, family="nbinomial", offset=log(pob), verbose = F,
#control.inla=list(strategy="gaussian"),
control.compute=list(config=T, dic=T, cpo=T, waic=T),
control.fixed = list(correlation.matrix=T),
control.predictor=list(link=1,compute=TRUE)
)
return(result)
}
inla.batch.safe <- possibly(inla.batch, otherwise = NA_real_)
P. vivax
#Create adjacency matrix
n2 <- nrow(vzla)
nb.map2 <- poly2nb(vzla.sf)
nb2INLA("./map2.graph",nb.map2)
codes2<-as.data.frame(vzla.sf)
codes2$index<-as.numeric(row.names(codes2))
codes2<-subset(codes2,select=c("MUNICIPI0","index"))
dt_final2<-merge(vzla,codes2,by="MUNICIPI0")
# MODEL
pv_22<-c(formula = cases ~ 1,
formula = cases ~ 1 + f(index, model = "bym", graph = "./map2.graph") +
f(year, model = "iid") + viirs ,
formula = cases ~ 1 + f(index, model = "bym", graph = "./map2.graph") +
f(year, model = "iid") + f(inla.group(viirs), model = "rw1"))
# ------------------------------------------
# PV estimation
# ------------------------------------------
names(pv_22)<-pv_22
INLA:::inla.dynload.workaround()
test_pv_22 <- pv_22 %>% purrr::map(~inla.batch.safe(formula = .))
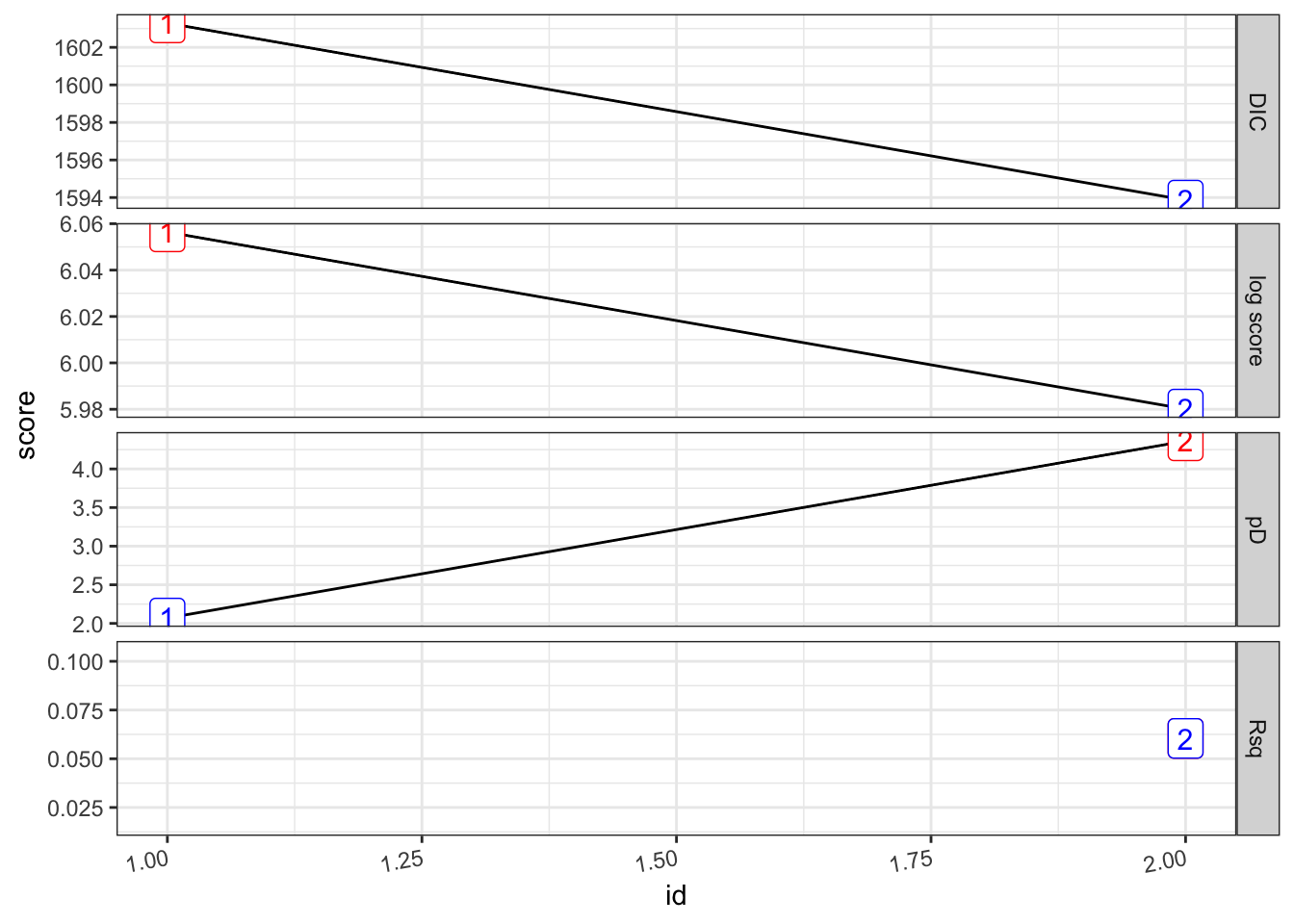
(pv_22.s <- test_pv_22 %>%
purrr::map(~Rsq.batch.safe(model = ., dic.null = test_pv_22[[1]]$dic, n = n2)) %>%
bind_rows(.id = "formula") %>% mutate(id = row_number()))
# Sumamry
summary(test_pv_22[[2]])
##
## Call:
## c("inla(formula = formula, family = \"nbinomial\", data = dat1, offset
## = log(pob), ", " verbose = F, control.compute = list(config = T, dic =
## T, ", " cpo = T, waic = T), control.predictor = list(link = 1, ", "
## compute = TRUE), control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.3, Running = 0.493, Post = 0.235, Total = 2.03
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -5.107 0.706 -6.539 -5.103 -3.698 -5.096 0
## viirs -0.461 0.249 -0.956 -0.461 0.034 -0.461 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -5.107 0.706 -6.539 -5.103 -3.698 -5.097 0
## viirs 2 -0.461 0.249 -0.963 -0.458 0.026 -0.453 0
##
## Random effects:
## Name Model
## index BYM model
## year IID model
##
## Model hyperparameters:
## mean sd
## size for the nbinomial observations (1/overdispersion) 1.390 0.273
## Precision for index (iid component) 1890.151 1876.880
## Precision for index (spatial component) 0.225 0.113
## Precision for year 0.856 0.599
## 0.025quant 0.5quant
## size for the nbinomial observations (1/overdispersion) 0.923 1.367
## Precision for index (iid component) 128.527 1335.227
## Precision for index (spatial component) 0.079 0.202
## Precision for year 0.165 0.711
## 0.975quant mode
## size for the nbinomial observations (1/overdispersion) 1.99 1.324
## Precision for index (iid component) 6863.22 351.987
## Precision for index (spatial component) 0.51 0.161
## Precision for year 2.40 0.439
##
## Expected number of effective parameters(stdev): 16.12(0.834)
## Number of equivalent replicates : 3.72
##
## Deviance Information Criterion (DIC) ...............: 746.60
## Deviance Information Criterion (DIC, saturated) ....: 92.30
## Effective number of parameters .....................: 17.13
##
## Watanabe-Akaike information criterion (WAIC) ...: 747.21
## Effective number of parameters .................: 14.67
##
## Marginal log-Likelihood: -410.29
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
##
## Call:
## c("inla(formula = formula, family = \"nbinomial\", data = dat1, offset
## = log(pob), ", " verbose = F, control.compute = list(config = T, dic =
## T, ", " cpo = T, waic = T), control.predictor = list(link = 1, ", "
## compute = TRUE), control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.39, Running = 0.586, Post = 0.343, Total = 2.32
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -5.569 0.68 -6.937 -5.568 -4.207 -5.567 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -5.569 0.68 -6.937 -5.568 -4.207 -5.566 0
##
## Random effects:
## Name Model
## index BYM model
## year IID model
## inla.group(viirs) RW1 model
##
## Model hyperparameters:
## mean sd
## size for the nbinomial observations (1/overdispersion) 1.38e+00 2.71e-01
## Precision for index (iid component) 1.91e+03 1.91e+03
## Precision for index (spatial component) 1.96e-01 9.20e-02
## Precision for year 8.13e-01 5.67e-01
## Precision for inla.group(viirs) 1.87e+04 1.85e+04
## 0.025quant 0.5quant
## size for the nbinomial observations (1/overdispersion) 0.915 1.35e+00
## Precision for index (iid component) 130.996 1.35e+03
## Precision for index (spatial component) 0.073 1.78e-01
## Precision for year 0.157 6.76e-01
## Precision for inla.group(viirs) 1268.790 1.33e+04
## 0.975quant mode
## size for the nbinomial observations (1/overdispersion) 1.98e+00 1.311
## Precision for index (iid component) 6.99e+03 358.525
## Precision for index (spatial component) 4.27e-01 0.146
## Precision for year 2.27e+00 0.419
## Precision for inla.group(viirs) 6.74e+04 3466.127
##
## Expected number of effective parameters(stdev): 16.05(0.771)
## Number of equivalent replicates : 3.74
##
## Deviance Information Criterion (DIC) ...............: 746.88
## Deviance Information Criterion (DIC, saturated) ....: 91.97
## Effective number of parameters .....................: 17.00
##
## Watanabe-Akaike information criterion (WAIC) ...: 747.57
## Effective number of parameters .................: 14.61
##
## Marginal log-Likelihood: -413.16
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
test_pv_22[[3]] %>%
plot_random3(exp = F, lab1 = "VIIRS", vars = "inla.group(viirs)")
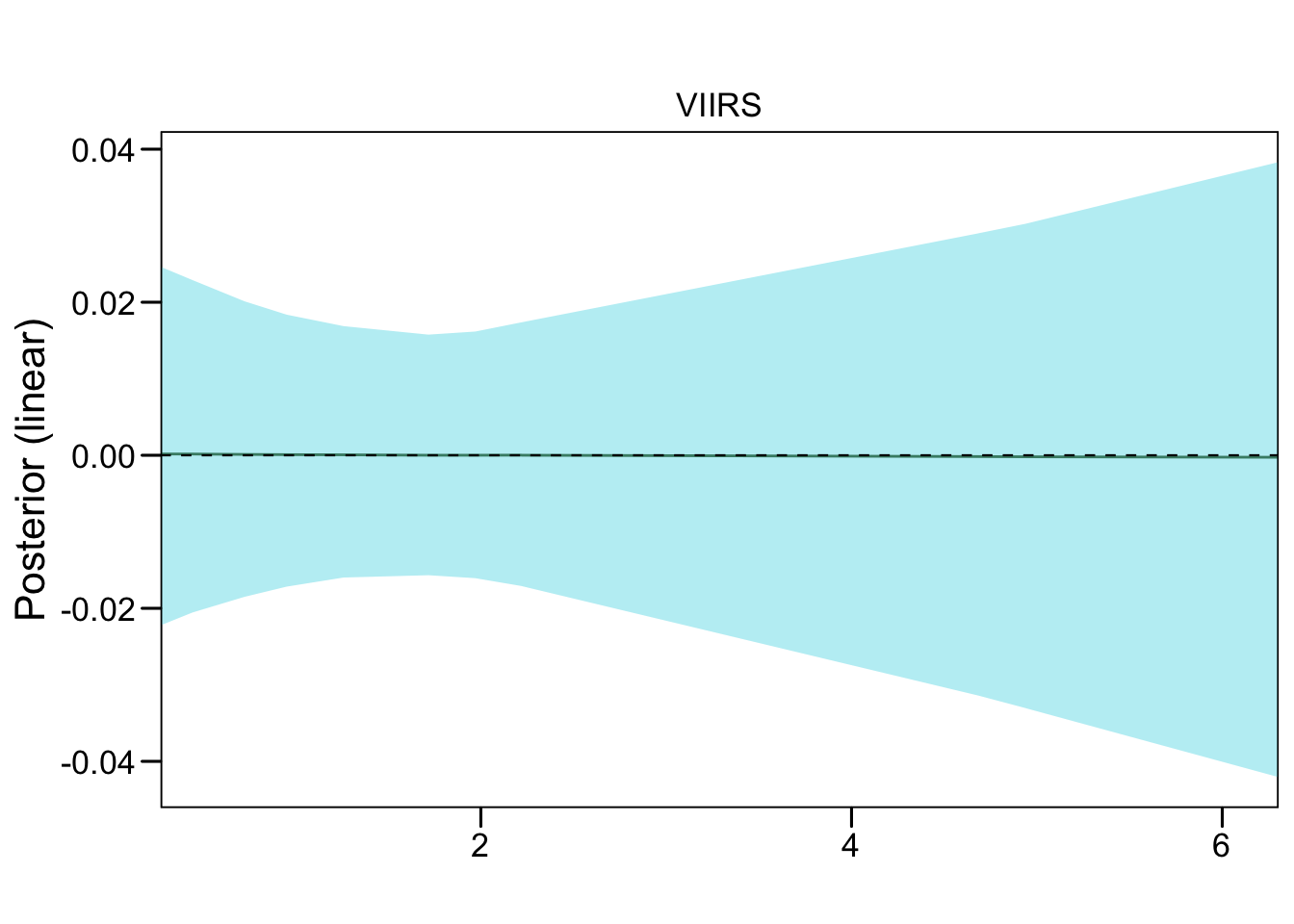
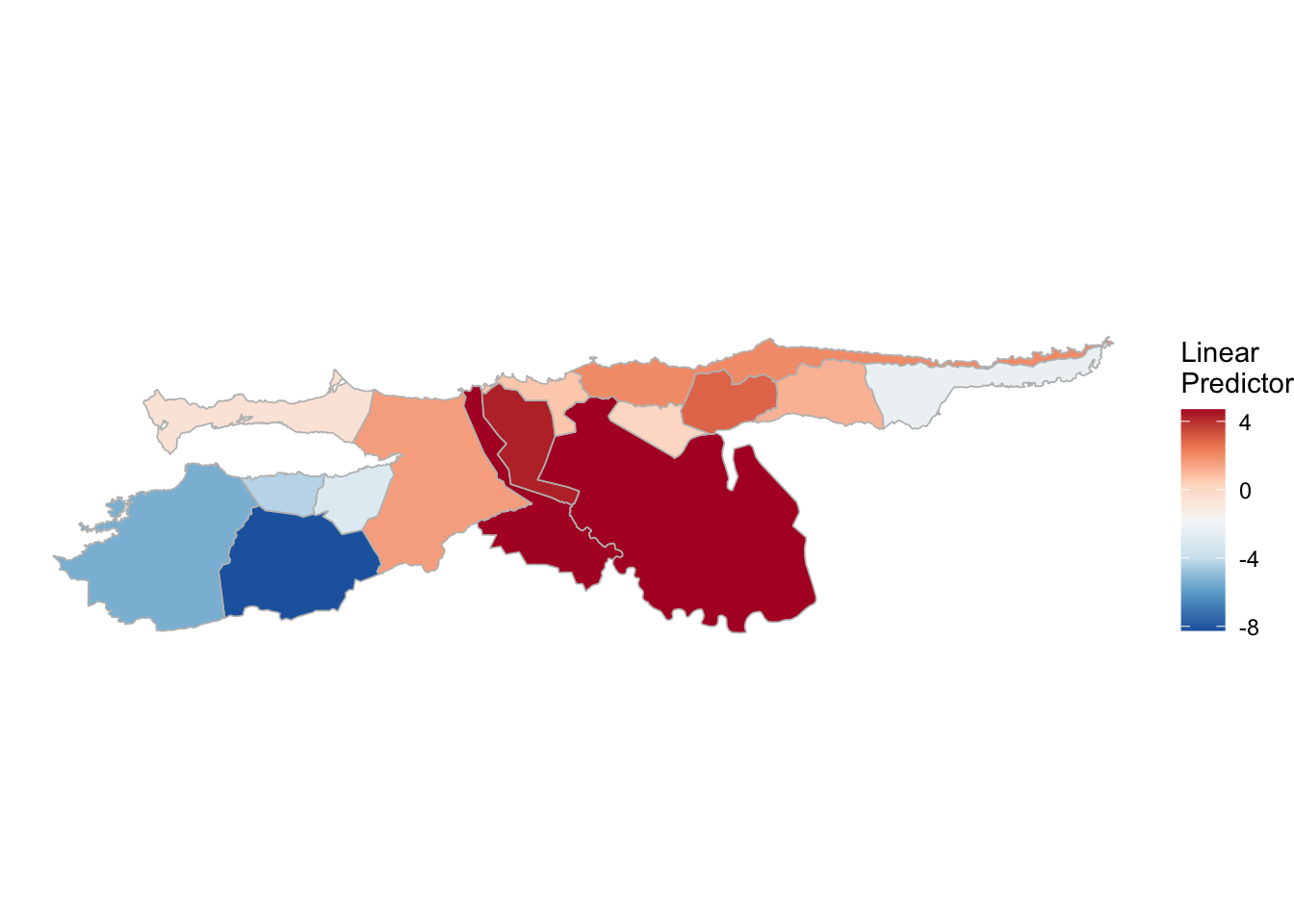
(rm_pv_1 <- test_pv_22[[2]] %>% plot_random_map2(map = vzla.sf, name= "", id = MUNICIPI0, col1 = "gray"))
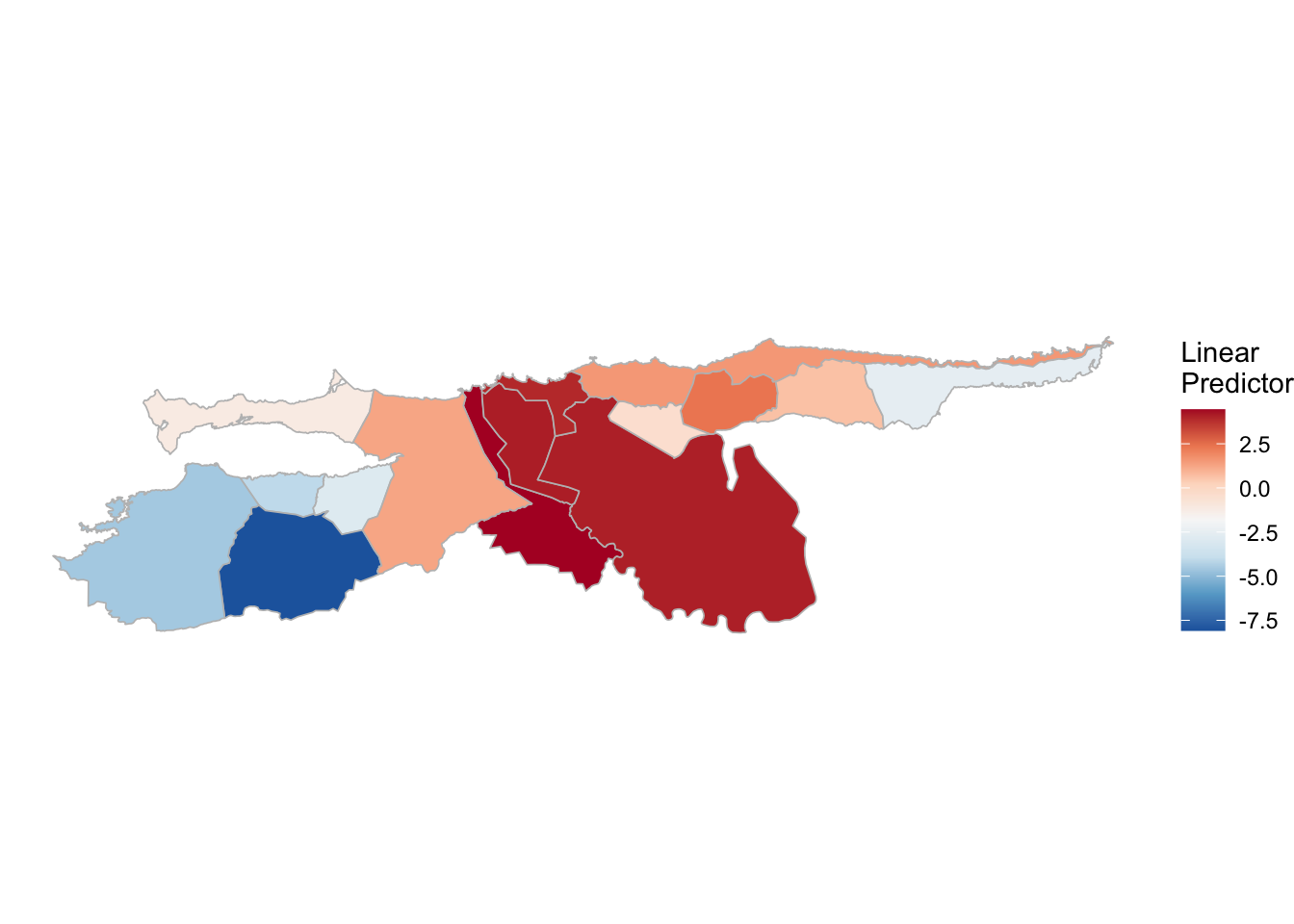
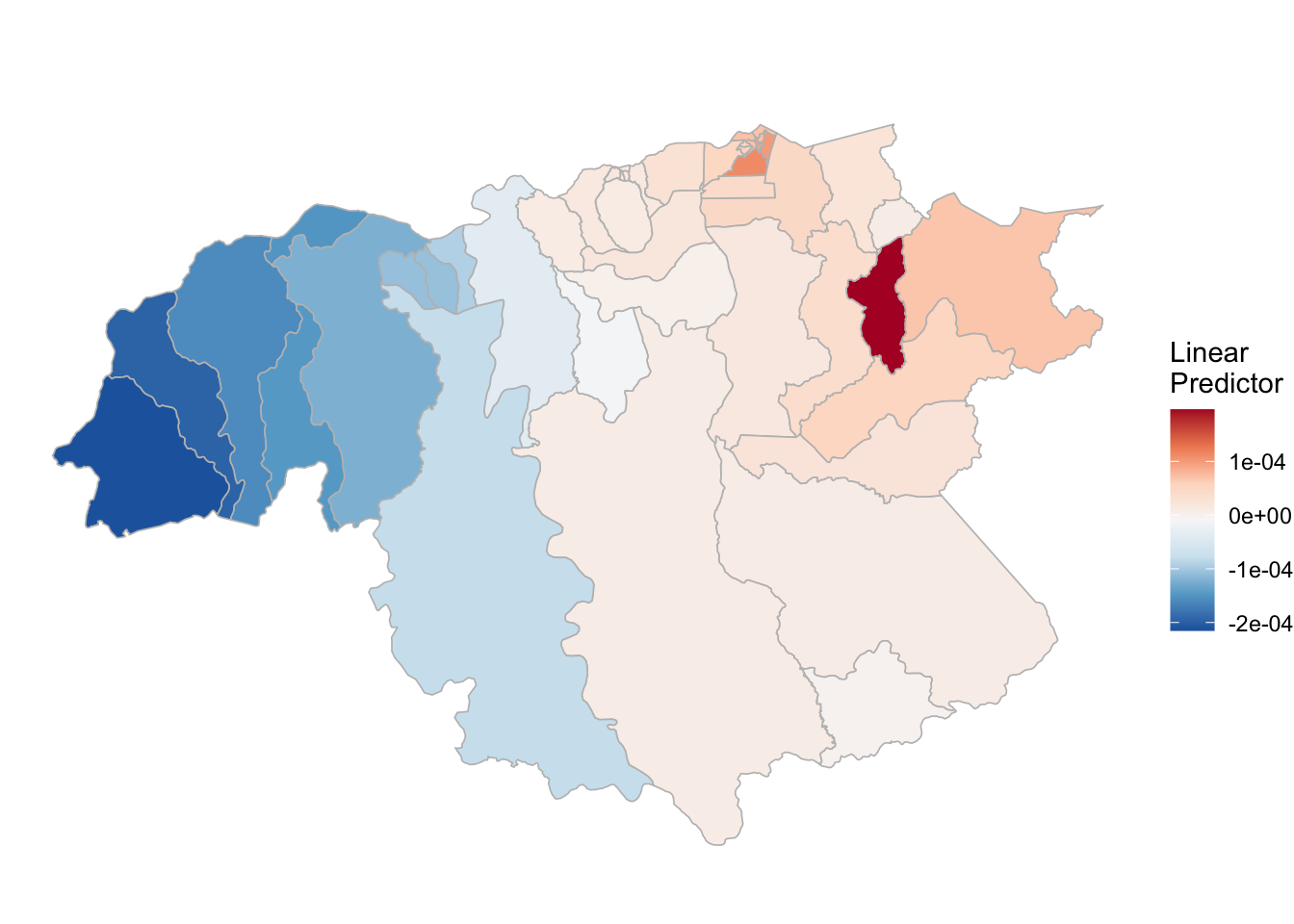
(rm_pv_2 <- test_pv_22[[3]] %>% plot_random_map2(map = vzla.sf, name= "", id = MUNICIPI0, col1 = "gray"))