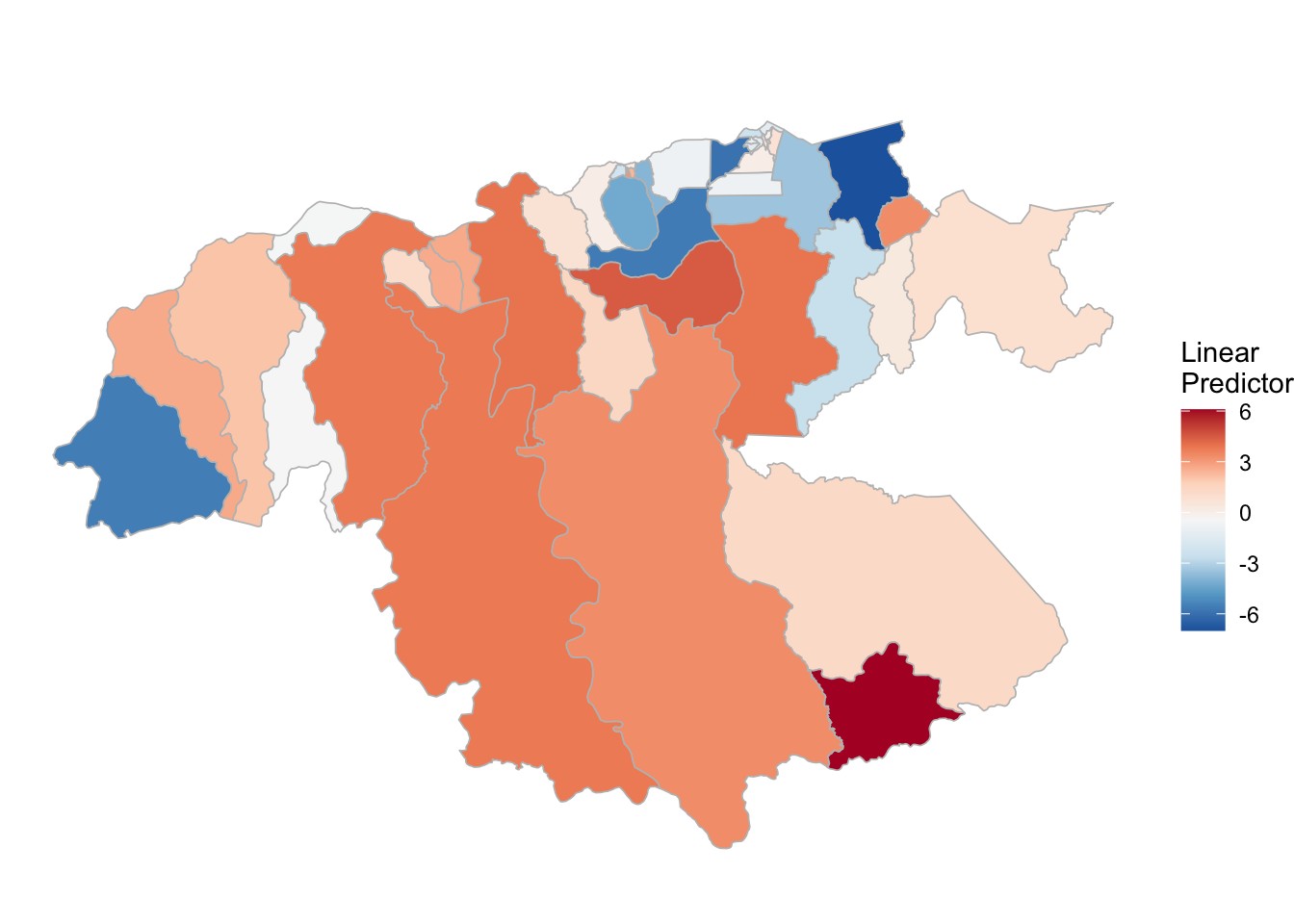
P. vivax
#Create adjacency matrix
n1 <- nrow(dt_final1)
nb.map1 <- poly2nb(area.bs1)
nb2INLA("./map1.graph",nb.map1)
codes1<-as.data.frame(area.bs1)
codes1$index<-as.numeric(row.names(codes1))
codes1<-subset(codes1,select=c("cod","index"))
dt_final1<-merge(d_v_yy,codes1,by="cod") %>%
mutate(ii = ifelse(cod == "SIFONTES-DALLA COSTA", 1, 0),
ii = ifelse(cod == "SIFONTES-SAN ISIDRO", 1, ii)) %>%
filter(ii!=1)
# MODEL
pv_21<-c(formula = PV ~ 1,
formula = PV ~ 1 + f(index, model = "bym", graph = "./map1.graph") +
f(Year, model = "iid") + viirs + mei,
formula = PV ~ 1 + f(index, model = "bym", graph = "./map1.graph") +
f(Year, model = "iid") + f(inla.group(viirs), model = "rw1") + mei)
# ------------------------------------------
# PV estimation
# ------------------------------------------
names(pv_21)<-pv_21
INLA:::inla.dynload.workaround()
test_pv_21 <- pv_21 %>% purrr::map(~inla.batch.safe(formula = .))
(pv_21.s <- test_pv_21 %>%
purrr::map(~Rsq.batch.safe(model = ., dic.null = test_pv_21[[1]]$dic, n = n1)) %>%
bind_rows(.id = "formula") %>% mutate(id = row_number()))
pv_21.s %>%
plot_score()
# Sumamry
summary(test_pv_21[[2]])
##
## Call:
## c("inla(formula = formula, family = \"nbinomial\", data = dat1, offset
## = log(pop), ", " verbose = F, control.compute = list(config = T, dic =
## T, ", " cpo = T, waic = T), control.predictor = list(link = 1, ", "
## compute = TRUE), control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.07, Running = 1.89, Post = 0.271, Total = 3.22
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -4.981 0.461 -5.868 -4.994 -4.023 -5.017 0
## viirs -0.270 0.018 -0.305 -0.270 -0.236 -0.269 0
## mei -0.970 0.275 -1.483 -0.980 -0.399 -0.999 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -4.981 0.461 -5.876 -4.991 -4.029 -5.006 0
## viirs 2 -0.270 0.018 -0.305 -0.270 -0.236 -0.269 0
## mei 3 -0.970 0.276 -1.512 -0.970 -0.429 -0.969 0
##
## Random effects:
## Name Model
## index BYM model
## Year IID model
##
## Model hyperparameters:
## mean sd
## size for the nbinomial observations (1/overdispersion) 0.203 0.018
## Precision for index (iid component) 1753.521 1780.363
## Precision for index (spatial component) 1723.368 1741.564
## Precision for Year 2.502 2.366
## 0.025quant 0.5quant
## size for the nbinomial observations (1/overdispersion) 0.169 0.203
## Precision for index (iid component) 99.900 1215.408
## Precision for index (spatial component) 100.544 1198.720
## Precision for Year 0.371 1.819
## 0.975quant mode
## size for the nbinomial observations (1/overdispersion) 0.242 0.202
## Precision for index (iid component) 6516.131 257.846
## Precision for index (spatial component) 6387.709 263.147
## Precision for Year 8.736 0.951
##
## Expected number of effective parameters(stdev): 6.16(0.528)
## Number of equivalent replicates : 34.90
##
## Deviance Information Criterion (DIC) ...............: 2153.24
## Deviance Information Criterion (DIC, saturated) ....: NaN
## Effective number of parameters .....................: 7.38
##
## Watanabe-Akaike information criterion (WAIC) ...: 2154.65
## Effective number of parameters .................: 7.57
##
## Marginal log-Likelihood: -1102.11
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
##
## Call:
## c("inla(formula = formula, family = \"nbinomial\", data = dat1, offset
## = log(pop), ", " verbose = F, control.compute = list(config = T, dic =
## T, ", " cpo = T, waic = T), control.predictor = list(link = 1, ", "
## compute = TRUE), control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.2, Running = 2.25, Post = 0.234, Total = 3.69
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -13.195 1.120 -15.414 -13.194 -10.989 -13.190 0
## mei -0.446 0.517 -1.460 -0.447 0.573 -0.449 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -13.196 1.120 -15.410 -13.196 -10.988 -13.194 0
## mei 2 -0.446 0.517 -1.461 -0.446 0.572 -0.447 0
##
## Random effects:
## Name Model
## index BYM model
## Year IID model
## inla.group(viirs) RW1 model
##
## Model hyperparameters:
## mean sd
## size for the nbinomial observations (1/overdispersion) 0.938 0.123
## Precision for index (iid component) 0.123 0.031
## Precision for index (spatial component) 1812.361 1745.562
## Precision for Year 0.667 0.424
## Precision for inla.group(viirs) 2.899 1.875
## 0.025quant 0.5quant
## size for the nbinomial observations (1/overdispersion) 0.717 0.930
## Precision for index (iid component) 0.072 0.119
## Precision for index (spatial component) 118.635 1299.631
## Precision for Year 0.164 0.568
## Precision for inla.group(viirs) 0.745 2.438
## 0.975quant mode
## size for the nbinomial observations (1/overdispersion) 1.201 0.917
## Precision for index (iid component) 0.194 0.113
## Precision for index (spatial component) 6411.274 321.702
## Precision for Year 1.764 0.395
## Precision for inla.group(viirs) 7.789 1.713
##
## Expected number of effective parameters(stdev): 51.45(1.65)
## Number of equivalent replicates : 4.18
##
## Deviance Information Criterion (DIC) ...............: 1889.96
## Deviance Information Criterion (DIC, saturated) ....: NaN
## Effective number of parameters .....................: 51.21
##
## Watanabe-Akaike information criterion (WAIC) ...: 1886.26
## Effective number of parameters .................: 38.29
##
## Marginal log-Likelihood: -1025.01
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
test_pv_21[[3]] %>%
plot_random3(exp = F, lab1 = "VIIRS", vars = "inla.group(viirs)")
(rm_pv_1 <- test_pv_21[[2]] %>% plot_random_map2(map = area.bs1, name= "", id = cod, col1 = "gray"))
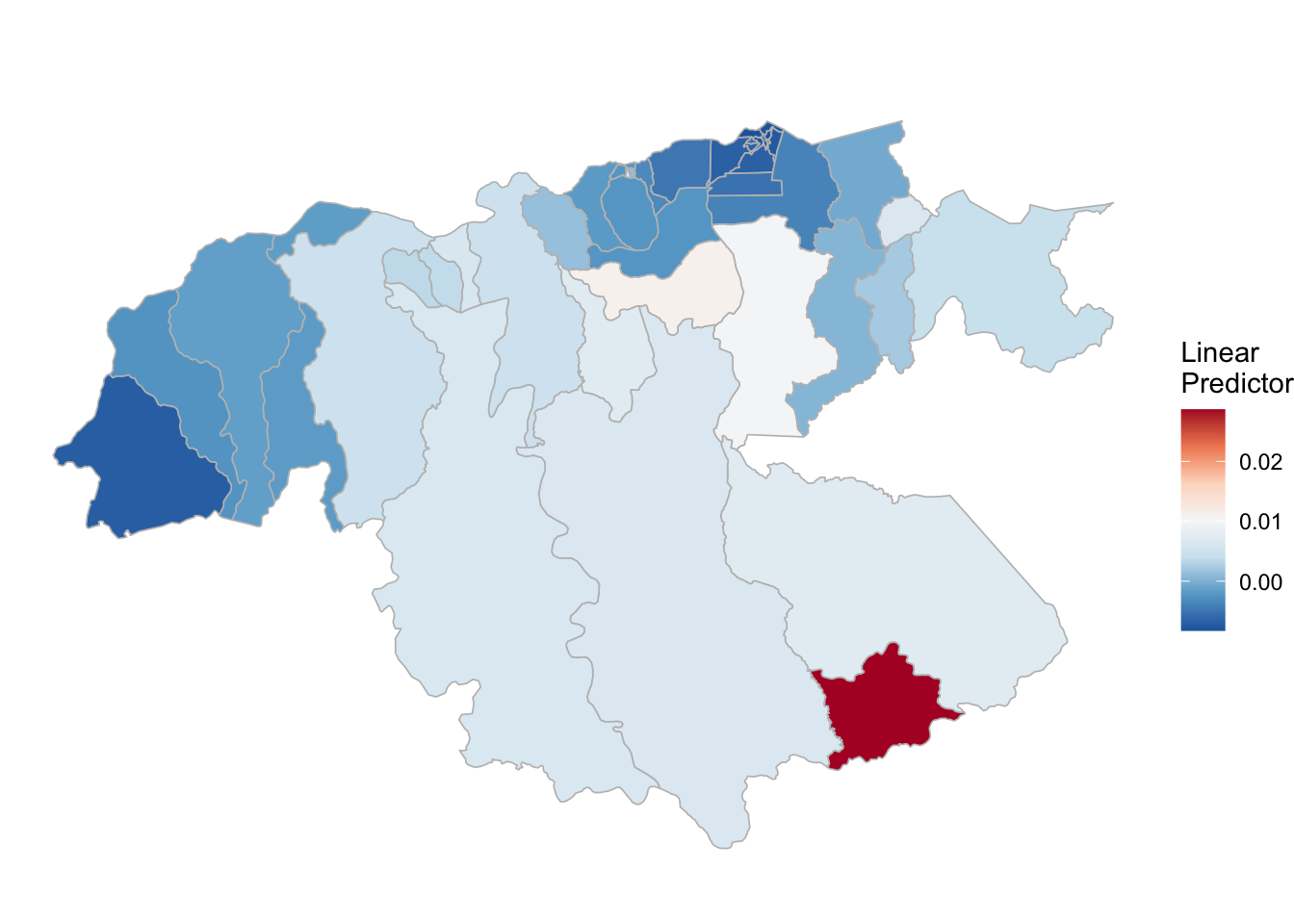
(rm_pv_2 <- test_pv_21[[3]] %>% plot_random_map2(map = area.bs1, name= "", id = cod, col1 = "gray"))
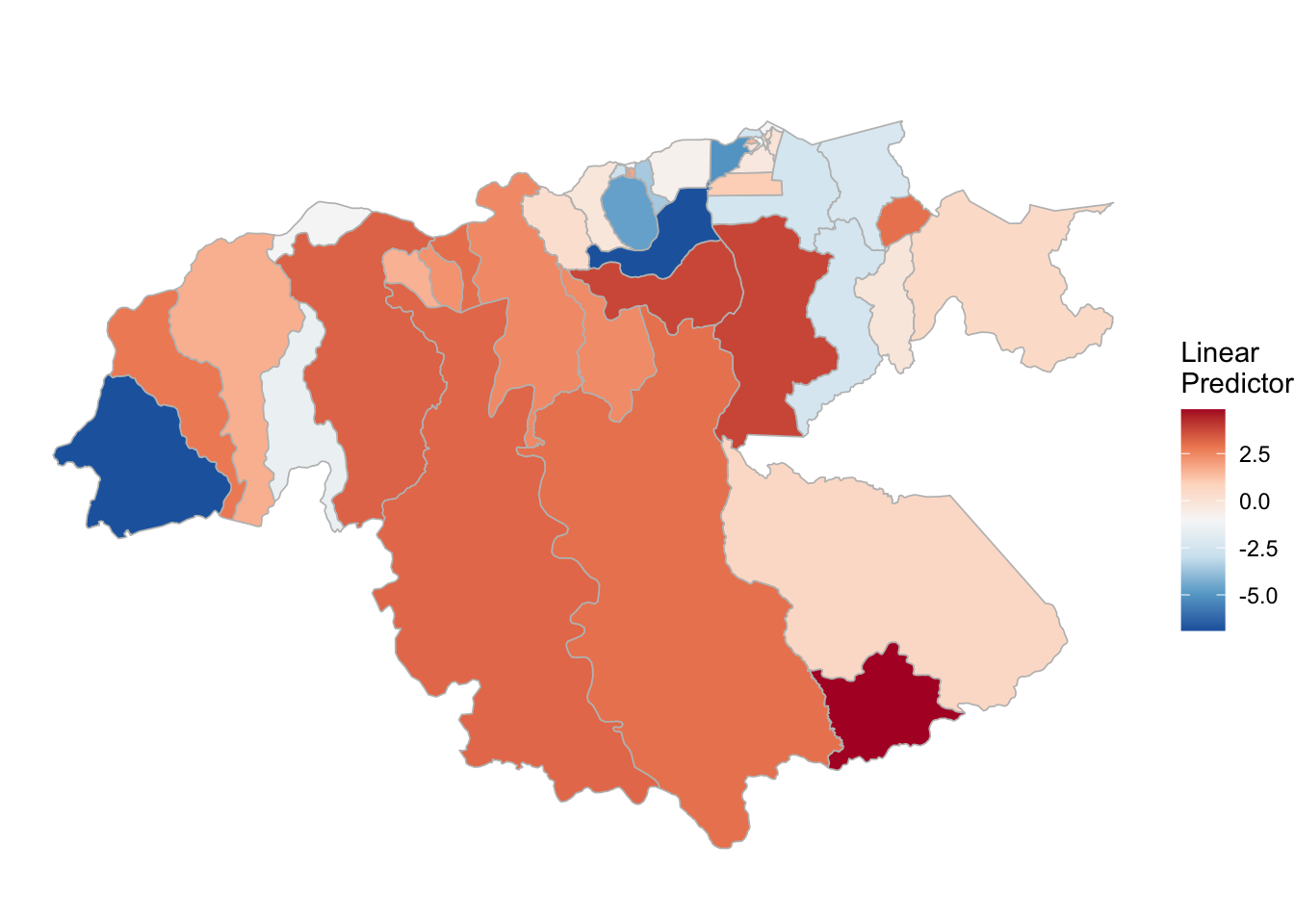
P. falciparum
# MODEL
pf_21<-c(formula = PF ~ 1,
formula = PF ~ 1 + f(index, model = "bym", graph = "./map1.graph") +
f(Year, model = "iid") + viirs + mei,
formula = PF ~ 1 + f(index, model = "bym", graph = "./map1.graph") +
f(Year, model = "iid") + f(inla.group(viirs), model = "rw1") + mei)
# ------------------------------------------
# PV estimation
# ------------------------------------------
names(pf_21)<-pf_21
INLA:::inla.dynload.workaround()
test_pf_21 <- pf_21 %>% purrr::map(~inla.batch.safe(formula = .))
(pf_21.s <- test_pf_21 %>%
purrr::map(~Rsq.batch.safe(model = ., dic.null = test_pf_21[[1]]$dic, n = n1)) %>%
bind_rows(.id = "formula") %>% mutate(id = row_number()))
pf_21.s %>%
plot_score()
# Sumamry
summary(test_pf_21[[2]])
##
## Call:
## c("inla(formula = formula, family = \"nbinomial\", data = dat1, offset
## = log(pop), ", " verbose = F, control.compute = list(config = T, dic =
## T, ", " cpo = T, waic = T), control.predictor = list(link = 1, ", "
## compute = TRUE), control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.1, Running = 9.99, Post = 0.223, Total = 11.3
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -1.650 0.443 -2.518 -1.651 -0.781 -1.651 0
## viirs -0.040 0.030 -0.099 -0.040 0.018 -0.040 0
## mei -0.918 0.440 -1.748 -0.930 -0.020 -0.955 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -1.650 0.443 -2.519 -1.65 -0.782 -1.650 0
## viirs 2 -0.040 0.030 -0.099 -0.04 0.018 -0.040 0
## mei 3 -0.918 0.440 -1.778 -0.92 -0.050 -0.924 0
##
## Random effects:
## Name Model
## index BYM model
## Year IID model
##
## Model hyperparameters:
## mean sd
## size for the nbinomial observations (1/overdispersion) 6.80e-02 7.00e-03
## Precision for index (iid component) 1.94e+03 1.91e+03
## Precision for index (spatial component) 1.46e+03 1.78e+03
## Precision for Year 1.80e+04 1.87e+04
## 0.025quant 0.5quant
## size for the nbinomial observations (1/overdispersion) 0.055 6.80e-02
## Precision for index (iid component) 132.436 1.38e+03
## Precision for index (spatial component) 89.860 9.17e+02
## Precision for Year 1255.251 1.24e+04
## 0.975quant mode
## size for the nbinomial observations (1/overdispersion) 8.20e-02 0.069
## Precision for index (iid component) 7.00e+03 360.880
## Precision for index (spatial component) 6.19e+03 241.454
## Precision for Year 6.80e+04 3444.573
##
## Expected number of effective parameters(stdev): 3.01(0.015)
## Number of equivalent replicates : 71.33
##
## Deviance Information Criterion (DIC) ...............: 1802.65
## Deviance Information Criterion (DIC, saturated) ....: NaN
## Effective number of parameters .....................: -0.172
##
## Watanabe-Akaike information criterion (WAIC) ...: 1804.16
## Effective number of parameters .................: 1.32
##
## Marginal log-Likelihood: -950.82
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
##
## Call:
## c("inla(formula = formula, family = \"nbinomial\", data = dat1, offset
## = log(pop), ", " verbose = F, control.compute = list(config = T, dic =
## T, ", " cpo = T, waic = T), control.predictor = list(link = 1, ", "
## compute = TRUE), control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.24, Running = 4.71, Post = 0.217, Total = 6.17
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -15.991 1.187 -18.373 -15.975 -13.686 -15.937 0
## mei 0.608 0.593 -0.588 0.620 1.741 0.642 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -15.991 1.186 -18.349 -15.983 -13.666 -15.961 0
## mei 2 0.609 0.594 -0.552 0.607 1.778 0.604 0
##
## Random effects:
## Name Model
## index BYM model
## Year IID model
## inla.group(viirs) RW1 model
##
## Model hyperparameters:
## mean sd
## size for the nbinomial observations (1/overdispersion) 1.238 0.346
## Precision for index (iid component) 0.118 0.025
## Precision for index (spatial component) 1650.189 1816.043
## Precision for Year 0.633 0.656
## Precision for inla.group(viirs) 2.668 1.899
## 0.025quant 0.5quant
## size for the nbinomial observations (1/overdispersion) 0.815 1.154
## Precision for index (iid component) 0.073 0.116
## Precision for index (spatial component) 111.319 1100.339
## Precision for Year 0.009 0.394
## Precision for inla.group(viirs) 0.579 2.185
## 0.975quant mode
## size for the nbinomial observations (1/overdispersion) 2.122 0.973
## Precision for index (iid component) 0.171 0.115
## Precision for index (spatial component) 6508.760 301.667
## Precision for Year 2.305 0.007
## Precision for inla.group(viirs) 7.634 1.418
##
## Expected number of effective parameters(stdev): 48.14(1.06)
## Number of equivalent replicates : 4.47
##
## Deviance Information Criterion (DIC) ...............: 1343.98
## Deviance Information Criterion (DIC, saturated) ....: NaN
## Effective number of parameters .....................: 45.91
##
## Watanabe-Akaike information criterion (WAIC) ...: 1336.63
## Effective number of parameters .................: 31.19
##
## Marginal log-Likelihood: -749.02
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
test_pf_21[[3]] %>%
plot_random3(exp = F, lab1 = "VIIRS", vars = "inla.group(viirs)")
(rm_pf_1 <- test_pf_21[[2]] %>% plot_random_map2(map = area.bs1, name= "", id = cod, col1 = "gray"))
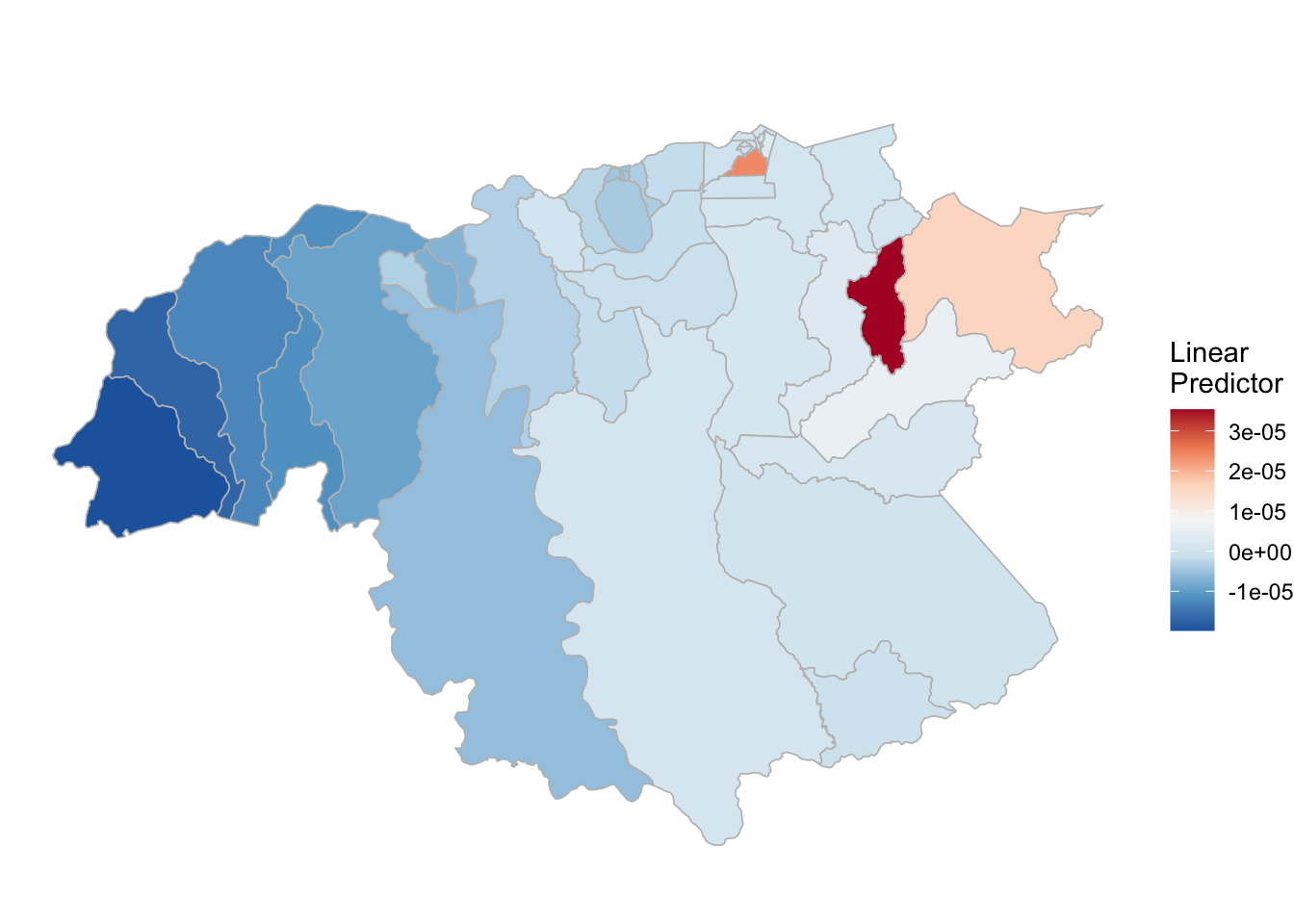
(rm_pf_2 <- test_pf_21[[3]] %>% plot_random_map2(map = area.bs1, name= "", id = cod, col1 = "gray"))