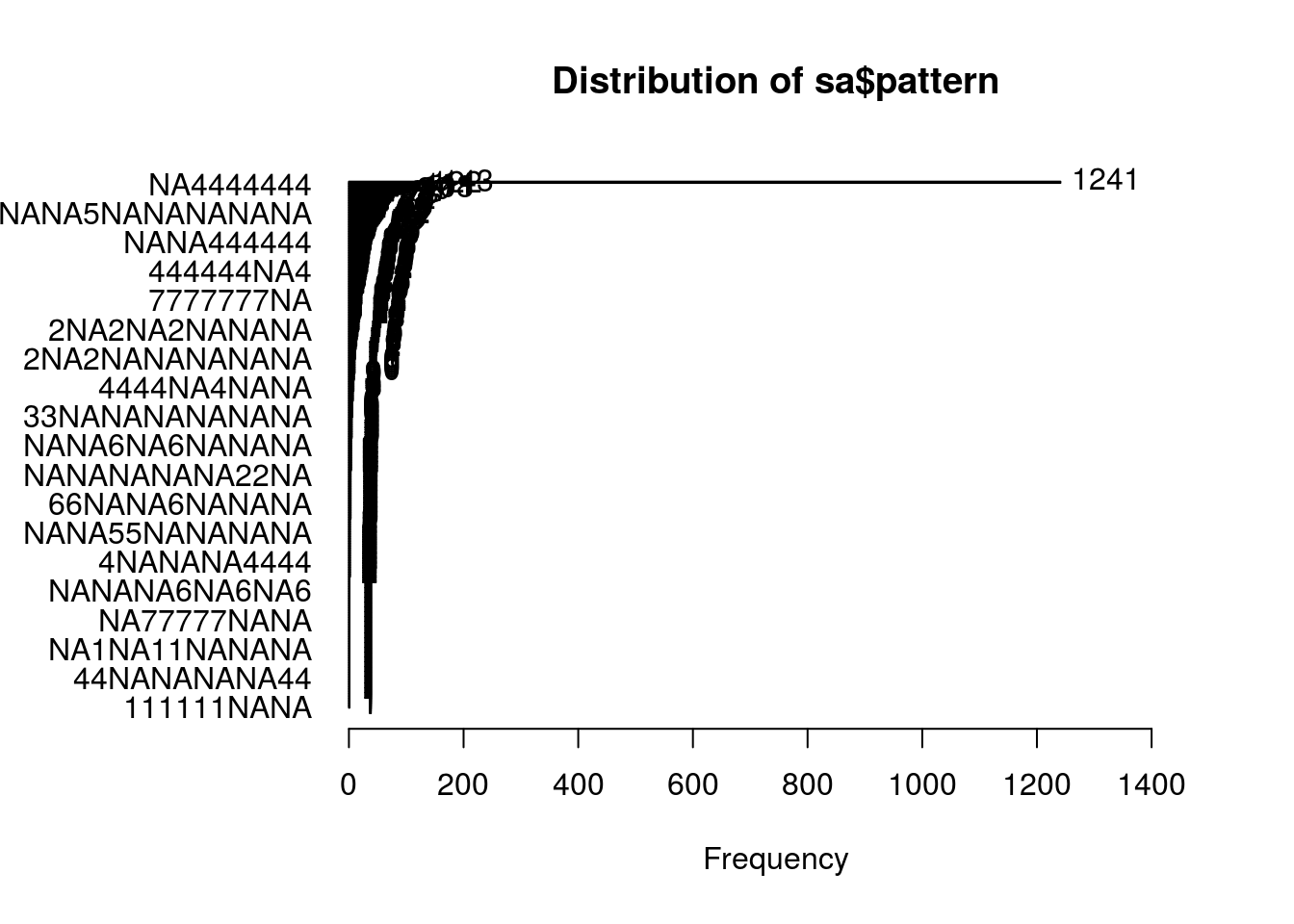
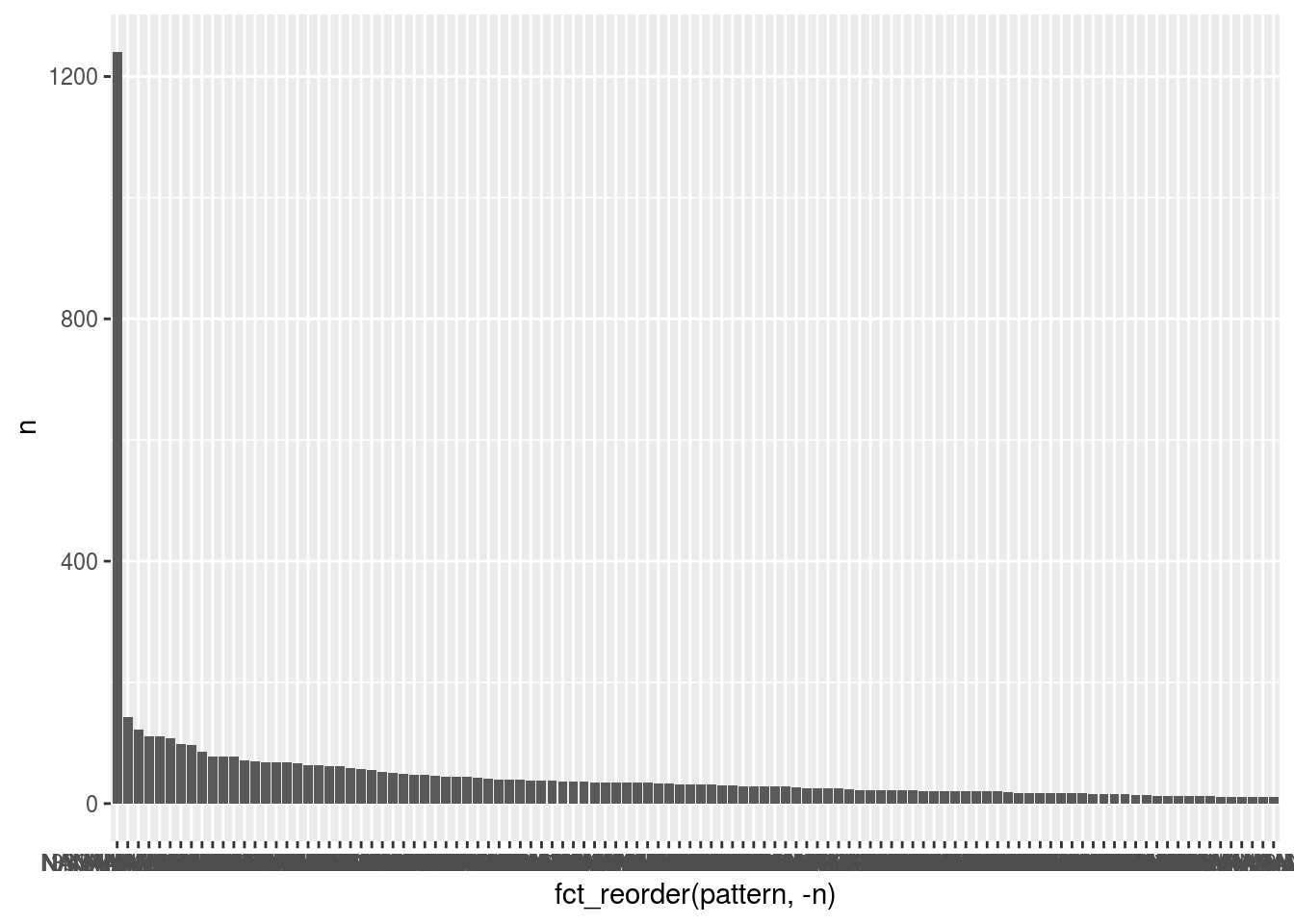
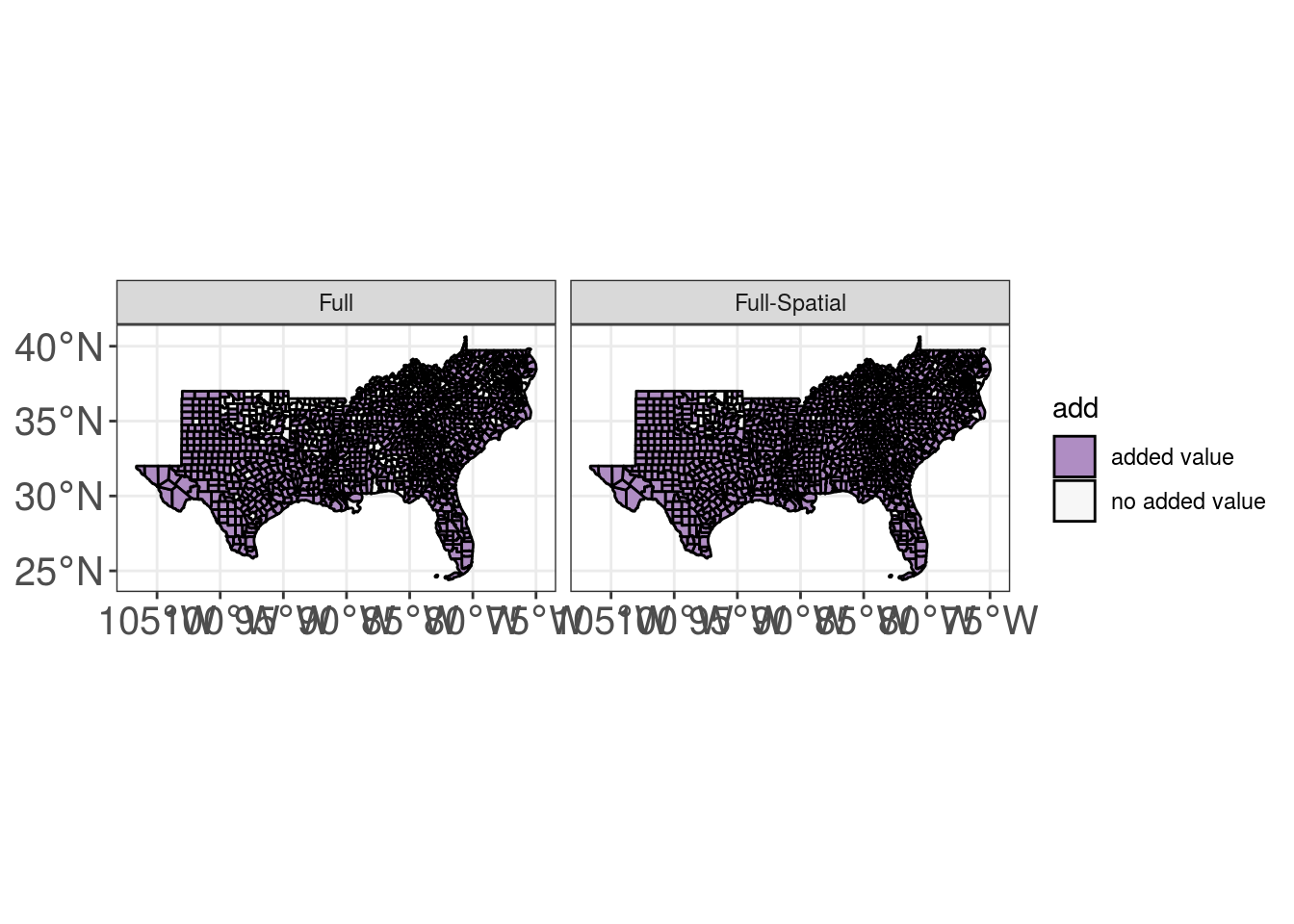
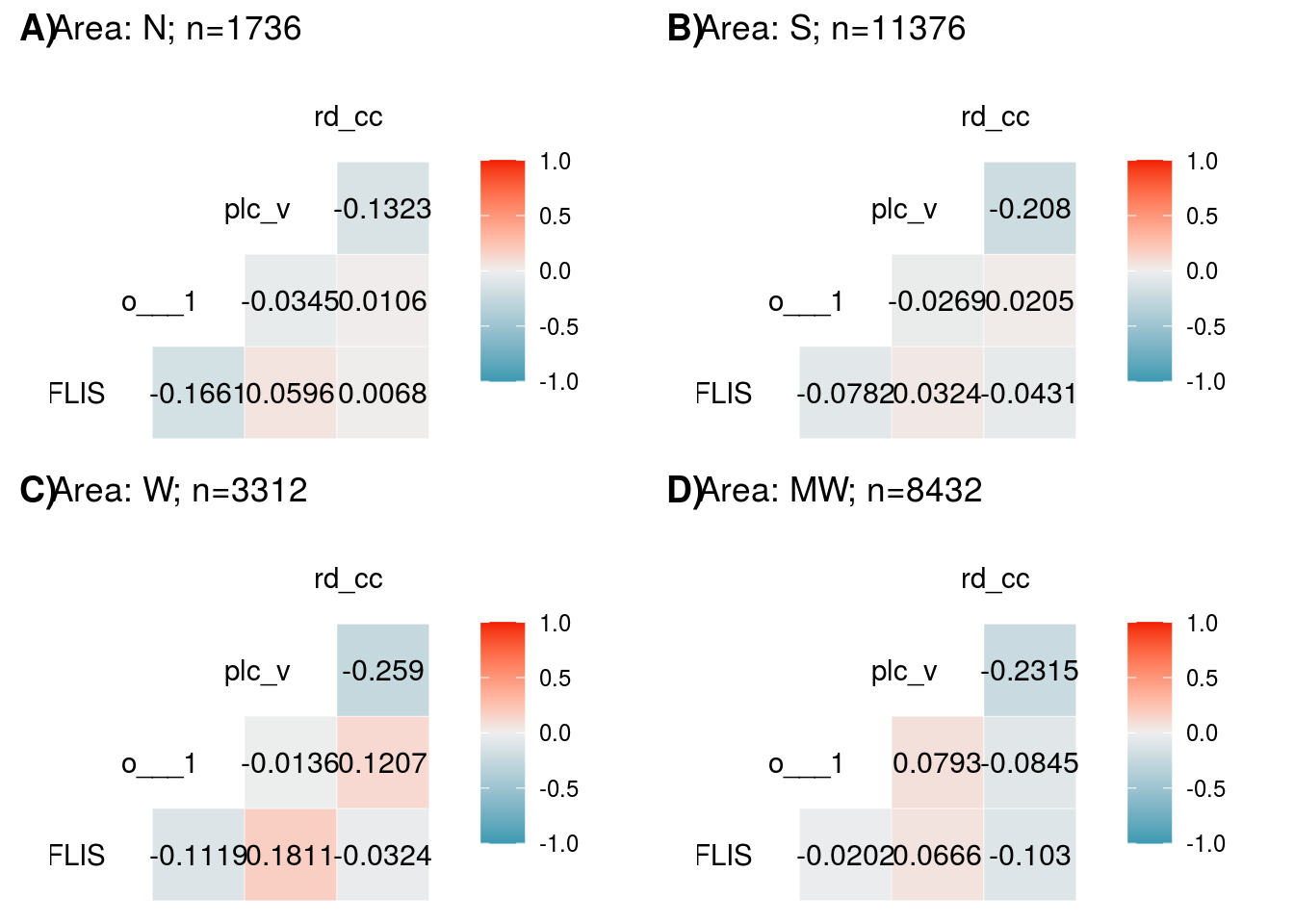
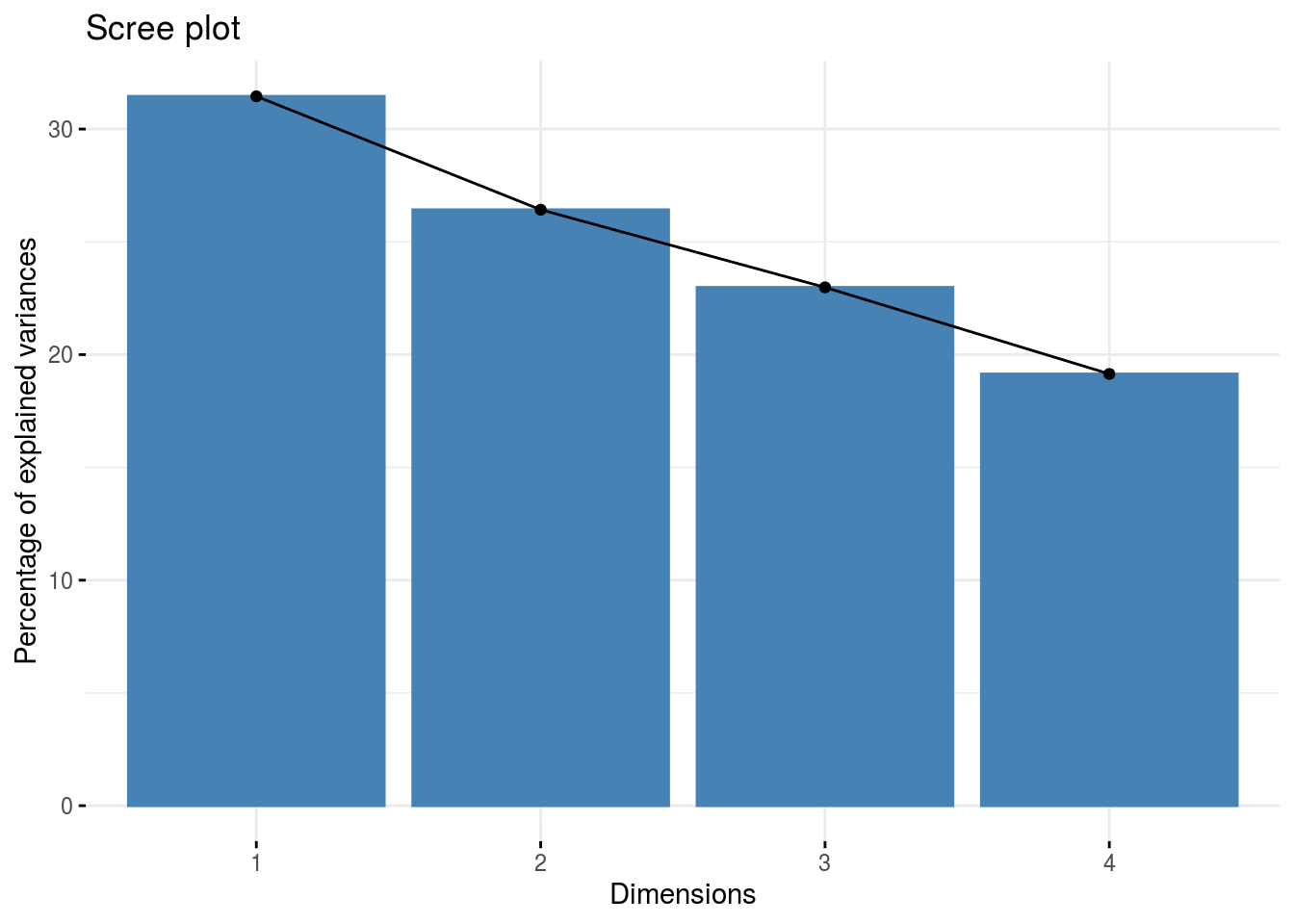
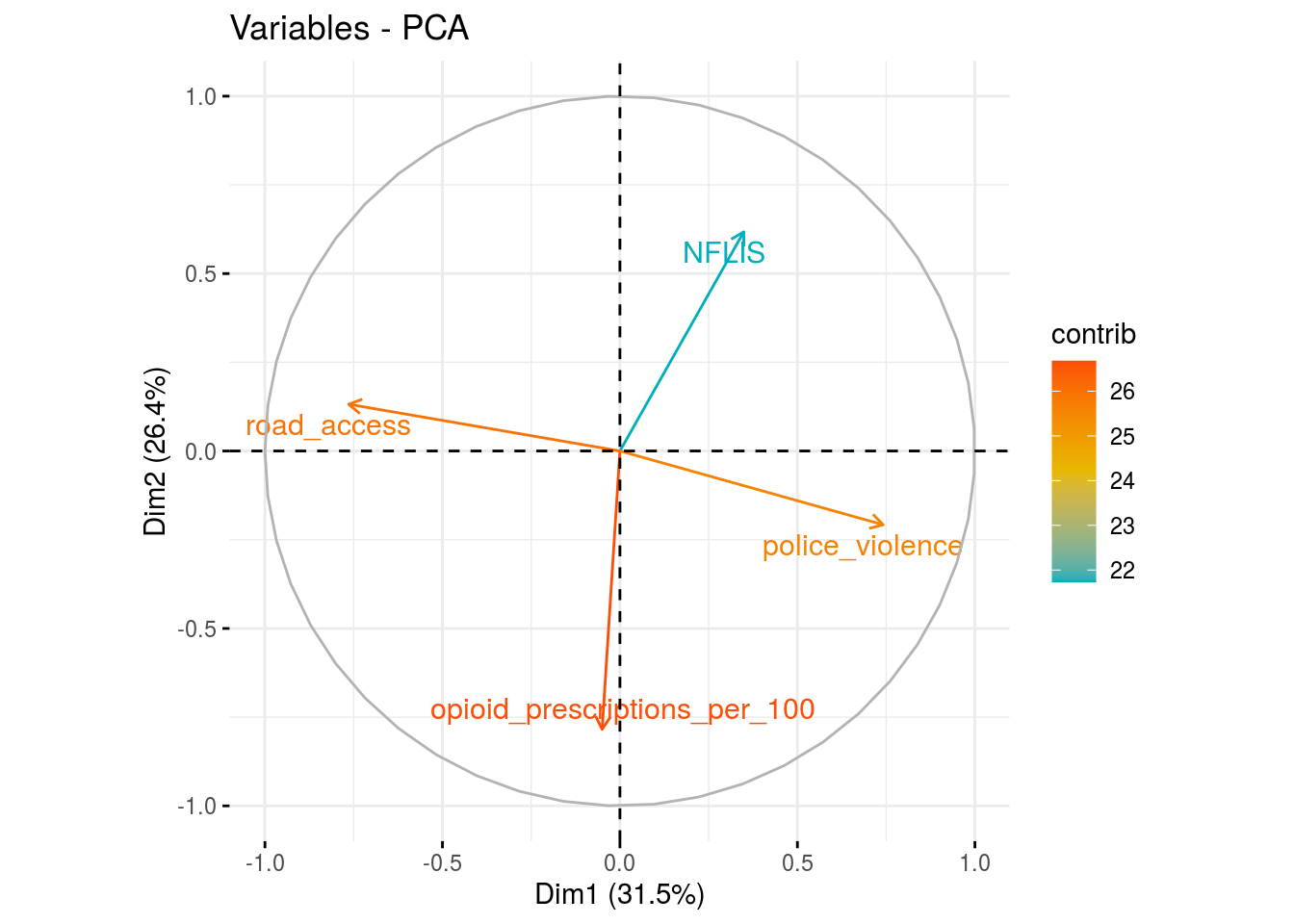
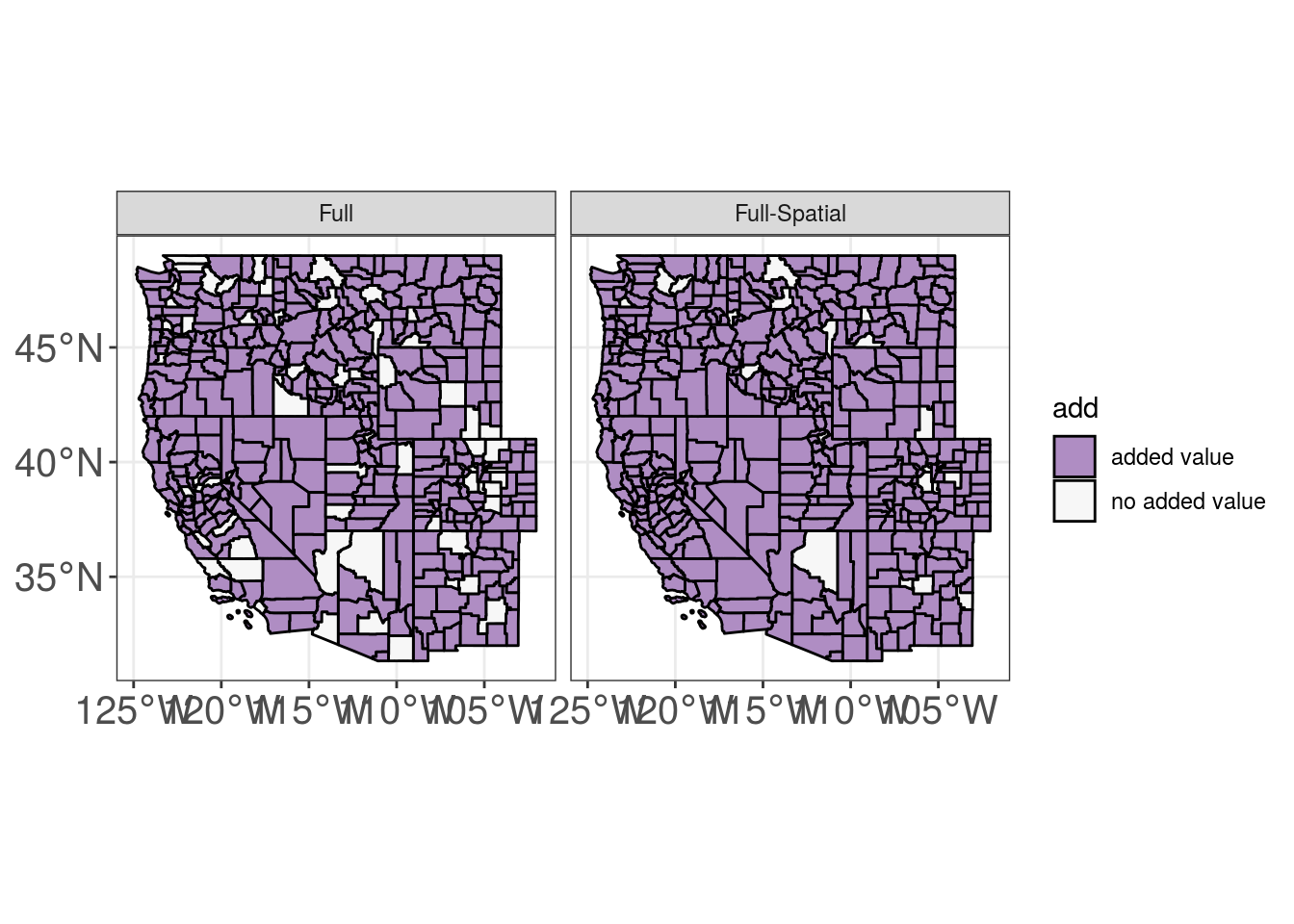
Year Random Effects
##
## Call:
## c("inla(formula = formula, family = \"binomial\", data = dat1, verbose
## = F, ", " control.compute = list(config = T, dic = T, cpo = T, waic =
## T), ", " control.predictor = list(link = 1, compute = TRUE),
## control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.57, Running = 6.87, Post = 4.75, Total = 13.2
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -5.595 0.713 -7.132 -5.522 -4.414 -5.485 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -5.595 0.713 -7.12 -5.539 -4.4 -5.57 0
##
## Random effects:
## Name Model
## index BYM model
## Year RW1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for index (iid component) 1863.210 1836.900 126.232 1321.073
## Precision for index (spatial component) 0.017 0.005 0.010 0.017
## Precision for Year 0.114 0.078 0.023 0.096
## 0.975quant mode
## Precision for index (iid component) 6721.569 344.814
## Precision for index (spatial component) 0.029 0.016
## Precision for Year 0.314 0.060
##
## Expected number of effective parameters(stdev): 161.05(5.42)
## Number of equivalent replicates : 8.08
##
## Deviance Information Criterion (DIC) ...............: Inf
## Deviance Information Criterion (DIC, saturated) ....: Inf
## Effective number of parameters .....................: Inf
##
## Watanabe-Akaike information criterion (WAIC) ...: 443.50
## Effective number of parameters .................: 85.78
##
## Marginal log-Likelihood: -323.28
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
##
## Call:
## c("inla(formula = formula, family = \"binomial\", data = dat1, verbose
## = F, ", " control.compute = list(config = T, dic = T, cpo = T, waic =
## T), ", " control.predictor = list(link = 1, compute = TRUE),
## control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 2.09, Running = 47.9, Post = 29, Total = 79.1
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -14.379 1.269 -17.047 -14.159 -12.294 -13.938 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -14.382 1.269 -17.045 -14.173 -12.29 -13.982 0
##
## Random effects:
## Name Model
## index BYM model
## Year RW1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for index (iid component) 1800.285 1796.984 121.839 1268.088
## Precision for index (spatial component) 0.013 0.002 0.009 0.013
## Precision for Year 0.187 0.127 0.036 0.158
## 0.975quant mode
## Precision for index (iid component) 6571.712 332.931
## Precision for index (spatial component) 0.018 0.013
## Precision for Year 0.513 0.097
##
## Expected number of effective parameters(stdev): 470.88(9.64)
## Number of equivalent replicates : 18.12
##
## Deviance Information Criterion (DIC) ...............: Inf
## Deviance Information Criterion (DIC, saturated) ....: Inf
## Effective number of parameters .....................: Inf
##
## Watanabe-Akaike information criterion (WAIC) ...: 950.71
## Effective number of parameters .................: 173.96
##
## Marginal log-Likelihood: -624.83
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
##
## Call:
## c("inla(formula = formula, family = \"binomial\", data = dat1, verbose
## = F, ", " control.compute = list(config = T, dic = T, cpo = T, waic =
## T), ", " control.predictor = list(link = 1, compute = TRUE),
## control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 1.97, Running = 12.8, Post = 12.1, Total = 26.8
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -9.42 1.35 -12.446 -9.23 -7.284 -9.096 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -9.422 1.35 -12.455 -9.25 -7.275 -9.187 0
##
## Random effects:
## Name Model
## index BYM model
## Year RW1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for index (iid component) 1326.825 1170.834 72.581 988.791
## Precision for index (spatial component) 0.032 0.012 0.015 0.030
## Precision for Year 1.081 0.881 0.174 0.845
## 0.975quant mode
## Precision for index (iid component) 4290.94 182.735
## Precision for index (spatial component) 0.06 0.027
## Precision for Year 3.42 0.466
##
## Expected number of effective parameters(stdev): 99.01(6.79)
## Number of equivalent replicates : 25.09
##
## Deviance Information Criterion (DIC) ...............: Inf
## Deviance Information Criterion (DIC, saturated) ....: Inf
## Effective number of parameters .....................: Inf
##
## Watanabe-Akaike information criterion (WAIC) ...: 267.75
## Effective number of parameters .................: 38.99
##
## Marginal log-Likelihood: -145.37
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed
##
## Call:
## c("inla(formula = formula, family = \"binomial\", data = dat1, verbose
## = F, ", " control.compute = list(config = T, dic = T, cpo = T, waic =
## T), ", " control.predictor = list(link = 1, compute = TRUE),
## control.fixed = list(correlation.matrix = T))" )
## Time used:
## Pre = 2.55, Running = 34.2, Post = 20.7, Total = 57.4
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) -12.445 1.156 -14.865 -12.335 -10.493 -12.169 0
##
## Linear combinations (derived):
## ID mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 1 -12.445 1.156 -14.865 -12.346 -10.48 -12.245 0
##
## Random effects:
## Name Model
## index BYM model
## Year RW1 model
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for index (iid component) 1899.686 1859.968 127.271 1351.493
## Precision for index (spatial component) 0.022 0.004 0.014 0.021
## Precision for Year 0.287 0.195 0.060 0.240
## 0.975quant mode
## Precision for index (iid component) 6.8e+03 346.524
## Precision for index (spatial component) 3.1e-02 0.020
## Precision for Year 7.9e-01 0.155
##
## Expected number of effective parameters(stdev): 273.83(8.98)
## Number of equivalent replicates : 23.09
##
## Deviance Information Criterion (DIC) ...............: Inf
## Deviance Information Criterion (DIC, saturated) ....: Inf
## Effective number of parameters .....................: Inf
##
## Watanabe-Akaike information criterion (WAIC) ...: 696.84
## Effective number of parameters .................: 115.55
##
## Marginal log-Likelihood: -414.44
## CPO and PIT are computed
##
## Posterior marginals for the linear predictor and
## the fitted values are computed