4 Text mining of multispecies interactions in published literature
The following workflow (4.1) is defined to explore the use of available text mining tools to mine published events of interaction between species. First, a comprehensive search phase looks to gather the most amount of potential articles to mine for species interactions. The second phase involves the analysis and classification of the downloaded articles based on the similarity and frequency of words mentioned in the article text. Additionally, specific entities such as scientific names, geographical locations or person names can be automatically identified and extracted from the article text. Posible product outputs from this workflow correspond to summarized versions of the text, a model to classify articles based on the particular words denoting the presence of a multispecies interaction, automatic taxonomic identification of species present in the text and an estimate of the geographical scope of the article, based on the detection and geocoding of geographical names.
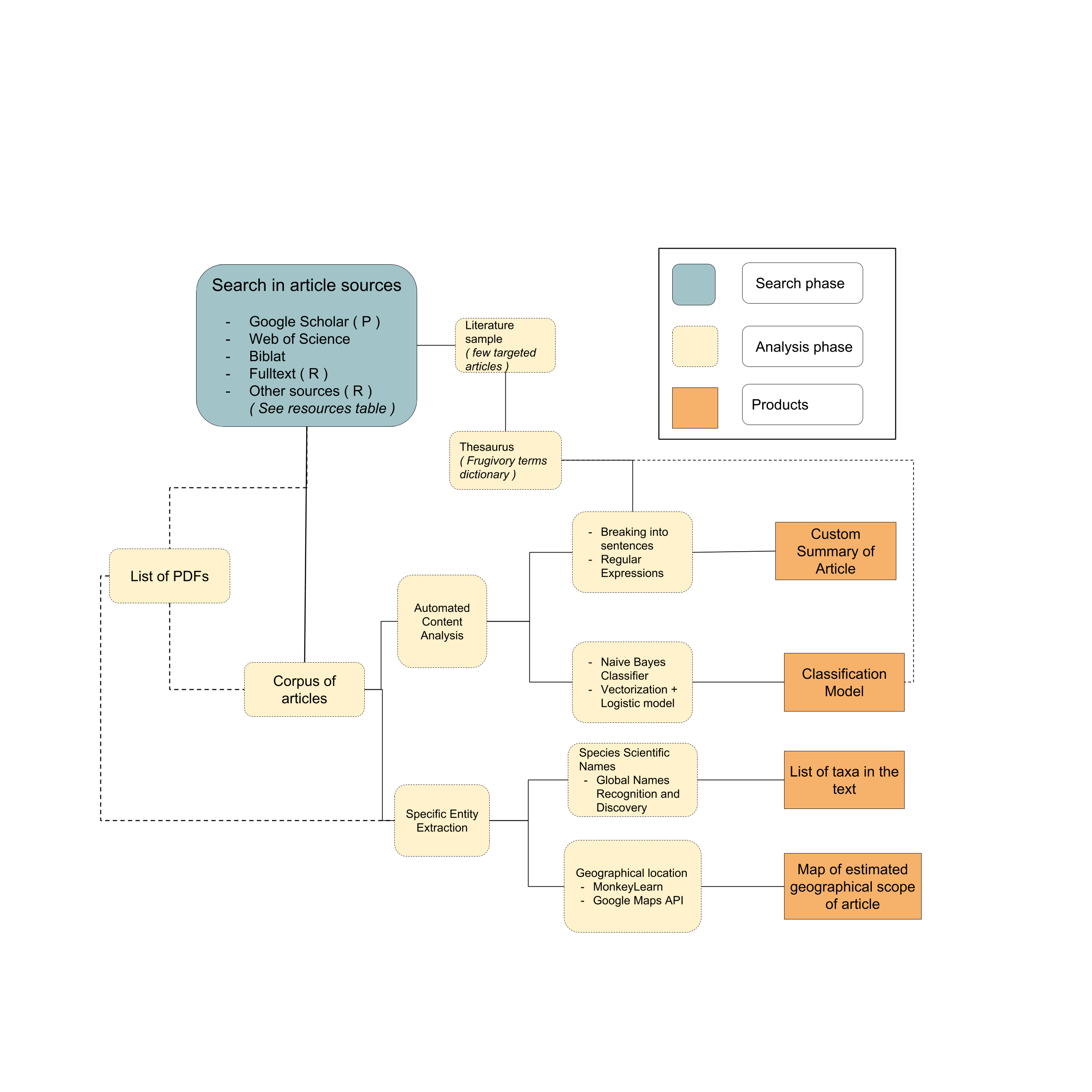
Figure 4.1: General proposed workflow for the search and classification of literature with species interactions records. Lines represent connections between steps of the workflow. Dashed lines represent optional ways or extra steps that could be avoided. Products can potentially be used together to design more specific workflows tailored to a researcher custom interests
4.1 Search phase
Articles in ecology are usually found in academic web databases (Appendix Table 3). Such databases provide different ways to access to the information, perhaps the most common way to access to them is through a user interface on the internet. A researcher will type some keywords of interest and the database will return a list of results matching the query terms (keywords). Academic web search engines usually provide results based on the title, abstract or a few keywords. Therefore, they are unlikely to discriminate the presence of interactions in the results provided. General web academic searches may produce thousands of often unrelated articles (Figure 4.2). In the other hand, the inclusion of many keywords will results in and underrepresentation of the available literature on the topic of interest. The first step towards gathering information on species interactions is to identify those sources and databases which hold ecological literature.
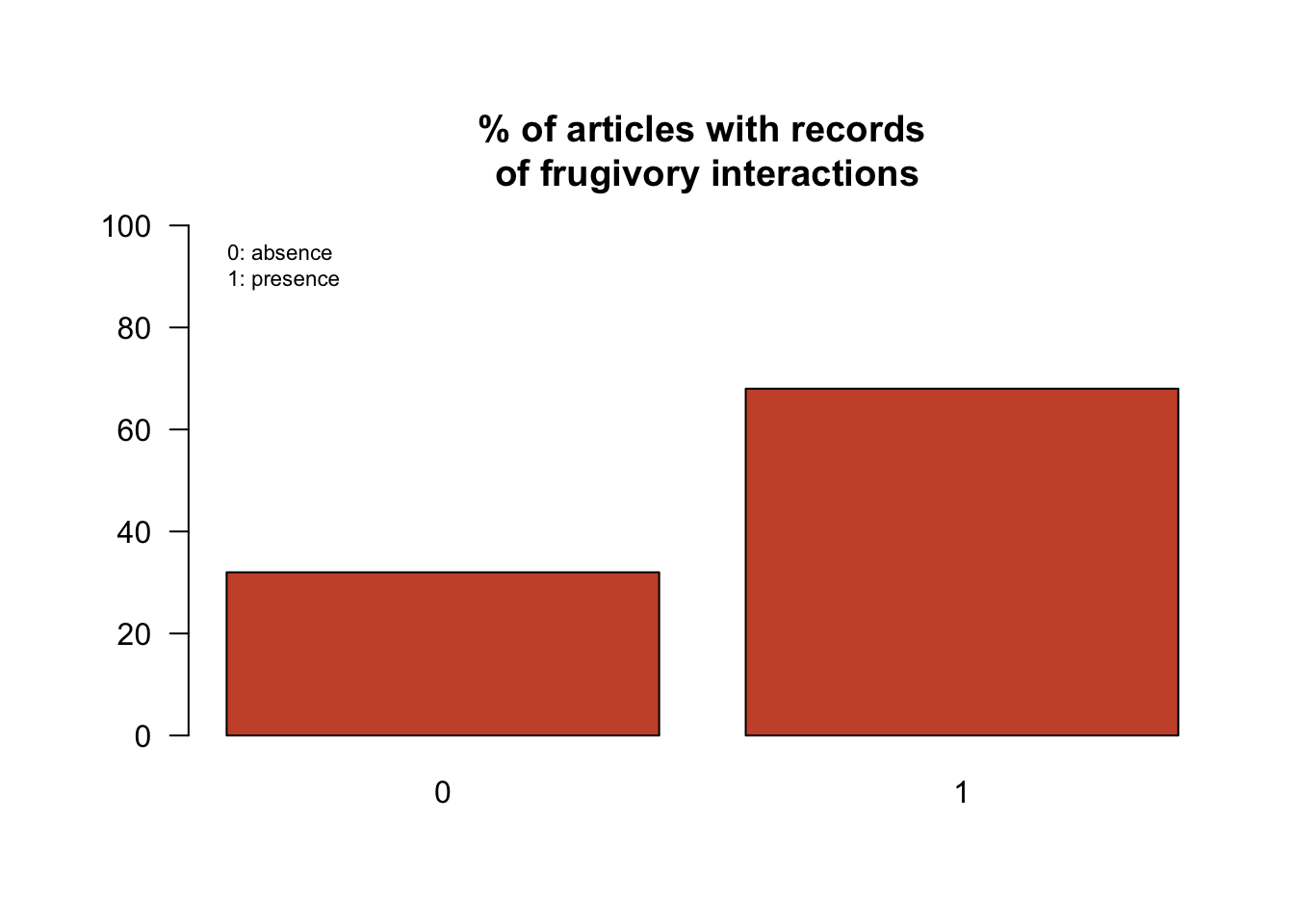
Figure 4.2: Barplots showing the presence of articles with frugivory interactions from in the first 100 articles retrieved from a single query in the Public Library of Science (PLOS) https://www.plos.org/ (keywords: frugiv + dispers + tropic)
Figure 4.2 shows the imprecision of articles retrieval relying only upon an initial search terms query. Time was spent to read and manage 30% of those articles retrieved only to find out that there are not any mentions of species interaction related to frugivory. This time could have been well spent in other research tasks such as data analysis or writing. In addition, the plots show only the presence of a frugivory interaction in the text regardless of taxa or geographic region, which are additional pieces of information key to classify documents based on the interest of the researcher (e.g. bat - fruit interactions in Colombia). Additional keywords can be included in the initial query (e.g. bat; Colombia) but this will most likely cause additional noise in the query results and it will require repetitions of queries including different combinations of keywords.
4.1.1 Programmatic queries and downloads
A typical literature review from a researcher in ecology usually involves the following steps: 1) go to the database webpage (e.g. Web of Science); 2) type the keywords; 3) navigate through the results output (usually of 10 - 20 articles per page); 4) select the article to download (based on the title, authors or a preview of the abstract); 5) download the article (if there is a fulltext version available or if he/she has credentials); 6) Read the article; 7) Keep of discard after reading (or screening) it; 8) Repeat. This process, aside from being time-consuming, has several flaws. The whole process is difficult to record, vaguely reported and dependent on the query abilities and credentials of the researcher. This makes literature reviews, inefficient, unreproducible, potentially biased and very susceptible to human error. Not all databases allow batch downloads articles in a machine readable format. Being this especially true for the largest scientific publishers (Elsevier, Wiley, Thomson Reuters) (LIBER 2014).
Open-Source databases usually provide API’s. Querying and downloading articles via API’s is a more much efficient process, but it requires some knowledge in SOAP (Simple Object Access Protocol) internet protocols to exchange data through the internet. Accessing a database through API’s significantly reduces clicking time as search (sometimes downloads) can be performed with a few lines of code. Scientific publications are indexed on the internet with a DOI (Digital Object Identifier). This identifier is unique to each publication and remains constant independent of the article URL, making it useful when managing articles downloaded in batch. Moreover, some API’s allows the direct download of the article by parts like only the figures, or just the main body of text. A direct download text is more useful than downloading PDF documents because the text downloaded is already machine readable, allowing to a direct incorporation from the search to posterior text mining and machine learning procedures. The fulltext (Chamberlain 2016) is a package developed within the RopenSci initiative. According to its authors, its goal is to act as a “single interface” of other R packages which allows the retrieval of fulltext data from scholarly journal articles, focusing in open access journals.
As it was previously mentioned, not all scientific databases provide means to perform a programmatic download. A first option is to manually download the PDFs one by one directly from the publisher page (assuming that you have the access). PDFs is a format that mimics the printed version of a document, being basically a digital print of it. This format designed and useful for human - readers is not the best for computers and locks valuable information contained in tables and figures. The mining of this type of information is outside the scope of this literature review but there are several packages like metagear (Lajeunesse 2016) with functions to extract information from tables and figures. Moreover, new initiatives, like Zenodo are being created as repositories to facilitate the mining of figures and tables from scientific articles in biodiversity and other fields of science.
Nevertheless, it is possible to “semi”- automatize (for some cases) the process of downloading articles. If a researcher is interested into programmatically query Google Scholar, there is an unofficial API sci-hub.py. This API search for articles in Google Scholar and download them from Sci-hub when there is not a free version available. This API only works for python and currently, the automatic download from Sci - Hub is blocked by captchas. However, the list of URLs directing to the articles from a particular query in Google scholar can be retrieved fairly easy. After doing some preprocessing on this list the URLs can be passed to the download.file implemented in base R for download the PDFs.
For the case of queries made to crossref, the fulltext can not be retrieved as crossref is a database of citations, however, a list of articles DOI’s can be retrieved. To every element of this DOIs list (from crossref of some other sources) the prefix "http://sci-hub.cc/" can be added to create a link to download via Sci-Hub. This link can be pasted directly into any browser, type the captcha and get the PDF. Sci-Hub is the largest repository of scientific articles and becomes very useful if a researcher does not have access to subscription databases or hits a paywall
4.1.2 OCR list of PDF’s
When a researcher has a list of PDFs of the articles of interest to mine, the content those articles have to be transformed into a machine readable format for applying text mining procedures. Optical character recognition (OCR) is the recognition of text characters from digital sources of which the text is not available in text format (e.g. pdf, scans, pictures).There have been developments in the OCR field since 1920, but arguably at this time one of the best tools for OCR is Tesseract an open source OCR software being developed actually by Google. Tesseract has the advantage of producing a high-quality output and is very good to recognize text from PDF regardless of the different formatting (very useful when the pdf comes from a scan of the original printed article) making the information available to mine.
Google Tesseract can be called with the package tesseract (Ooms 2016). Tesseract produces a high-quality output of text with the function the function pdf_render_page. The limitations are the initial transformation of PDFs to images and pdf_render_page converts pdf to tiff page by page. However, these limitations can be overcome with a little of extra coding.
Another function to handle text included in PDFs is the function pdf_text included in the pdftools (Ooms 2016) package. The output of this function is less accurate as the output from the tesseract OCR, nevertheless, there is an advantage of being faster and recognizing all the pdf at once. Yet another function to perform OCR on a list of PDFs is ft_extract included also in the fulltext package. Both functions, at the difference of Google Tesseract (which is slower but with a better output), can be used to perform extractions of text from large quantities of articles, in the initial path a list of pdf articles can be used as input.
The text output from the list of PDFs can be now used to create a corpus and perform the desired mining procedures.
4.1.3 Corpus of text and preprocessing
A document corpus is a structured set containing the desired documents to which text - mining techniques can be applied (Fortuna et al. 2005). A corpus can be created with the text version of articles extracted from different sources. A corpus can be permanent (stored in the physical memory) or a virtual (stored in virtual memory). Once the corpus is created, to facilitate even more the machine readability of the text, it is necessary to perform some preprocessing to the corpus. This preprocessing includes removing punctuation marks, numbers, common word endings, and stop words and extra white spaces. The package tm (Feinerer and Hornik. 2015) provides the functions VectorSource and Corpus to facilitate these tasks.
4.2 Analysis phase
4.2.1 Building Thesaurus for frugivory, plant - animal interactions
In order to identify articles containing relevant ecological information species interactions, we must first be able to correctly identify documents talking about frugivory. Researchers use a variety specific words to describe this particular ecological event. Among others; diet, frugivory, seed dispersal, interaction are terms that are used frequently in articles to describe frugivory. The list of possible terms for describing frugivory interactions is certainly finite, but for a single researcher (even for a specialist) recalling all possible related terms can become an uneasy task. Therefore, the best option is to look up for those terms directly in the literature. To perform this task, a corpus can be created with a targeted sample of articles previously known to contain interactions on frugivory. Once the corpus is created and preprocessed, a document - term matrix - Matrix with columns representing the articles and rows as terms - can be calculated with the function DocumentTermMatrix from the tm package. The frequency of terms per article can then be calculated as the colSums of the mentioned matrix. Those significant terms are located between a variable margin of not too general, not too specific terms Figure 4.3.
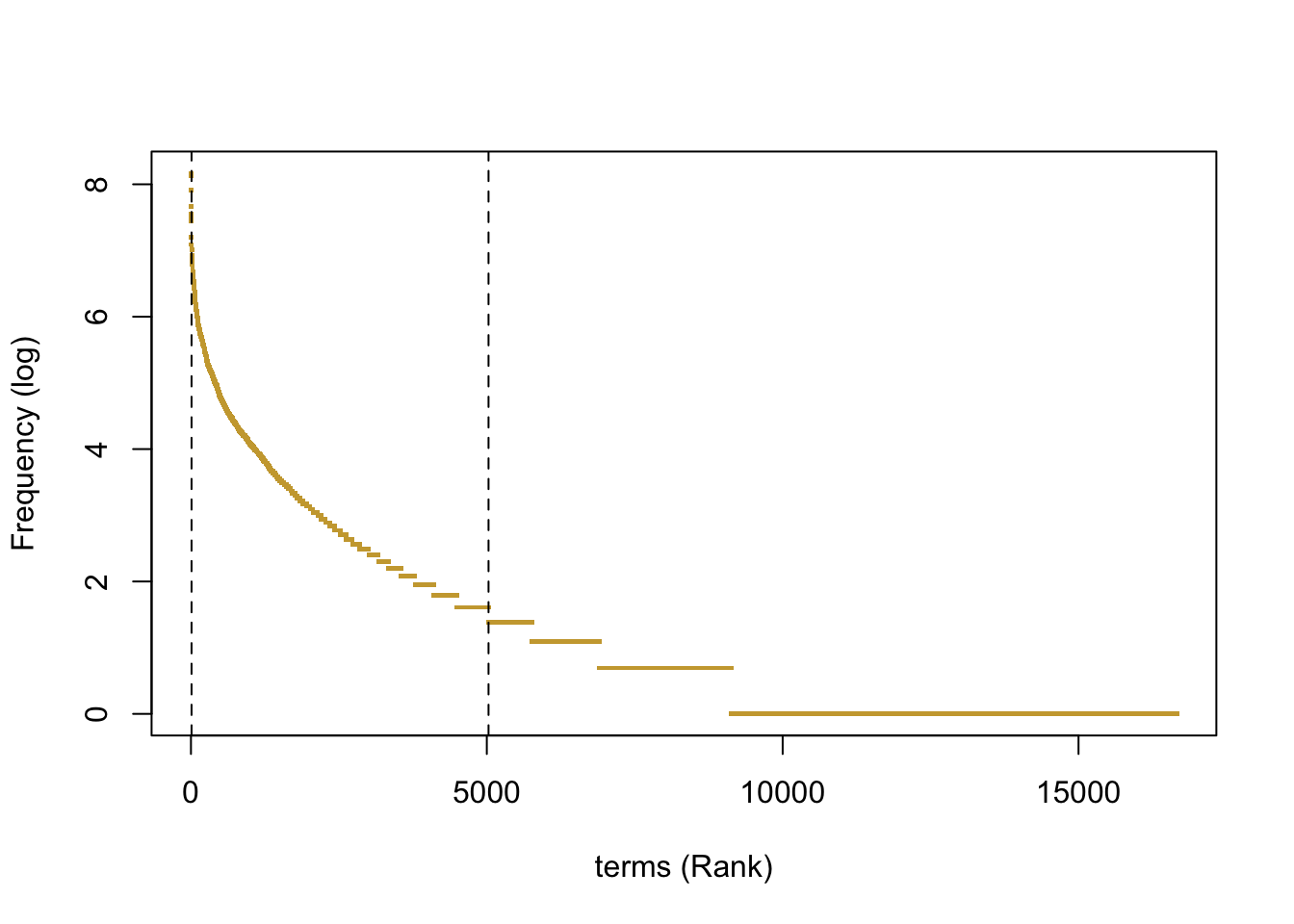
Figure 4.3: Frequency - rank plot of terms in the dtm used to build the thesaurus. Frequencies of words included in the thesaurus falled between the dashed lines.)
After setting those upper and lower frequency limits, terms can be manually classified into (Y / N) categories accordingly to its relation with the concept of frugivory. Y will represent a positive relation and N non -relation.
4.2.2 Automated Content Analysis
Due to its large and diverse applications, research on text mining and machine learning has produced a variety of algorithms and frameworks designed to extract desired information from texts (Thessen et al. 2012; 2014; 2016, Nuñez - Mair et al. 2016). Automated Content Analysis (ACA) and information extraction pipelines have been recently developed to produce and optimize qualitative and quantitative literature synthesis focused on Biological Sciences (Thessen 2012; Nuñes - Mir et al. 2016). Automated Content Analysis are capable to discover and describe concepts in large bodies of text (Blei 2012) transforming individual terms as variables to identify clusters of strongly correlated words in the body of literature that are likely to represent a determined concept or idea (Krippendorff et al. 2013 ; Nuñez - Mir et al. 2016). One of the principal advantages of ACA text mining frameworks is the training of a particular model like the naive Bayes classifier model (Peng and Schuurmans 2003) using a text input (Figure @ref(fig|aca) - Stage 3 (left side)). This becomes useful when handling a large collection of articles as a researcher could only provide a few sample training articles. However, one limitation of current ACA frameworks is not focusing on developing a programmatic way to search and retrieve of articles used as input for the framework. Using ACA and computational text mining tools enhance the reproducibility of a traditional literature search as it involves the creation of a framework, which automatically stores metadata about the work performed and can be easily shared as code.
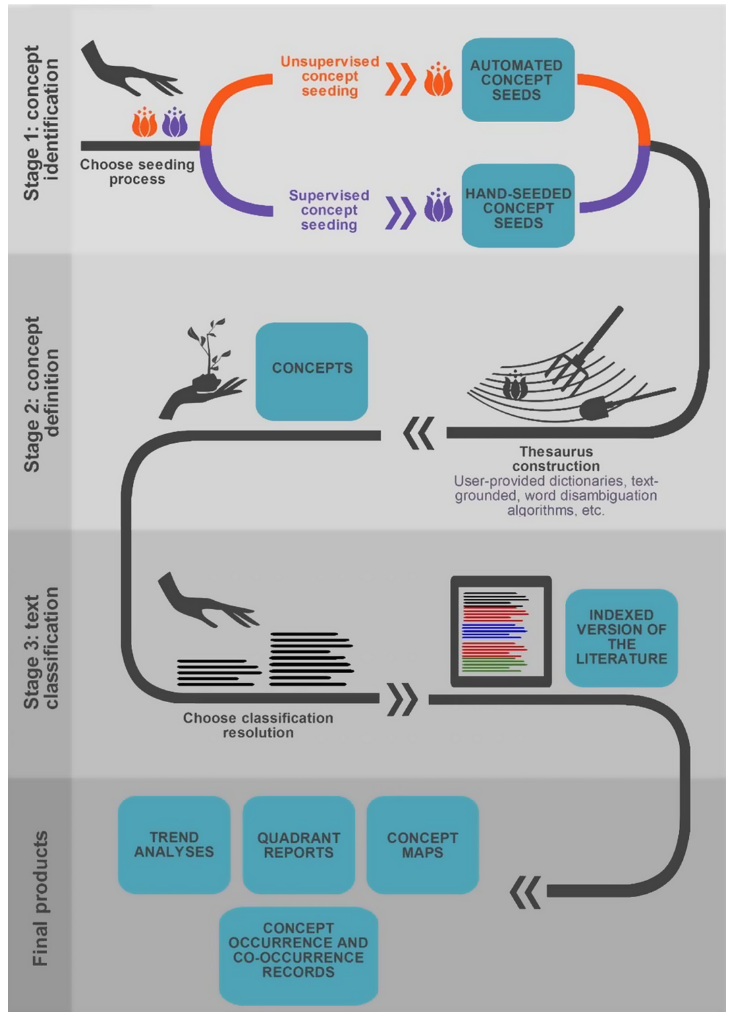
Figure 4.4: Automatic Content Analisys conceptual map, representing the three stages proposed in Nuñez - Mir et al. (2016) (obtained from Nuñez - Mair et al. (2016)
4.2.3 Text classification using Naive Bayes Classifier
The Naive - Bayes classification model is a conditional probabilistic model which assumes independence of the features used as variables to train the model (in this case the terms). Hence, the probability of finding an interaction in a text depends on the common occurrences of individual terms determined in the training dataset (Peng and Schuurmans 2003). Regardless of the general simplicity of the model and independence assumptions, naive Bayes classifiers have been extensively used in text classification frameworks, to build search engines and spam filters (Zhang et al 2007). For R there are packages like caret (Kuhn 2016) and e1071 (Meyer et al. 2015) with functions to train a naive Bayes model.
4.2.4 Text classification using Vectorization
4.2.4.1 The bag of words model
The bag of words is a classification model used for Natural Language Processing (Joachims 2002). In this model, each document of the large collection of documents is categorized by the specific subset of pieces containing text (e.g. individual words, terms or sentences) in relation of the total multi-set (bag) representing all collective pieces of text (Kosala & Blockeel 2000). The bag of words model has several applications in document classification techniques and has found its way even to applications such as computer vision models. In text mining applications, typically a bag of words is constructed from a corpus of text (i.e. A structured set containing the desired documents to which text - mining techniques can be applied). The corpus first is preprocessed to eliminate common words that appear frequently in all texts like “the”, “a”, “to”; removing punctuation marks, stemming words, etc (Gurusamy and Kannan 2014). A document term sparse matrix representing the relations between the documents and the each of the terms contained in the bag of words can be calculated. The matrix (Figure 4.5 can be filled with different metrics such as the frequencies of each term on a document.

Figure 4.5: Matrix showing the frequencies of 6 example terms on a set of 20 hypothetical articles (obtained from http://www.rdatamining.com)
Taking a closer look to the example above, every document now can be now represented as an sparse vector represented by each matrix column. Therefore:
Doc(1) = [1,0,0,0,0,0]
Doc(2) = [1,0,0,0,0,0]
.
.
Doc(19) = [0,0,0,0,1,0]
Doc(20) = [0,0,1,0,1,0] Each vector represents the determined frequency of terms in the document, in the first case, for Doc(1) the vector is showing the one-time presence of the term data in the text. The vector also provides information about the absence of the other terms. The matrix can also be transposed, transformed and simplified, depending on the specific needs in the model building phase. Additionally, terms can not necessarily comprise individual words, terms can also be pairs or n-pairs of words (N-gram model). However, calculations based only on the frequency and co-occurrence of uni-gram (words) terms (Turney et al. 2010; Pennington et al. 2014) within the document are often enough to achieve high-quality predictions on document classification tasks like Sentiment Analysis (Medhat et al. 2014).
text2vec is an R package which provides an API for text analysis and natural language processing (Selivanov 2016). One of the many advantages of this API is their memory efficiency as it uses streams and iterators, avoiding the excessive use of RAM memory. In addition, it is designed to be faster and is equipped with additional functions to create vocabularies and document term matrices. The vector resulting from the dtm matrix can be used to fit classifier models, such as logistic regression models. This last step can be performed with the function cv.glmnet from the package glmnet (Friedman et al. 2010). The function cv.glmnet also allows to performs cross-validation and L-norm penalization.
4.2.5 Building customized summaries
Regular expressions are patterns of sequential characters that a program used to match in input text (Microsoft 2017). Regular expressions allow to identify and extract specific portions of text included within a larger pool of text. Within this framework, regular expressions can be used to find and extract those portions of the articles (such as sentences) that contain specific custom defined terms. In a scientific text, each sentence tends to correspond to defined idea. The sent_detect function included in the qdap package (Rinker 2013) allows breaking the text of articles into sentences using endmark boundaries (such as “.”) to separate a continuous text into a list of sentences. Then using a list of terms (defined thesaurus, scientific names of taxa, geographical locations, etc.) with the regular expression function grepl provided in base R, a user can retrieve those sentences that match the terms provided. The result will be a summarized version of the text based on specific terms of interest for the researcher.
4.3 Specific Entity Extraction
Researchers in ecology sometimes are interested in the ecological interactions from particular taxa or for a particular location. Therefore, the taxonomic and geographical scope of an article becomes of interest for a posterior classification based on the particular interests of the researcher.
4.3.1 Extracting scientific names from text
Specific algorithms such as Neti Neti (Akella et al. 2012) and Taxon Finder (Leary 2014) have been designed with the purpose of recognizing scientific names on a given text. For example: when visualizing the digital versions of scanned documents published in the Biodiversity Heritage Library (BHL) webpage (http://www.biodiversitylibrary.org/) already include box on which the scientific names on the page are showed with the respective link to the taxon page on the Encyclopedia of Life (EOL) (http://eol.org/), this extra available information is already useful to researchers, interested on a particular species or taxon, to quickly discriminate useful pages on which to search for information.
Global Names Recognition and Discovery (GNRD) provides a web-based API that combines the names discovery engines in one framework. Global Names Recognition and Discovery tool can be queried directly with GET or POST request via R, Python or Perl. Additionally, a request option to the GNRD has been recently implemented in the package taxize. As an additional feature, GNRD could recognize names directly from PDF, txt and html formats, however, to send directly single or collection of pdf files to the API an independent request GET or POST outside the package taxize works faster. GNRD is called by the function scrapenames in the package taxize:
The taxize package (Chamberlain et al. 2016) acts as a hub of web API’s created to manage all sort of things related with scientific names and taxonomy. For instance, one could retrieve the parent taxonomical information, resolve names and connect with EOL and GBIF pages. The functionalities of this package are wide and for ecologist dealing with large lists of scientific names is a great package. I definitely recommend to explore more specific functions. ?taxize
4.3.2 Extracting location information from text.
Similarly to species scientific names, geographical information contained in the article is a valuable classifier depending on the needs of the ecologist. MonkeyLearn is a Machine Learning API specialized on extracting entities from a text. Such entities can be locations inside a text. MonkeyLearn provides an R package and although is not 100% free has a free option with limited speed and a maximum of 50.000 queries per month. (http://monkeylearn.com/pricing/). To use this API the user has to register for free in their web page and get an API key. The entity classifier id for geographical locations is ex_isnnZRbS, there are other classifiers such as US addresses, phone numbers, emails, etc. However, entity extraction is the one that fits the needs of this framework.