Automatic classification of articles.
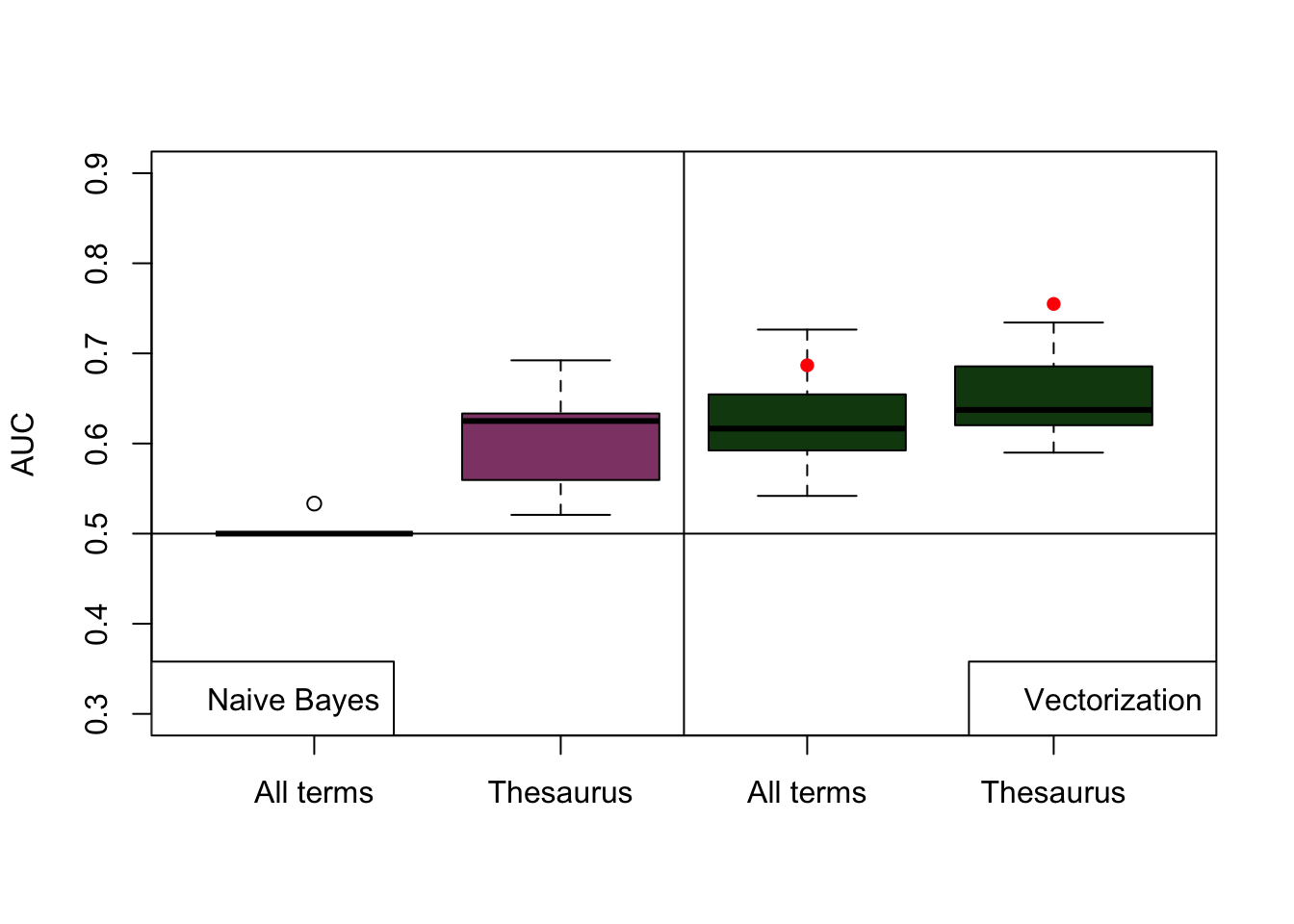
When applying naive Bayes classifier with all terms the model fails to predict the presence of interactions in frugivory in articles. The major drawback when trying to predict interactions from large pieces of text with the naive Bayes model is the underlying assumptions of independence of features of the model. naive Bayes classifier relies upon in particular the presence only of terms contained in the training corpus, the model is not making predictions based on the possible correlations between terms. Therefore, if a portion of the terms describing articles with the presence of interactions is not present in the training, the results of the predictions will not be accurate or if they are is mostly due to random factors. Using the thesaurus, as it reduces the number of variables to train the model improved its predictive capabilities. However, the model is still not accurate enough as it has a high variance in AUC from the test articles. This shows that the predictive capabilities of naive Bayes using the thesaurus as training largely depends on the type of articles used to test the model (Figure 5.1).
Vectorization methods deal better with large matrices and the vector space incorporates the variation due to the correlation between terms present in the corpus of articles. AUC from cross-validation (k = 3)of the logistic model is in overall higher than the AUC’s from naive Bayes classification (Figure 5.1). AUC from testing the model in a different set of articles are in between the ranges of the AUC obtained with cross-validation. It is necessary to note that the training of both models (naive Bayes and logistic regression) was performed with a set of 80 articles a test with 20 articles. This implies that classification models such as the ones tested in this literature review are still to uncertain to automatically classify articles and they require a relatively large training sample. The lower number of training articles are likely to reduce the predictive capabilities of the model. This limits the use of the automatic classification of literature only to a large corpus of articles. The classification models presented in this literature review are not the only options to predict the occurrence of species interactions in a text and further research is needed.
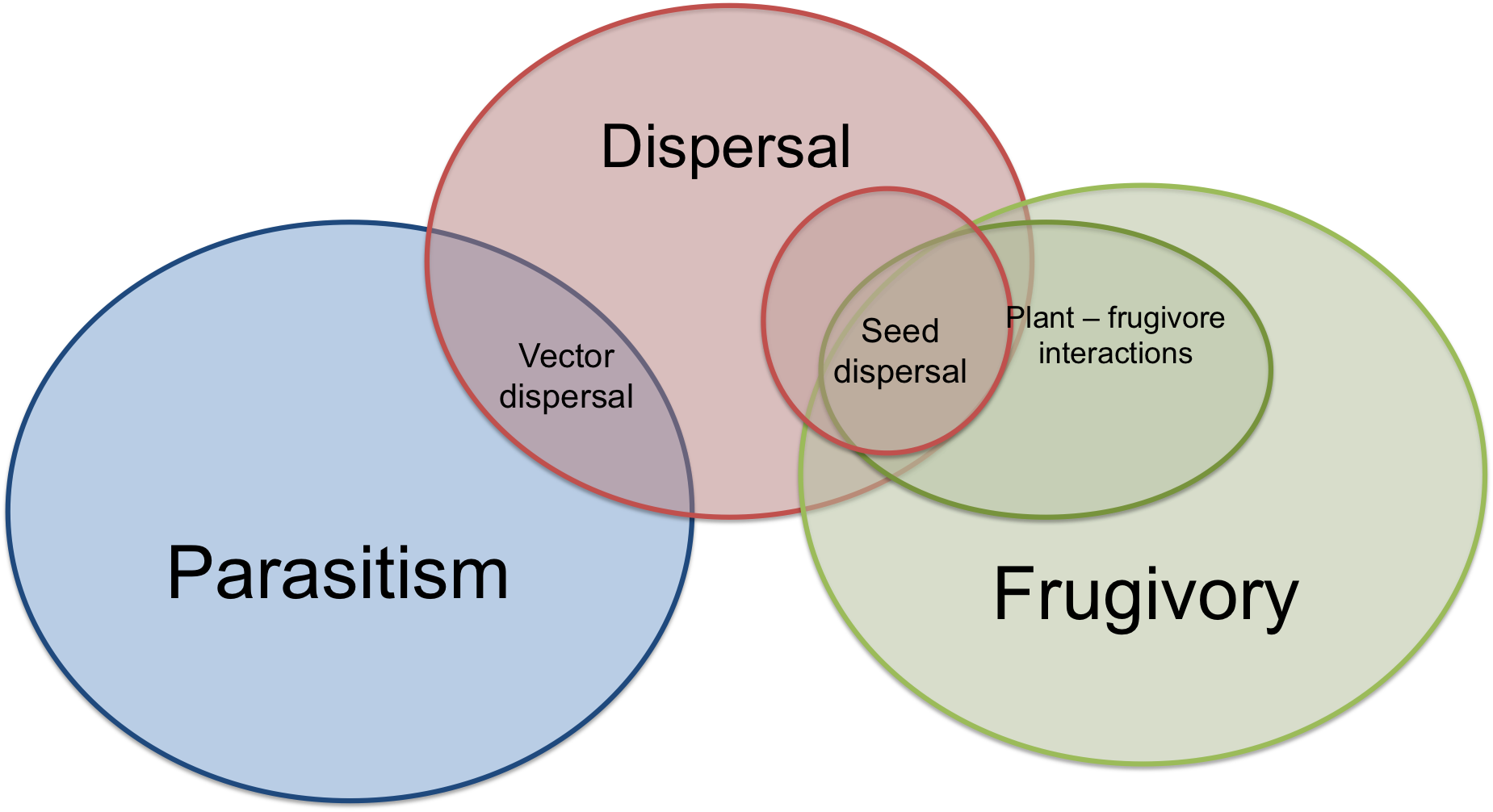
All articles present in the corpus to train/test the models are written with a scientific jargon, so they are highly semantically related. Frugivory - related terms semantically related with terms used to describe other ecological processes such as publications on host - parasite interactions or seed dispersal, which share similar terminology. This absence of pattern makes difficult the categorization of articles based on the individual terms or the vector space represented by the terms related to frugivory (Figure 5.2).
Open Source vs non - open source
Scholarly journals which provide API’s to access their content have major advantages over the ones which provide only articles as PDF format. Bulk download allows the direct incorporation of articles into computational frameworks. PDF format locks the information contained inside the article and prevent (or add extra steps) to the mining procedures. Open source journals have different schemes of publication process and fees, but all share in common that the reader is not charged to access to their content. Their infrastructure is available to all web users on the open web (usually in XML format), their content can be easily retrieved in bulk and by parts (LIBER 2014). In addition, this allows the use of researcher - developed tools and algorithms to mine and retrieve the content of interest. Open source journals have reported that the demand placed on its servers, coming from robots designed for content mining is negligible and any increase in that demand will be easy to manage (LIBER 2014). Mining licenses, provided by publisher houses (like Elsevier TDM license) aside of coming at an additional cost, explicitly prohibits the use of any automated programs to download its content, even more, by using the publisher API’s researchers are being imposed a license on their research output preventing the impact and re - use of publicly funded research (Hartgerink 2015). This has already caused limitations on researchers using mining techniques to complete their own research (see Elseiver stopped me doing my research ; Wiley also stopped me doing my research.
The right to read is the right to mine (LIBER 2014), the application of custom frameworks for text and data mining is far easier when applied on open source journals. However, this causes a limitation as it will imply that the information collected after the mining process is technically biased. However, until publishing houses let go their current way of delivering information and embrace the venues of openly published data, individual researchers (with and without institutional access) have to find ways to bypass the system and continue with the development of science. This problem is not new and there are several options available to get those papers locked behind pay walls. LibGen, BooksSC and Sci-Hub are all initiatives created with the intention to breach the gap in knowledge distribution imposed by publisher houses and made scientific knowledge available to every person in the world and promote the Open Access to Science (Sci - Hub 2016). A recent work published in Science shows that request of downloads of scientific articles to Sci - Hub servers is coming all around the world, regardless of country income or access to an institutional subscription to publisher houses (Bohannon 2016; Kramer 2016).
The debate on whether publishers should charge individual researchers for the right to read the work of peer researchers around the world is still hot (Harzing et al. 2016; Spezi et al. 2016). Given that the core of publishing scientific literature (research, editing, and peer reviewing) is done by scientists which often do not get any profit money in return; publishing houses do not usually fund research; and science advances building on the work made by others, purchasing access to read (basically) their own work seems like a really unfair deal from the researchers point of view. Publisher houses like Elsevier or Wiley charge universities and institutions to access the content published in their journals, it is estimated that European universities spend around 2 billion euros per year on subscriptions for Scientific Technical and Medical published content (LIBER 2014). To a single researcher without institutional access, reaching to information may cost up to 1000 USD per month, then “[…] Why should they (publishers) receive anything more than a small amount for managing the journal?” (Bohannon 2016).
Summarizing the article content and extracting specific features.
The number of sentences present in the summarized version of the articles is correlated with the presence of interactions. Generally, a lower number of sentences in the summary means the absence of an interaction in the article. For this corpus, summarized articles with less than 50 sentences tend to correspond to articles without records of frugivory interactions in the text. With combining programmatic searches and download of articles with the creation of customized summaries a single researcher could potentially improve the time spend into dig into the literature on a particular topic. Results of web academic engines retrieved only with few keywords can not represent the specific interest of ecologists for determined articles related to their research. Humans are prone to tiredness, customized summaries could allow to researchers to read large portions of literature with less effort. Further discrimination of relevant literature will then be made based on the semantic characteristics of the article itself, rather than titles or general abstracts. The custom summaries can also be parsed to the abstract_screener function from metagear (Lajaunesse 2016) package to manually classify articles to perform classification of articles for a meta-analysis.
Extracting locations and scientific names from the text is a good classifier of articles providing that the researcher has a previous knowledge of the geographical location or taxa of interest to perform a literature review. However, the mine of those entities is relatively slow, creating some limitations when extracting entities from corpus made with a large number of articles (500+ articles). In addition, when locations are extracted from OCR pdfs the function recognizes the locations present in the author affiliations and references sections (e.g. “Ithaca, NY” from an author coming from Cornell, or “Berlin” in references from Springer). This problem is reduced when mining articles extracted from open sources APIs (like PLOS) as it only uses the body of text as input. Nevertheless, the output of the function still has some noise produced by mentions of locations unrelated to the research location inside the text (e.g. “CO 2 was sampled by a six-port rotary valve (Valco, Houston, Texas, USA”).