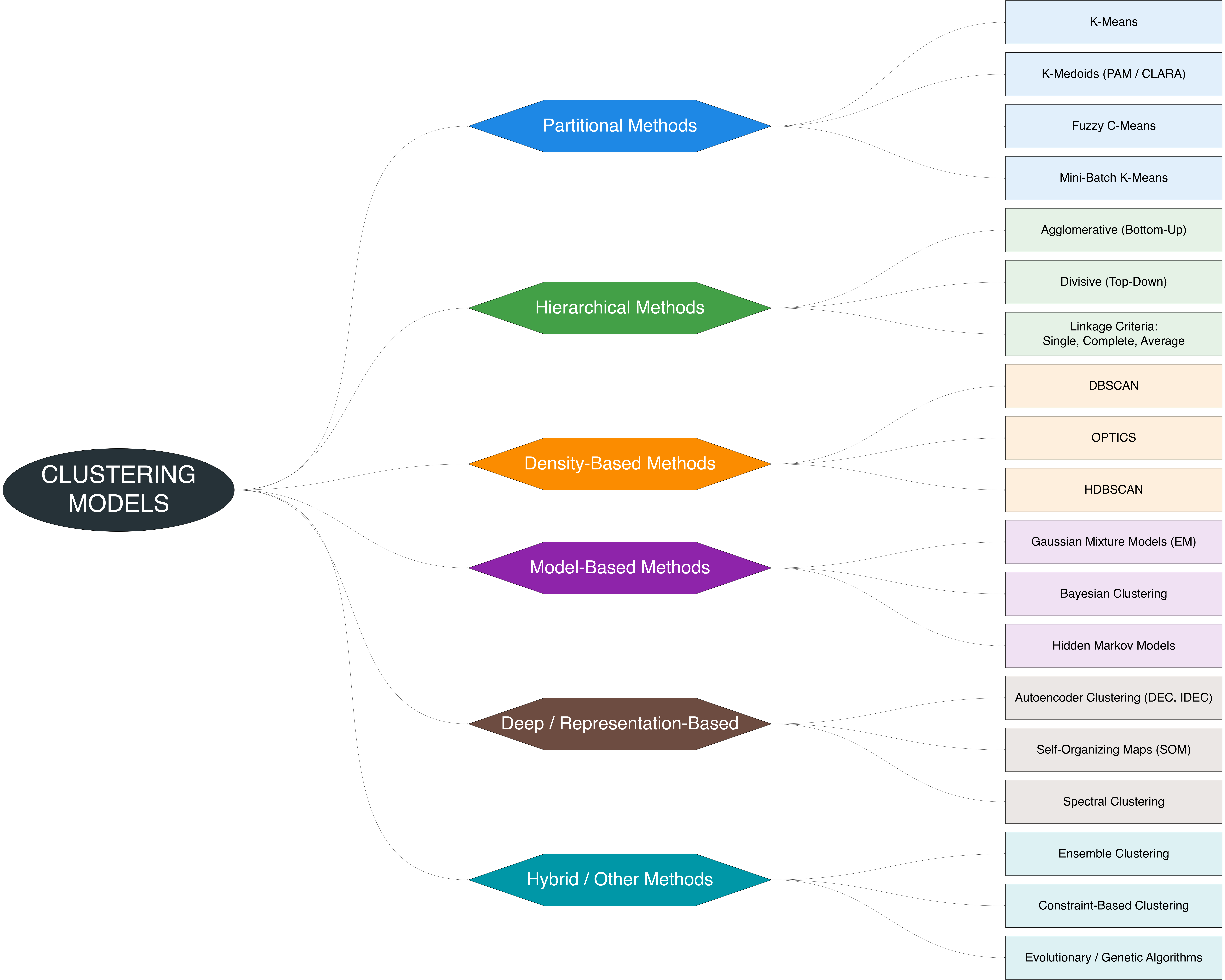
4 Clustering Models
Clustering is one of the fundamental techniques in unsupervised learning, designed to group objects based on their similarities without relying on predefined labels. This method uncovers hidden patterns, natural structures, and meaningful relationships within data across diverse applications—from customer segmentation and anomaly detection to sensor behavior analysis in industrial systems. The mind map provided offers a comprehensive overview of major clustering methodologies, including partition-based algorithms, hierarchical models, density-based approaches, probabilistic frameworks, deep learning–based representations, and hybrid techniques. This visual guide helps readers grasp the broader landscape of clustering algorithms and understand the key distinctions among them, supporting more informed decisions when selecting the most appropriate method for various analytical tasks. For additional detail, refer to Figure 4.1.
4.1 Intro to Clustering
To build an initial understanding of clustering, it is helpful to begin with a clear visual explanation. The short video below provides an accessible introduction to the core idea behind clustering—how data points are grouped based on similarity, why this process is useful, and where it is commonly applied. This foundational overview serves as a starting point before exploring the more detailed concepts and the comprehensive mind map presented in the next sections.
4.2 Partition-Based
Partition-based clustering is a fundamental technique in unsupervised learning that divides a dataset into a predefined number of non-overlapping clusters. Each data point is assigned to exactly one cluster based on a similarity or distance measure, most commonly the Euclidean distance. The objective is to create clusters that are internally cohesive and externally well separated.
In this approach, the number of clusters \(k\) must be specified in advance. The algorithm then iteratively assigns data points to clusters and updates the cluster centers (or representatives) until a stable solution is reached. Owing to its simplicity, computational efficiency, and strong performance on well-structured datasets, partition-based clustering is widely used in pattern recognition, customer segmentation, image analysis, and industrial analytics.
Representative algorithms include k-Means, which minimizes the sum of squared distances to cluster centroids; k-Medoids, which uses actual data points as cluster centers to enhance robustness against outliers; k-Means++, which improves the initialization process; and CLARA, which adapts k-Medoids for large datasets through sampling. Partition-based methods perform best when clusters are approximately spherical, similar in size, relatively free of extreme outliers, and when the value of \(k\) is known or can be estimated reliably. Although these methods can be sensitive to initialization and struggle with irregularly shaped clusters, they remain among the most widely used clustering techniques in contemporary data analysis.
4.3 Hierarchical
Hierarchical clustering is an unsupervised learning method that organizes data into a hierarchy of nested clusters, typically visualized through a dendrogram. Unlike partition-based methods, it does not require predefining the number of clusters; the structure emerges naturally from the data. It is chosen for its interpretability and ability to reveal multi-level relationships within a dataset. Analysts can observe cluster formation at various similarity thresholds, making it ideal for exploratory analysis and pattern discovery.
Researchers and practitioners in fields such as data science, bioinformatics, text mining, and anomaly detection frequently employ hierarchical clustering to analyze structured and semi-structured data. It is most effective for moderate-sized datasets where one aims to explore hierarchical structure, evaluate similarity relationships, or derive cluster insights without repeated algorithm initialization. The method is widely available in statistical and machine-learning libraries, such as R (hclust, dendextend) and Python (scipy.cluster.hierarchy, sklearn).
Hierarchical clustering operates through two strategies:
- gglomerative (bottom-up): each point starts as its own cluster, and the most similar clusters are merged step-by-step based on linkage criteria (single, complete, average, Ward).
- Divisive (top-down): all points begin in one cluster, which is recursively divided into smaller groups.
Clusters are finally obtained by cutting the dendrogram at the desired distance or similarity threshold.
4.4 Density-Based
Density-Based Clustering is a clustering approach that groups data points based on the density of observations within a region, where areas with high concentrations of points form clusters and sparse regions are treated as noise or outliers. This method matters because it can identify clusters of arbitrary shapes—irregular, elongated, or non-convex—that are difficult for partition-based methods like K-Means to detect. It is also advantageous because it does not require predefining the number of clusters, making it particularly valuable for exploratory analysis when the underlying structure of the data is unknown.
This technique is well suited for analysts and researchers working with spatial data, sensor data, operational measurements, or datasets with uneven point distribution. Density-based methods are especially useful when the dataset contains many outliers, when cluster boundaries are not well defined, or when the analysis involves anomaly detection. In practical applications, it is widely used in geospatial analysis to identify hotspots, in industrial monitoring to detect abnormal sensor patterns, in cybersecurity to flag suspicious activities, and in finance for identifying unusual transactions.
Operationally, Density-Based Clustering functions by defining a search radius (ε) and a minimum number of points (MinPts) required to form a dense region. Points with enough neighbors are classified as core points, points that are within the neighborhood of a core point are considered border points, and all remaining points are labeled as noise. Clusters emerge through chains of connected dense regions, allowing complex and natural patterns in the data to be captured without forcing rigid geometric shapes.
While the method has limitations—most notably sensitivity to parameter selection and reduced performance in high-dimensional spaces—its ability to model real-world cluster structures and detect outliers effectively makes Density-Based Clustering, especially DBSCAN, one of the most powerful and widely used techniques in modern data analysis.
4.5 Probabilistic
Probabilistic clustering is a method of unsupervised learning where clusters are defined by probability distributions rather than fixed boundaries. It addresses what the structure of the data looks like by estimating how likely each data point belongs to each cluster, instead of forcing a hard assignment. This approach is used when datasets exhibit overlapping groups, hidden latent structure, or uncertainty in boundaries—conditions where deterministic clustering fails to capture the true relationships. It is applied where soft assignments are valuable, such as text mining, image classification, medical diagnostics, and anomaly detection. The technique relies on why probabilistic modeling is effective: it naturally handles ambiguity, provides richer information through membership probabilities, and supports principled model comparison using likelihood-based criteria. Methods such as Gaussian Mixture Models (GMM) and Expectation–Maximization (EM) clustering represent who performs the clustering—the probabilistic components that model each group as a distribution. Finally, the process describes how clustering is performed: by estimating the parameters of each distribution, computing membership probabilities for every point, and iteratively updating these values until the likelihood converges. This distribution-driven, uncertainty-aware perspective makes probabilistic clustering a powerful tool for understanding complex, real-world data.
4.6 Deep Learning-Based
Deep learning–based clustering is an advanced approach that uses deep neural networks to learn meaningful representations of high-dimensional or unstructured data before performing clustering. Unlike traditional methods that rely on raw features, this technique transforms data into a latent space where important patterns are emphasized and noise is reduced. It is used because many real-world datasets—such as images, text, audio, sensor streams, and industrial signals—contain complex, non-linear structures that classical algorithms like k-Means or DBSCAN cannot model effectively. This approach benefits researchers and practitioners in fields such as computer vision, natural language processing, bioinformatics, and intelligent analytics, where discovering hidden structure is essential. It is applied in tasks like image grouping, document organization, anomaly detection, user-behavior modeling, and representation learning. Deep learning–based clustering is most suitable when the dataset is large, high-dimensional, or difficult to separate using traditional techniques, especially when feature engineering becomes challenging. It works by training a neural network—typically an autoencoder or embedding model—to map data into a compact latent space, then clustering those embeddings using algorithms such as k-Means or Gaussian Mixture Models. More advanced methods, such as Deep Embedded Clustering (DEC), integrate representation learning and clustering into a single end-to-end optimization process, producing more coherent and well-separated clusters.
4.7 Hybrid
Hybrid clustering combines two or more clustering approaches to leverage their complementary strengths and overcome the limitations of individual methods. It is used when datasets are complex, noisy, high-dimensional, or contain clusters with irregular shapes that cannot be captured effectively by a single technique. Hybrid clustering is important because real-world data often exhibits mixed structures—some regions may form dense clusters, others may follow hierarchical patterns, while some require partitioning for refinement—making hybrid solutions more flexible and accurate. It is applied in domains such as bioinformatics, customer segmentation, image processing, anomaly detection, and large-scale industrial analytics, where robust and adaptive clustering is essential. Hybrid clustering works by combining methods such as density-based algorithms (e.g., DBSCAN) to detect core structures, hierarchical clustering to capture multi-level relationships, and partition-based methods like k-Means to refine cluster boundaries. More advanced hybrid models may also integrate spectral clustering, probabilistic approaches, or deep-learning-based embeddings to improve performance. This approach is most suitable when the data contains both dense and sparse regions, varies in cluster size or shape, or when a single method consistently fails to capture the underlying structure. By integrating complementary strategies, hybrid clustering produces more stable, interpretable, and high-quality clustering results across diverse data scenarios.
4.8 Applied of Clustering
4.8.1 Partition Based
Load Libraries & Dataset
library(factoextra)
library(cluster)
library(e1071) # Fuzzy C-Means
library(ClusterR) # Mini-Batch KMeans
library(plotly)
library(dplyr)Load dataset
data(iris)
iris_data <- iris[,1:4]
iris_labels <- iris$Species
# Scale data
iris_data_scale <- scale(iris_data)
set.seed(123)Determine Optimal Number of Clusters
Elbow Method (WSS):
wss <- function(k) kmeans(iris_data_scale, k, nstart=25)$tot.withinss
k.values <- 1:10
wss_values <- sapply(k.values, wss)
fig_elbow <- plot_ly(
x=k.values, y=wss_values, type='scatter', mode='lines+markers',
marker=list(size=8, color='#1E88E5'), line=list(color='#1E88E5')
) %>% layout(title="Elbow Method (WSS)", xaxis=list(title="Number of Clusters"), yaxis=list(title="Total WSS"))
fig_elbowGap Statistic:
gap_stat <- clusGap(iris_data_scale, FUN=kmeans, nstart=25, K.max=10, B=50)
gap_df <- data.frame(
k=1:10,
gap=gap_stat$Tab[, "gap"],
SE=gap_stat$Tab[, "SE.sim"]
)
fig_gap <- plot_ly(
gap_df, x=~k, y=~gap, type='scatter', mode='lines+markers',
error_y=list(type="data", array=gap_df$SE),
marker=list(size=8, color='#2CA02C'), line=list(color='#2CA02C')
) %>% layout(title="Gap Statistic", xaxis=list(title="Number of Clusters"), yaxis=list(title="Gap Value"))
fig_gapSilhouette Method:
sil_width <- sapply(2:10, function(k){
km <- kmeans(iris_data_scale, centers=k, nstart=25)
ss <- silhouette(km$cluster, dist(iris_data_scale))
mean(ss[,3])
})
fig_sil <- plot_ly(
x=2:10, y=sil_width, type='scatter', mode='lines+markers',
marker=list(size=8, color='#D62728'), line=list(color='#D62728')
) %>% layout(title="Silhouette Method", xaxis=list(title="Number of Clusters"), yaxis=list(title="Average Silhouette Width"))
fig_sil4.8.1.1 Apply Clustering Algorithms (k=3)
# ----- 1. K-Means -----
km <- kmeans(iris_data_scale, centers=3, nstart=25)
iris$KMeans <- as.factor(km$cluster)# ----- 2. K-Medoids (PAM) -----
pam_res <- pam(iris_data_scale, k=3)
iris$KMedoids <- as.factor(pam_res$clustering)# ----- 3. Fuzzy C-Means -----
fcm <- cmeans(iris_data_scale, centers=3, m=2)
iris$FuzzyCMeans <- as.factor(apply(fcm$membership,1,which.max))library(ClusterR)
set.seed(123)
# Data preparation
iris_data <- iris[, 1:4]
iris_data_scale <- scale(iris_data)
iris_mat <- as.matrix(iris_data_scale)
# Train Mini-Batch K-means
mb <- MiniBatchKmeans(
data = iris_mat,
clusters = 3,
batch_size = 20,
num_init = 5,
max_iters = 30
)
# New recommended way:
# Use 'predict' S3 method for MiniBatchKmeans
pred <- predict(
object = mb,
newdata = iris_mat,
fuzzy = FALSE # ensures hard clusters, not probabilities
)
# Add cluster labels
iris$MiniBatch <- as.factor(pred)
# Check cluster distribution
table(iris$MiniBatch)
1 2 3
50 49 51 2D PCA Visualization (Plotly)
library(dplyr)
library(plotly)
# PCA
pca <- prcomp(iris_data_scale)
pca_df <- data.frame(
PC1 = pca$x[, 1],
PC2 = pca$x[, 2],
KMeans = iris$KMeans,
KMedoids = iris$KMedoids,
FuzzyCMeans = iris$FuzzyCMeans,
MiniBatch = iris$MiniBatch,
Species = iris_labels
)
# Plot function (clean + safe)
plot_cluster <- function(df, cluster_col, title) {
plot_ly(
df,
x = ~PC1,
y = ~PC2,
color = as.formula(paste0("~", cluster_col)),
symbol = ~Species,
symbols = c("circle", "diamond", "square"),
type = "scatter",
mode = "markers",
marker = list(size = 8, opacity = 0.85)
) %>%
layout(title = title)
}
# Figures
fig_km <- plot_cluster(pca_df, "KMeans", "K-Means")
fig_kmed <- plot_cluster(pca_df, "KMedoids", "K-Medoids")
fig_fcm <- plot_cluster(pca_df, "FuzzyCMeans", "Fuzzy C-Means")
fig_mbkm <- plot_cluster(pca_df, "MiniBatch", "Mini-Batch K-Means")Combined Plotly Dashboard
library(plotly)
# ----- Dashboard 1: Evaluation Methods -----
dashboard_eval <- subplot(
fig_elbow, fig_gap, fig_sil,
nrows = 3,
margin = 0.05,
shareX = FALSE, shareY = FALSE,
titleX = TRUE, titleY = TRUE
) %>% layout(
title = list(
text = "<b>Clustering Evaluation Summary</b>",
font = list(size = 20)
)
)
# ----- Dashboard 2: PCA Cluster Visualization -----
dashboard_clusters <- subplot(
fig_km, fig_kmed,
fig_fcm, fig_mbkm,
nrows = 2,
margin = 0.05,
shareX = FALSE, shareY = FALSE,
titleX = TRUE, titleY = TRUE
) %>% layout(
title = list(
text = "<b>PCA Visualization of Clustering Results</b>",
font = list(size = 20)
)
)
dashboard_evaldashboard_clustersNotes:
- Row 1: Cluster selection methods (Elbow / Gap / Silhouette)
- Rows 2-3: Four clustering algorithms with 2D PCA
Cluster Evaluation
cat("K-Means vs Species:\n"); table(iris$KMeans, iris_labels)K-Means vs Species: iris_labels
setosa versicolor virginica
1 0 11 36
2 50 0 0
3 0 39 14cat("\nK-Medoids vs Species:\n"); table(iris$KMedoids, iris_labels)
K-Medoids vs Species: iris_labels
setosa versicolor virginica
1 50 0 0
2 0 9 36
3 0 41 14cat("\nFuzzy C-Means vs Species:\n"); table(iris$FuzzyCMeans, iris_labels)
Fuzzy C-Means vs Species: iris_labels
setosa versicolor virginica
1 50 0 0
2 0 39 13
3 0 11 37cat("\nMini-Batch K-Means vs Species:\n"); table(iris$MiniBatch, iris_labels)
Mini-Batch K-Means vs Species: iris_labels
setosa versicolor virginica
1 50 0 0
2 0 37 12
3 0 13 38Analysis:
- Setosa: typically forms a clear cluster in all algorithms
- Versicolor & Virginica: overlapping → Fuzzy C-Means shows memberships
- Mini-Batch K-Means: similar to regular K-Means but faster
- Gap / Silhouette: help to validate if k=3 is optimal
4.9 Summary Clustering Models
The table below (Table 4.1) provides a comprehensive summary of the major categories of clustering methods, the algorithms contained within each category, their core formulas, and commonly used packages for implementation in both R and Python. This presentation is intended to help readers understand the conceptual distinctions among the approaches while also identifying practical tools available for real-world applications.
| Category | Algorithms | Key Formula | R Packages | Python Packages |
|---|---|---|---|---|
| Partition-Based | k-Means, k-Medoids | \(\min \sum_{i=1}^k \sum_{x \in C_i} \|x - \mu_i\|^2\) | stats::kmeans; cluster::pam | sklearn: KMeans, KMedoids |
| Partition-Based | k-Means++ | Probabilistic center initialization: \(P(x)=\frac{D(x)^2}{\sum D(x)^2}\) | ClusterR::KMeans_rcpp | sklearn: KMeans (k-means++) |
| Partition-Based | CLARA (Clustering Large Applications) | Sampling + PAM applied to large datasets | cluster::clara | pyclustering |
| Hierarchical | Agglomerative (Bottom-Up) | Distance linkage: single, complete, average | stats::hclust; dendextend | sklearn: AgglomerativeClustering |
| Hierarchical | Divisive (Top-Down) | Recursive splitting by maximal dissimilarity | stats::hclust | scipy.cluster.hierarchy |
| Density-Based | DBSCAN | Core rule: \(|N_\varepsilon(x)| \geq MinPts\) | dbscan::dbscan | sklearn.cluster.DBSCAN |
| Density-Based | HDBSCAN | Density hierarchy via Minimum Spanning Tree | dbscan::hdbscan | hdbscan |
| Probabilistic | Gaussian Mixture Models (GMM) | \(p(x)=\sum_k \pi_k \mathcal{N}(x|\mu_k,\Sigma_k)\) | mclust | sklearn.mixture.GaussianMixture |
| Probabilistic | EM Clustering (Expectation–Maximization) | Iterative E-step & M-step until convergence | EMCluster | sklearn.mixture |
| Deep Learning-Based | Autoencoder-Based Clustering | Latent code: \(z = f_\theta(x)\) | keras; torch | TensorFlow; PyTorch |
| Deep Learning-Based | Deep Embedded Clustering (DEC) | Joint reconstruction + clustering loss | keras; torch | TensorFlow; PyTorch |
| Hybrid | Spectral Clustering | Graph Laplacian: \(L = D - W\) | kernlab::specc | sklearn.cluster.SpectralClustering |
| Hybrid | Affinity Propagation | Similarity-based message passing | apcluster | sklearn.cluster.AffinityPropagation |
