6 Exercise 2
6.1 Exercise 2 part 1 - Transforming Y with ln
Note that the default understanding of log in R is \(log_e\) = ln = log.
(a) Perform analysis the data frame SIMDATAST to create and display six graphs using a 2 by 3 layout.
- Produce a scatterplot of \(y_1\) versus \(x_1\).
# Begin by setting up the 2x3 grid
par(mfrow = c(2, 3))
# Plot y1 vs x1
plot(y1 ~ x1, data = SIMDATAST)- Plot the residuals versus fits for the model created by regressing \(y_1\) on \(x_1\) (call this model modx1). Based on the first two graphs, does a logarithmic transformation for the response variable make sense?
# Define the model
modx1 <- lm(y1 ~ x1, data = SIMDATAST)
# Plot the residual graphs
plot(modx1, which = c(1,2))
#This plot function will produce many analytical graphs so using c(1,2) selects that you want the fitted residual plot and the Q-Q plot as they are the first 2 it produces.
(iii) In the second row of graphs, create a scatterplot of \(ln(y_1\)) versus \(x_1\).
# Plot y1 vs ln(x1)
plot(log(y1) ~ x1, data = SIMDATAST)
(iv) Create a plot of the residuals versus the fits for the model and the Q-Q normal plot for log(\(y_1\)) ~ \(x_1\).
# Use the plot function:
plot(lm(log(y1) ~ x1, data = SIMDATAST), which = c(1,2))
# when Par is run along with the rest of this code it will put the 6 graphs in the grid.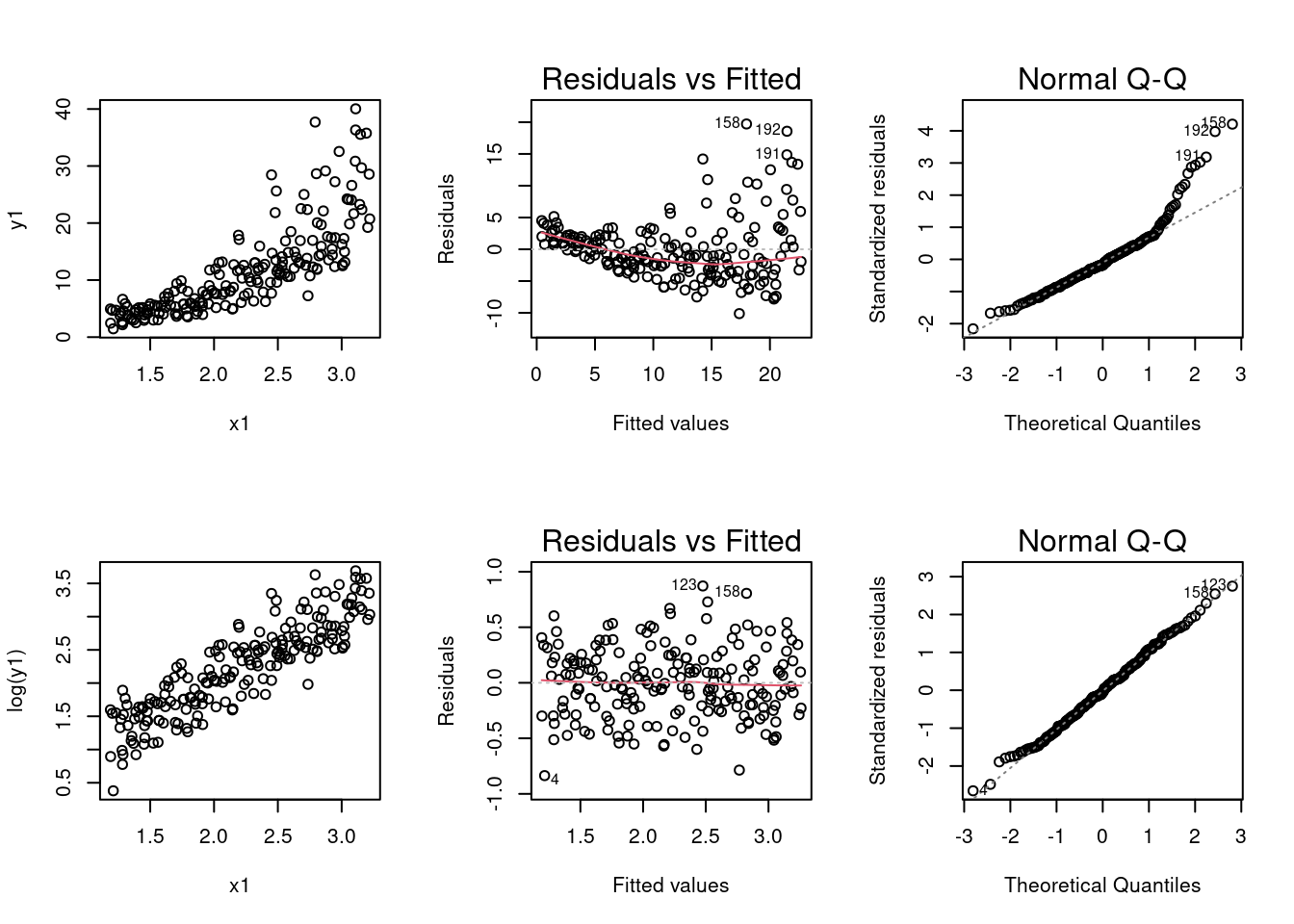
- Based on the second row of graphs, do the assumptions for the normal error model seem to be satisfied for the model log(\(y_1\)) ~ \(x_1\)?
6.2 Exercise 2 part 2 - Transforming Y with a reciprical
(a) Perform analysis the data frame SIMDATAST to create and display six graphs using a 2 by 3 layout.
- Produce a scatterplot of \(y_2\) versus \(x_2\).
# Begin by setting up the 2x3 grid
par(mfrow = c(2, 3))
# Plot y1 vs x1
plot(y2 ~ x2, data = SIMDATAST)- Plot the residuals versus fits for the model created by regressing \(y_2\) on \(x_2\) (call this model modx2).
# Define the model
modx2 <- lm(y2 ~ x2, data = SIMDATAST)
# Plot the residual graphs
plot(modx2, which = c(1,2))
#This plot function will produce many analytical graphs so using c(1,2) selects that you want the fitted residual plot and the Q-Q plot as they are the first 2 it produces.Based on the first two graphs, does a reciprocal transformation for the response variable make sense?
In the second row of graphs, create a scatterplot of \(y_2^{-1}\) versus\(x_2\).
# Plot y1 vs ln(x1)
plot((y2)^-1 ~ x2, data = SIMDATAST)- Create a plot of the residuals versus the fits and the Q-Q normal plot for \(y_2^{-1} \sim x_2\).
# Use the plot function:
plot(lm((y2)^-1 ~ x2, data = SIMDATAST), which = c(1,2))
# when Par is run along with the rest of this code it will put the 6 graphs in the grid.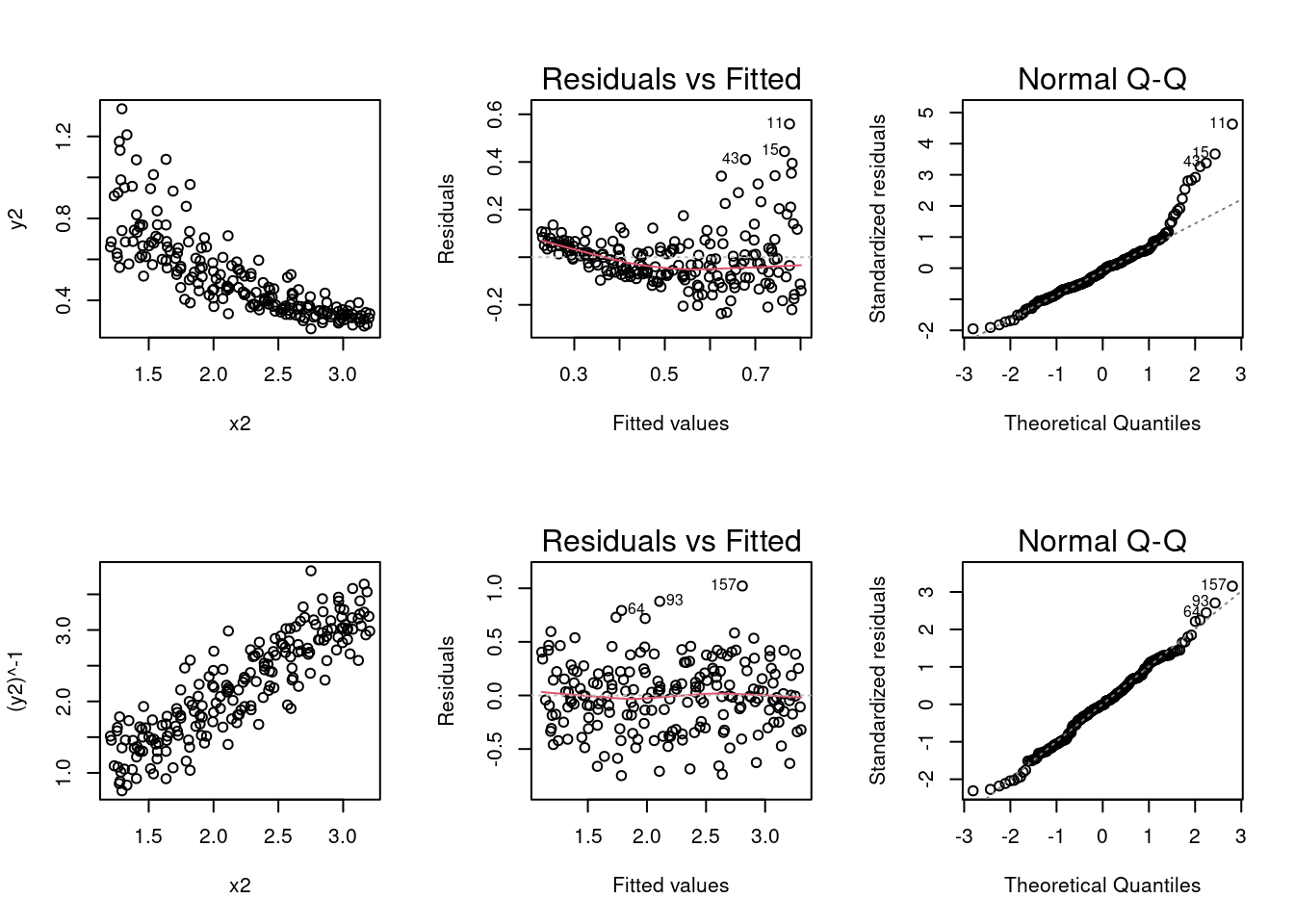
- Based on the second row of graphs, do the assumptions for the normal error model seem to be satisfied for the model \(y_2^{-1} \sim x_2\)?