Chapter 9 The dplyr package (week 6)
9.1 What is dplyr?
When you import your own data into R (probably using the
readrpackage intidyverse), it is rare that you get the data in the desired form you need for your data visualization and data modeling. Thedplyrpackage provides a set of functions to transform or manipulate your data into the right form you need.The official tidyverse website (https://dplyr.tidyverse.org/index.html) introduces the
dplyrpackage as follows:
"dplyr is a grammar of data manipulation, providing a consistent set of verbs that help you solve the most common data manipulation challenges:
mutate()adds new variables that are functions of existing variablesselect()picks variables based on their names.filter()picks cases based on their values.summarise()reduces multiple values down to a single summary.arrange()changes the ordering of the rows.
These all combine naturally with
group_by()which allows you to perform any operation “by group”. You can learn more about them invignette("dplyr"). As well as these single-table verbs, dplyr also provides a variety of two-table verbs, which you can learn about invignette("two-table").
If you are new to dplyr, the best place to start is the data transformation chapter in R for data science."
- usage
verb(a data frame, what to do with the data frame)- e.g.,
filter(flights, month == 1)# filter theflightsdata frame to select all flights that departed on Jan.
9.2 Cheat Sheet
- The cheatsheet for the
dplyrpackage provides nice diagrams illustrating the functionality of various functions in thedplyrpackage.
9.3 Tibbles
- In Base R, we often use a data frame to store our data. A tibble is the tidyverse version of the data frame in Base R. Tidyverse functions take both the data frame and tibble. Often you may want to explicitly convert a data frame to a tibble using
as_tibble():
## # A tibble: 150 x 5
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <dbl> <dbl> <dbl> <dbl> <fct>
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## # ... with 140 more rowsFor more details about a tibble, you can check here.
9.4 Select columns with select()
select()subsets columns
## [1] "year" "month" "day" "dep_time"
## [5] "sched_dep_time" "dep_delay" "arr_time" "sched_arr_time"
## [9] "arr_delay" "carrier" "flight" "tailnum"
## [13] "origin" "dest" "air_time" "distance"
## [17] "hour" "minute" "time_hour"## # A tibble: 336,776 x 3
## year month day
## <int> <int> <int>
## 1 2013 1 1
## 2 2013 1 1
## 3 2013 1 1
## 4 2013 1 1
## 5 2013 1 1
## 6 2013 1 1
## 7 2013 1 1
## 8 2013 1 1
## 9 2013 1 1
## 10 2013 1 1
## # ... with 336,766 more rowsTidy selection provides many different ways for selecting variables. You can find more details about the tidy selection here.
:for selecting a range of consecutive variables
## # A tibble: 336,776 x 3
## year month day
## <int> <int> <int>
## 1 2013 1 1
## 2 2013 1 1
## 3 2013 1 1
## 4 2013 1 1
## 5 2013 1 1
## 6 2013 1 1
## 7 2013 1 1
## 8 2013 1 1
## 9 2013 1 1
## 10 2013 1 1
## # ... with 336,766 more rows!for taking the complement of a set of variables.
## # A tibble: 336,776 x 16
## dep_time sched_dep_time dep_delay arr_time sched_arr_time arr_delay carrier
## <int> <int> <dbl> <int> <int> <dbl> <chr>
## 1 517 515 2 830 819 11 UA
## 2 533 529 4 850 830 20 UA
## 3 542 540 2 923 850 33 AA
## 4 544 545 -1 1004 1022 -18 B6
## 5 554 600 -6 812 837 -25 DL
## 6 554 558 -4 740 728 12 UA
## 7 555 600 -5 913 854 19 B6
## 8 557 600 -3 709 723 -14 EV
## 9 557 600 -3 838 846 -8 B6
## 10 558 600 -2 753 745 8 AA
## # ... with 336,766 more rows, and 9 more variables: flight <int>,
## # tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>, distance <dbl>,
## # hour <dbl>, minute <dbl>, time_hour <dttm>Selection helper functions select variables by matching patterns in their names:
starts_with(): Starts with a prefix.
ends_with(): Ends with a suffix.
contains(): Contains a literal string.
matches(): Matches a regular expression.
num_range(): Matches a numerical range like x01, x02, x03.
## # A tibble: 336,776 x 2
## dep_time dep_delay
## <int> <dbl>
## 1 517 2
## 2 533 4
## 3 542 2
## 4 544 -1
## 5 554 -6
## 6 554 -4
## 7 555 -5
## 8 557 -3
## 9 557 -3
## 10 558 -2
## # ... with 336,766 more rows## # A tibble: 336,776 x 5
## dep_time sched_dep_time arr_time sched_arr_time air_time
## <int> <int> <int> <int> <dbl>
## 1 517 515 830 819 227
## 2 533 529 850 830 227
## 3 542 540 923 850 160
## 4 544 545 1004 1022 183
## 5 554 600 812 837 116
## 6 554 558 740 728 150
## 7 555 600 913 854 158
## 8 557 600 709 723 53
## 9 557 600 838 846 140
## 10 558 600 753 745 138
## # ... with 336,766 more rows## # A tibble: 336,776 x 6
## dep_time sched_dep_time arr_time sched_arr_time air_time time_hour
## <int> <int> <int> <int> <dbl> <dttm>
## 1 517 515 830 819 227 2013-01-01 05:00:00
## 2 533 529 850 830 227 2013-01-01 05:00:00
## 3 542 540 923 850 160 2013-01-01 05:00:00
## 4 544 545 1004 1022 183 2013-01-01 05:00:00
## 5 554 600 812 837 116 2013-01-01 06:00:00
## 6 554 558 740 728 150 2013-01-01 05:00:00
## 7 555 600 913 854 158 2013-01-01 06:00:00
## 8 557 600 709 723 53 2013-01-01 06:00:00
## 9 557 600 838 846 140 2013-01-01 06:00:00
## 10 558 600 753 745 138 2013-01-01 06:00:00
## # ... with 336,766 more rows# select all columns starts with "dep", contains "time", and between year and day (inclusive)
select(flights, starts_with("dep"), contains("time"), year:day)## # A tibble: 336,776 x 10
## dep_time dep_delay sched_dep_time arr_time sched_arr_time air_time
## <int> <dbl> <int> <int> <int> <dbl>
## 1 517 2 515 830 819 227
## 2 533 4 529 850 830 227
## 3 542 2 540 923 850 160
## 4 544 -1 545 1004 1022 183
## 5 554 -6 600 812 837 116
## 6 554 -4 558 740 728 150
## 7 555 -5 600 913 854 158
## 8 557 -3 600 709 723 53
## 9 557 -3 600 838 846 140
## 10 558 -2 600 753 745 138
## # ... with 336,766 more rows, and 4 more variables: time_hour <dttm>,
## # year <int>, month <int>, day <int>## # A tibble: 336,776 x 3
## year day sched_dep_time
## <int> <int> <int>
## 1 2013 1 515
## 2 2013 1 529
## 3 2013 1 540
## 4 2013 1 545
## 5 2013 1 600
## 6 2013 1 558
## 7 2013 1 600
## 8 2013 1 600
## 9 2013 1 600
## 10 2013 1 600
## # ... with 336,766 more rows# billboard is a dataset in the tidyverse package
# billboard contains song rankings for billboard top 100 in 2000
billboard## # A tibble: 317 x 79
## artist track date.entered wk1 wk2 wk3 wk4 wk5 wk6 wk7 wk8
## <chr> <chr> <date> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2 Pac Baby~ 2000-02-26 87 82 72 77 87 94 99 NA
## 2 2Ge+h~ The ~ 2000-09-02 91 87 92 NA NA NA NA NA
## 3 3 Doo~ Kryp~ 2000-04-08 81 70 68 67 66 57 54 53
## 4 3 Doo~ Loser 2000-10-21 76 76 72 69 67 65 55 59
## 5 504 B~ Wobb~ 2000-04-15 57 34 25 17 17 31 36 49
## 6 98^0 Give~ 2000-08-19 51 39 34 26 26 19 2 2
## 7 A*Tee~ Danc~ 2000-07-08 97 97 96 95 100 NA NA NA
## 8 Aaliy~ I Do~ 2000-01-29 84 62 51 41 38 35 35 38
## 9 Aaliy~ Try ~ 2000-03-18 59 53 38 28 21 18 16 14
## 10 Adams~ Open~ 2000-08-26 76 76 74 69 68 67 61 58
## # ... with 307 more rows, and 68 more variables: wk9 <dbl>, wk10 <dbl>,
## # wk11 <dbl>, wk12 <dbl>, wk13 <dbl>, wk14 <dbl>, wk15 <dbl>, wk16 <dbl>,
## # wk17 <dbl>, wk18 <dbl>, wk19 <dbl>, wk20 <dbl>, wk21 <dbl>, wk22 <dbl>,
## # wk23 <dbl>, wk24 <dbl>, wk25 <dbl>, wk26 <dbl>, wk27 <dbl>, wk28 <dbl>,
## # wk29 <dbl>, wk30 <dbl>, wk31 <dbl>, wk32 <dbl>, wk33 <dbl>, wk34 <dbl>,
## # wk35 <dbl>, wk36 <dbl>, wk37 <dbl>, wk38 <dbl>, wk39 <dbl>, wk40 <dbl>,
## # wk41 <dbl>, wk42 <dbl>, wk43 <dbl>, wk44 <dbl>, wk45 <dbl>, wk46 <dbl>,
## # wk47 <dbl>, wk48 <dbl>, wk49 <dbl>, wk50 <dbl>, wk51 <dbl>, wk52 <dbl>,
## # wk53 <dbl>, wk54 <dbl>, wk55 <dbl>, wk56 <dbl>, wk57 <dbl>, wk58 <dbl>,
## # wk59 <dbl>, wk60 <dbl>, wk61 <dbl>, wk62 <dbl>, wk63 <dbl>, wk64 <dbl>,
## # wk65 <dbl>, wk66 <lgl>, wk67 <lgl>, wk68 <lgl>, wk69 <lgl>, wk70 <lgl>,
## # wk71 <lgl>, wk72 <lgl>, wk73 <lgl>, wk74 <lgl>, wk75 <lgl>, wk76 <lgl>## [1] "artist" "track" "date.entered" "wk1" "wk2"
## [6] "wk3" "wk4" "wk5" "wk6" "wk7"
## [11] "wk8" "wk9" "wk10" "wk11" "wk12"
## [16] "wk13" "wk14" "wk15" "wk16" "wk17"
## [21] "wk18" "wk19" "wk20" "wk21" "wk22"
## [26] "wk23" "wk24" "wk25" "wk26" "wk27"
## [31] "wk28" "wk29" "wk30" "wk31" "wk32"
## [36] "wk33" "wk34" "wk35" "wk36" "wk37"
## [41] "wk38" "wk39" "wk40" "wk41" "wk42"
## [46] "wk43" "wk44" "wk45" "wk46" "wk47"
## [51] "wk48" "wk49" "wk50" "wk51" "wk52"
## [56] "wk53" "wk54" "wk55" "wk56" "wk57"
## [61] "wk58" "wk59" "wk60" "wk61" "wk62"
## [66] "wk63" "wk64" "wk65" "wk66" "wk67"
## [71] "wk68" "wk69" "wk70" "wk71" "wk72"
## [76] "wk73" "wk74" "wk75" "wk76"## # A tibble: 317 x 4
## wk20 wk21 wk22 wk23
## <dbl> <dbl> <dbl> <dbl>
## 1 NA NA NA NA
## 2 NA NA NA NA
## 3 14 12 7 6
## 4 70 NA NA NA
## 5 NA NA NA NA
## 6 94 NA NA NA
## 7 NA NA NA NA
## 8 86 NA NA NA
## 9 4 5 5 6
## 10 89 NA NA NA
## # ... with 307 more rows9.5 Filter rows with filter()
filter()subsets observations based on their valuesmultiple arguments to filter are combined using logical operators
&and|or!not
By default, multiple conditions are combined with
&
# filter the flights departed on Jan 1.
# Here month == 1 & day == 1
filter(flights, month == 1, day == 1) ## # A tibble: 842 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## 7 2013 1 1 555 600 -5 913 854
## 8 2013 1 1 557 600 -3 709 723
## 9 2013 1 1 557 600 -3 838 846
## 10 2013 1 1 558 600 -2 753 745
## # ... with 832 more rows, and 11 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>## # A tibble: 55,403 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 11 1 5 2359 6 352 345
## 2 2013 11 1 35 2250 105 123 2356
## 3 2013 11 1 455 500 -5 641 651
## 4 2013 11 1 539 545 -6 856 827
## 5 2013 11 1 542 545 -3 831 855
## 6 2013 11 1 549 600 -11 912 923
## 7 2013 11 1 550 600 -10 705 659
## 8 2013 11 1 554 600 -6 659 701
## 9 2013 11 1 554 600 -6 826 827
## 10 2013 11 1 554 600 -6 749 751
## # ... with 55,393 more rows, and 11 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>## # A tibble: 7,198 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 623 627 -4 933 932
## 4 2013 1 1 728 732 -4 1041 1038
## 5 2013 1 1 739 739 0 1104 1038
## 6 2013 1 1 908 908 0 1228 1219
## 7 2013 1 1 1028 1026 2 1350 1339
## 8 2013 1 1 1044 1045 -1 1352 1351
## 9 2013 1 1 1114 900 134 1447 1222
## 10 2013 1 1 1205 1200 5 1503 1505
## # ... with 7,188 more rows, and 11 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm># filter the flights with arr_delay <= 120 or dep_delay > 120
filter(flights, arr_delay <= 120 | dep_delay > 120) ## # A tibble: 325,773 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## 7 2013 1 1 555 600 -5 913 854
## 8 2013 1 1 557 600 -3 709 723
## 9 2013 1 1 557 600 -3 838 846
## 10 2013 1 1 558 600 -2 753 745
## # ... with 325,763 more rows, and 11 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>9.6 Add new variables with mutate()
mutate()create a new variable.
## # A tibble: 336,776 x 22
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## 7 2013 1 1 555 600 -5 913 854
## 8 2013 1 1 557 600 -3 709 723
## 9 2013 1 1 557 600 -3 838 846
## 10 2013 1 1 558 600 -2 753 745
## # ... with 336,766 more rows, and 14 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>,
## # gain <dbl>, hours <dbl>, gain_per_hour <dbl>mutate()executes the transformations iteratively so that later transformations can use the columns created by earlier transformations.
## # A tibble: 336,776 x 22
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## 7 2013 1 1 555 600 -5 913 854
## 8 2013 1 1 557 600 -3 709 723
## 9 2013 1 1 557 600 -3 838 846
## 10 2013 1 1 558 600 -2 753 745
## # ... with 336,766 more rows, and 14 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>,
## # gain <dbl>, hours <dbl>, gain_per_hour <dbl>transmute()creates a new variable but drops others.
transmute(flights, gain = arr_delay - dep_delay, hours = air_time / 60, gain_per_hour = gain / hours)## # A tibble: 336,776 x 3
## gain hours gain_per_hour
## <dbl> <dbl> <dbl>
## 1 9 3.78 2.38
## 2 16 3.78 4.23
## 3 31 2.67 11.6
## 4 -17 3.05 -5.57
## 5 -19 1.93 -9.83
## 6 16 2.5 6.4
## 7 24 2.63 9.11
## 8 -11 0.883 -12.5
## 9 -5 2.33 -2.14
## 10 10 2.3 4.35
## # ... with 336,766 more rowsThe scoped variant of
mutate()andtransmute()help us to apply the same transformation to multiple variables.mutate_all()andtransmute_all()affect every variablemutate_at()andtransmute_at()affect variables selected with a character vectormutate_if()andtransmute_if()affect variables selected with a predicate function (a function that returns TRUE or FALSE)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa# apply log() to the columns that contain "Sepal"
mutated <- mutate_at(iris, vars(contains("Sepal")), log)
head(mutated) ## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 1.629241 1.252763 1.4 0.2 setosa
## 2 1.589235 1.098612 1.4 0.2 setosa
## 3 1.547563 1.163151 1.3 0.2 setosa
## 4 1.526056 1.131402 1.5 0.2 setosa
## 5 1.609438 1.280934 1.4 0.2 setosa
## 6 1.686399 1.360977 1.7 0.4 setosa## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 1.629241 1.252763 0.3364722 -1.6094379 setosa
## 2 1.589235 1.098612 0.3364722 -1.6094379 setosa
## 3 1.547563 1.163151 0.2623643 -1.6094379 setosa
## 4 1.526056 1.131402 0.4054651 -1.6094379 setosa
## 5 1.609438 1.280934 0.3364722 -1.6094379 setosa
## 6 1.686399 1.360977 0.5306283 -0.9162907 setosaYou will find more details about the scoped variants of
mutate()andtransmute()here.Tidyverse changes rapidly. To indicate the lifecycle of packages (or functions), Tidyverse uses the lifecycle badges.
Now,
across()supersedes the family of “scoped variants” like summarise_at(), summarise_if(), and summarise_all(). It means that you will see moreacross()and less_at(),_if(),_all()functions in the future.- You can check the following links for more details about
across()
- You can check the following links for more details about
9.7 Arrange rows with arrange()
arrange()reorders rows in ascending order by default- use
desc()to order in descending order
- use
## # A tibble: 336,776 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 1 1 517 515 2 830 819
## 2 2013 1 1 533 529 4 850 830
## 3 2013 1 1 542 540 2 923 850
## 4 2013 1 1 544 545 -1 1004 1022
## 5 2013 1 1 554 600 -6 812 837
## 6 2013 1 1 554 558 -4 740 728
## 7 2013 1 1 555 600 -5 913 854
## 8 2013 1 1 557 600 -3 709 723
## 9 2013 1 1 557 600 -3 838 846
## 10 2013 1 1 558 600 -2 753 745
## # ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>## # A tibble: 336,776 x 19
## year month day dep_time sched_dep_time dep_delay arr_time sched_arr_time
## <int> <int> <int> <int> <int> <dbl> <int> <int>
## 1 2013 12 1 13 2359 14 446 445
## 2 2013 12 1 17 2359 18 443 437
## 3 2013 12 1 453 500 -7 636 651
## 4 2013 12 1 520 515 5 749 808
## 5 2013 12 1 536 540 -4 845 850
## 6 2013 12 1 540 550 -10 1005 1027
## 7 2013 12 1 541 545 -4 734 755
## 8 2013 12 1 546 545 1 826 835
## 9 2013 12 1 549 600 -11 648 659
## 10 2013 12 1 550 600 -10 825 854
## # ... with 336,766 more rows, and 11 more variables: arr_delay <dbl>,
## # carrier <chr>, flight <int>, tailnum <chr>, origin <chr>, dest <chr>,
## # air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>9.8 Grouped summaries with summarize()
summarize()collapses a tibble to a single row for summary (imagine the sum and average function in Excel)summarize()is not terribly useful unless we pair it withgroup_by().group_by()takes an existing tibble and converts it into a grouped tibble where operations are performed “by group”.- Using
group_by(), most data operations are done on groups defined by variables. - Together
group_by(),summarize()will produce one row for each group group_by()andsummarize()provide one of the tools that you’ll use most commonly when working with dplyr: grouped summaries.- summarize(data, …)
- … = Name-value pairs of summary functions. The name will be the name of the variable in the result. The value should be an expression that returns a single value like
min(x),n(), orsum(is.na(y)).
- … = Name-value pairs of summary functions. The name will be the name of the variable in the result. The value should be an expression that returns a single value like
- Summary functions takes a vector as an input and return a single value as an output.
- Counts
- Locations
- Position/
# name = delay, value = mean(dep_delay, na.rm = TRUE)
summarize(flights, delay_mean = mean(dep_delay, na.rm = TRUE), delay_sd = sd(dep_delay, na.rm = TRUE))## # A tibble: 1 x 2
## delay_mean delay_sd
## <dbl> <dbl>
## 1 12.6 40.2# Together `group_by()`, `summarize()` will produce one row for each group
by_day <- group_by(flights, year, month, day)
summarize(by_day, delay = mean(dep_delay, na.rm = TRUE))## `summarise()` regrouping output by 'year', 'month' (override with `.groups` argument)## # A tibble: 365 x 4
## # Groups: year, month [12]
## year month day delay
## <int> <int> <int> <dbl>
## 1 2013 1 1 11.5
## 2 2013 1 2 13.9
## 3 2013 1 3 11.0
## 4 2013 1 4 8.95
## 5 2013 1 5 5.73
## 6 2013 1 6 7.15
## 7 2013 1 7 5.42
## 8 2013 1 8 2.55
## 9 2013 1 9 2.28
## 10 2013 1 10 2.84
## # ... with 355 more rows9.9 The pipe operator %>%
You will find
%>%in almost all tidyverse codes. The pipe operator%>%from themagrittrpackage was developed to help you write code that is easier to read and understand or to enhance the readability of your code. Since the tidyverse automatically load%>%, you don’t need to manually load themagrittrpackage to use%>%. Here’s what the pipe operator%>%does:x %>% f(y)turns into f(x, y)x %>% f(y) %>% g(z)turns into g(f(x,y), z)
The pipe operator
%>%focuses on the the transformations, not what’s being transformed. You can read it as a series of imperative statements: do f() and “then” do g(). A good way to pronounce%>%when reading the code is “then”.Let’s see a concrete example.
by_dest <- group_by(flights, dest)
delay <- summarize(by_dest, count = n(), dist = mean(distance, na.rm = TRUE), delay = mean( arr_delay, na.rm = TRUE))
delay <- filter(delay, count > 20, dest != "HNL")
delay## # A tibble: 96 x 4
## dest count dist delay
## <chr> <int> <dbl> <dbl>
## 1 ABQ 254 1826 4.38
## 2 ACK 265 199 4.85
## 3 ALB 439 143 14.4
## 4 ATL 17215 757. 11.3
## 5 AUS 2439 1514. 6.02
## 6 AVL 275 584. 8.00
## 7 BDL 443 116 7.05
## 8 BGR 375 378 8.03
## 9 BHM 297 866. 16.9
## 10 BNA 6333 758. 11.8
## # ... with 86 more rows# It looks like delays increase with distance up to ~ 750 miles and then decrease. Maybe as flights get longer there's more ability to make up delays in the air?
ggplot(data = delay, mapping = aes(x = dist, y = delay)) +
geom_point( aes( size = count), alpha = 1/3) +
geom_smooth(se = FALSE)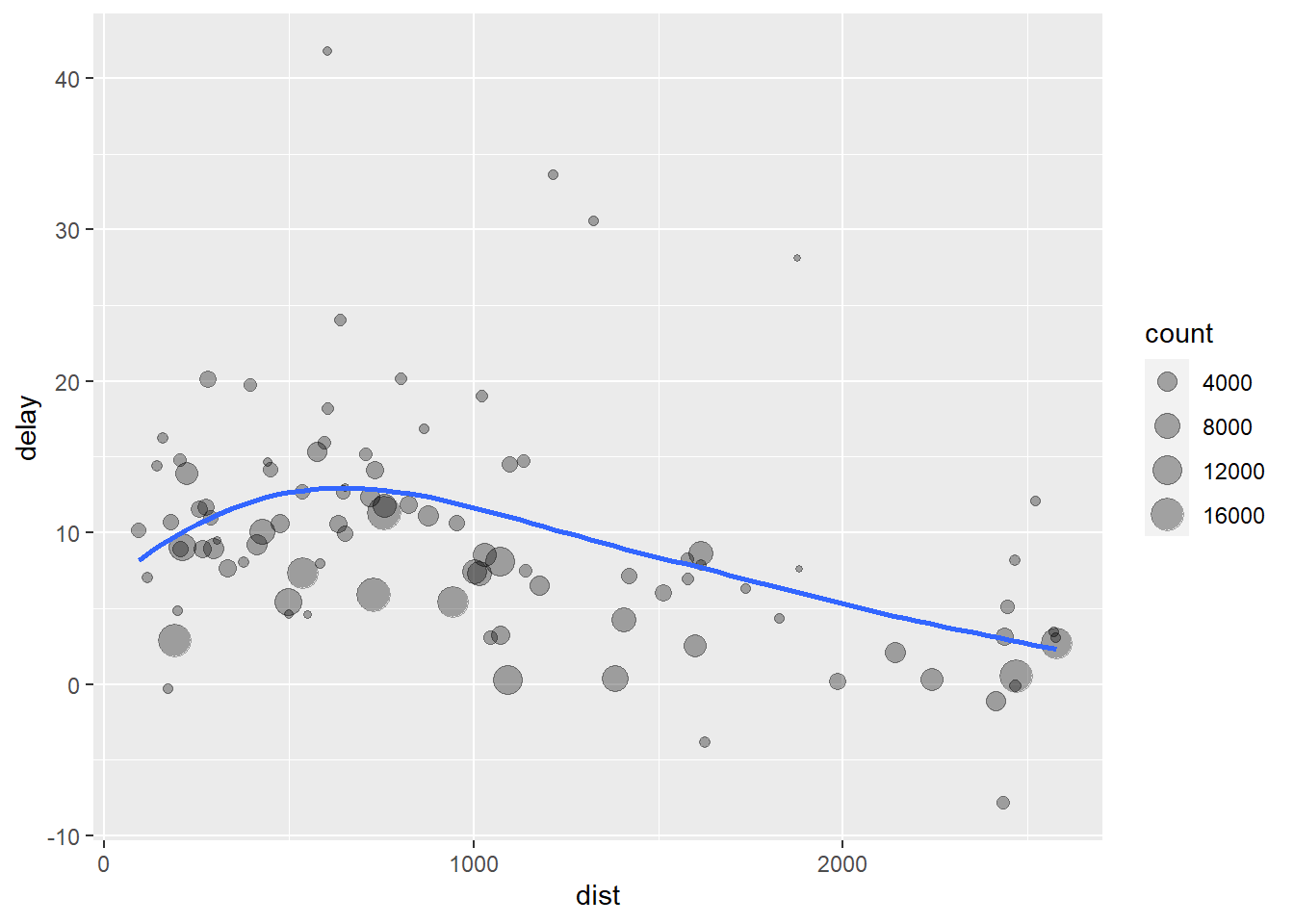
- The code above is a little frustrating to write because we have to give each intermediate data frame a name, even though we don’t care about it. Naming things is hard, so this slows down our analysis.The code below focuses on the transformations, not what’s being transformed, which makes the code easier to read. You can read it as a series of imperative statements: group, then summarize, then filter, then ggplot. As suggested by this reading, a good way to pronounce
%>%when reading code is “then.”
flights %>%
group_by(dest) %>%
summarize(count = n(), dist = mean( distance, na.rm = TRUE), delay = mean(arr_delay, na.rm = TRUE)) %>%
filter(count > 20, dest != "HNL") %>%
ggplot(mapping = aes(x = dist, y = delay)) +
geom_point(aes(size = count), alpha = 1/3) +
geom_smooth(se = FALSE)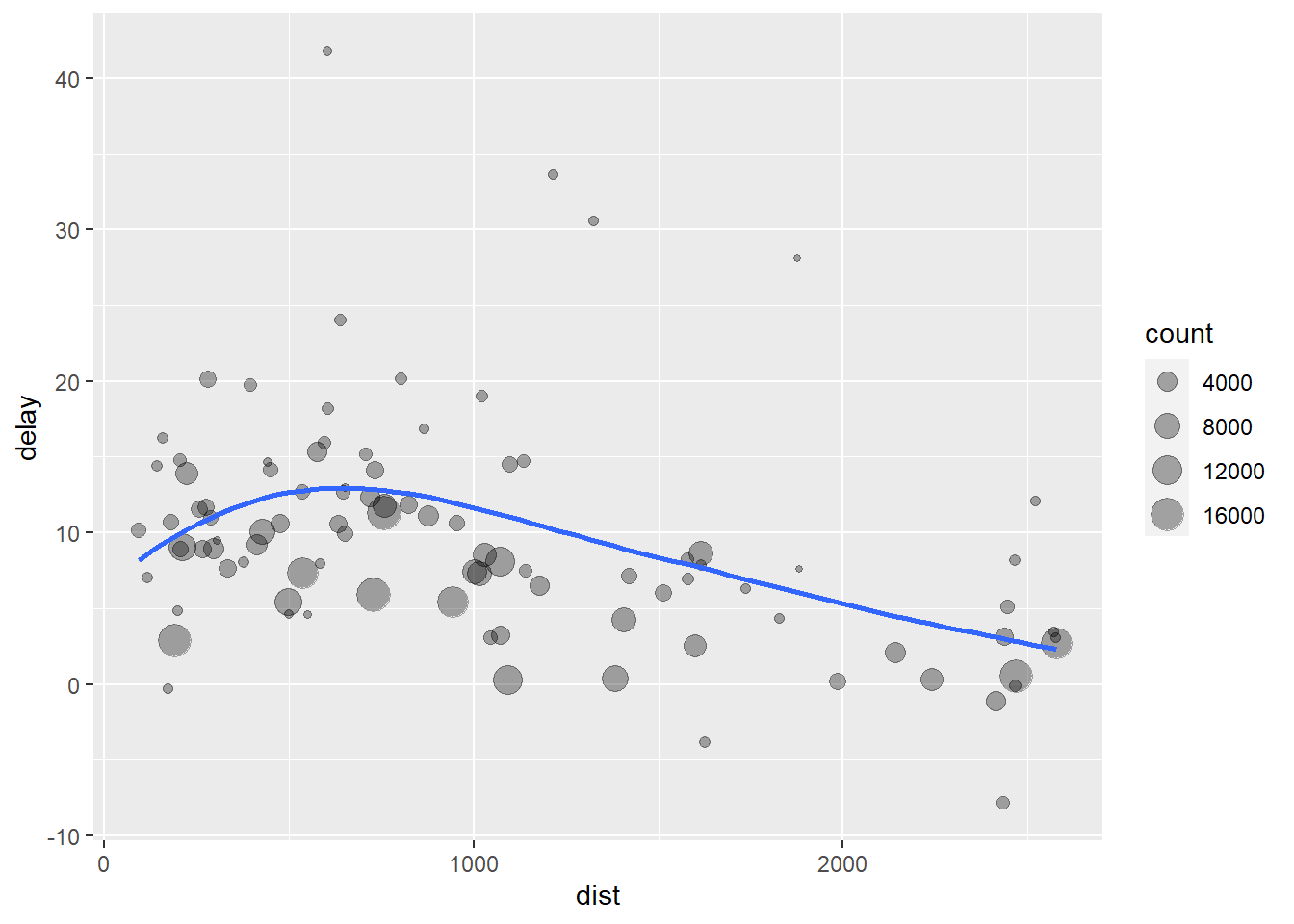
9.10 Style Guide
- It is important to make your codes easy to read by human (especially you). In order to enhance the readability of your codes, it is often recommended to
- add comments (
#in R) - follow programming style guides
- add comments (
- Programming style guides are a set of guidelines used when writing the source code for a specific programming language. Your R codes will still work even though you don’t follow the style guide. However, style guides will enhance the readability of your code. R has a couple of style guides:
9.11 Grouped summaries with summarize() (continue)
irisis a built-in dataset in R and gives the measurements in centimeters of the variables sepal length and width and petal length and width, respectively, for 50 flowers from each of 3 species of iris. The species are Iris setosa, versicolor, and virginica. You can find the pictures of three species here.
## # A tibble: 150 x 5
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## <dbl> <dbl> <dbl> <dbl> <fct>
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
## 7 4.6 3.4 1.4 0.3 setosa
## 8 5 3.4 1.5 0.2 setosa
## 9 4.4 2.9 1.4 0.2 setosa
## 10 4.9 3.1 1.5 0.1 setosa
## # ... with 140 more rows# without the pipe operator %>%
iris_grouped <- group_by(iris, Species)
summarise_all(iris_grouped, mean)## # A tibble: 3 x 5
## Species Sepal.Length Sepal.Width Petal.Length Petal.Width
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 setosa 5.01 3.43 1.46 0.246
## 2 versicolor 5.94 2.77 4.26 1.33
## 3 virginica 6.59 2.97 5.55 2.03# with the pipe operator %>%
# with summarise_all() - superseded by across()
iris %>%
group_by(Species) %>%
summarise_all(mean)## # A tibble: 3 x 5
## Species Sepal.Length Sepal.Width Petal.Length Petal.Width
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 setosa 5.01 3.43 1.46 0.246
## 2 versicolor 5.94 2.77 4.26 1.33
## 3 virginica 6.59 2.97 5.55 2.03# with the pipe operator %>%
# selection helper functions are useful for selecting columns
iris %>%
group_by(Species) %>%
summarise(across(starts_with("Sepal"), mean))## `summarise()` ungrouping output (override with `.groups` argument)## # A tibble: 3 x 3
## Species Sepal.Length Sepal.Width
## <fct> <dbl> <dbl>
## 1 setosa 5.01 3.43
## 2 versicolor 5.94 2.77
## 3 virginica 6.59 2.97- The High School and Beyond(HSB) dataset is a dataset in the
candiscpackage. The High School and Beyond Project was a longitudinal study of students in the U.S. carried out in 1980 by the National Center for Education Statistics. Data were collected from 58,270 high school students (28,240 seniors and 30,030 sophomores) and 1,015 secondary schools. The HSB data frame is sample of 600 observations, of unknown characteristics, originally taken from Tatsuoka (1988).
# as_tibble() converts a dataframe to a tibble
# a tibble prints the first part of a dataset by default
# We need head() to prints the first part of a dataframe
HSB_tbl <- as_tibble(HSB)
HSB_tbl## # A tibble: 600 x 15
## id gender race ses sch prog locus concept mot career read write
## <dbl> <fct> <fct> <fct> <fct> <fct> <dbl> <dbl> <dbl> <fct> <dbl> <dbl>
## 1 55 female hisp~ low publ~ gene~ -1.78 0.560 1 prof1 28.3 46.3
## 2 114 male afri~ midd~ publ~ acad~ 0.240 -0.350 1 opera~ 30.5 35.9
## 3 490 male white midd~ publ~ voca~ -1.28 0.340 0.330 prof1 31 35.9
## 4 44 female hisp~ low publ~ voca~ 0.220 -0.760 1 servi~ 31 41.1
## 5 26 female hisp~ midd~ publ~ acad~ 1.12 -0.740 0.670 servi~ 31 41.1
## 6 510 male white midd~ publ~ voca~ -0.860 1.19 0.330 opera~ 33.6 28.1
## 7 133 female afri~ low publ~ voca~ -0.230 0.440 0.330 school 33.6 35.2
## 8 213 female white low publ~ gene~ 0.0400 -0.470 0.670 cleri~ 33.6 59.3
## 9 548 female white midd~ priv~ acad~ 0.470 0.340 0.670 prof2 33.6 43
## 10 309 female white high publ~ gene~ 0.320 0.900 0.670 prof2 33.6 51.5
## # ... with 590 more rows, and 3 more variables: math <dbl>, sci <dbl>, ss <dbl># names() displays the column names of the HSB data
# ?HSB will give you more details about HSB data
names(HSB_tbl)## [1] "id" "gender" "race" "ses" "sch" "prog" "locus"
## [8] "concept" "mot" "career" "read" "write" "math" "sci"
## [15] "ss"## tibble [600 x 15] (S3: tbl_df/tbl/data.frame)
## $ id : num [1:600] 55 114 490 44 26 510 133 213 548 309 ...
## $ gender : Factor w/ 2 levels "male","female": 2 1 1 2 2 1 2 2 2 2 ...
## $ race : Factor w/ 4 levels "hispanic","asian",..: 1 3 4 1 1 4 3 4 4 4 ...
## $ ses : Factor w/ 3 levels "low","middle",..: 1 2 2 1 2 2 1 1 2 3 ...
## $ sch : Factor w/ 2 levels "public","private": 1 1 1 1 1 1 1 1 2 1 ...
## $ prog : Factor w/ 3 levels "general","academic",..: 1 2 3 3 2 3 3 1 2 1 ...
## $ locus : num [1:600] -1.78 0.24 -1.28 0.22 1.12 ...
## $ concept: num [1:600] 0.56 -0.35 0.34 -0.76 -0.74 ...
## $ mot : num [1:600] 1 1 0.33 1 0.67 ...
## $ career : Factor w/ 17 levels "clerical","craftsman",..: 9 8 9 15 15 8 14 1 10 10 ...
## $ read : num [1:600] 28.3 30.5 31 31 31 ...
## $ write : num [1:600] 46.3 35.9 35.9 41.1 41.1 ...
## $ math : num [1:600] 42.8 36.9 46.1 49.2 36 ...
## $ sci : num [1:600] 44.4 33.6 39 33.6 36.9 ...
## $ ss : num [1:600] 50.6 40.6 45.6 35.6 45.6 ...## `summarise()` ungrouping output (override with `.groups` argument)## # A tibble: 4 x 2
## race n
## <fct> <int>
## 1 hispanic 71
## 2 asian 34
## 3 african-amer 58
## 4 white 437## # A tibble: 4 x 2
## race n
## <fct> <int>
## 1 hispanic 71
## 2 asian 34
## 3 african-amer 58
## 4 white 437## # A tibble: 8 x 3
## race gender n
## <fct> <fct> <int>
## 1 hispanic male 37
## 2 hispanic female 34
## 3 asian male 15
## 4 asian female 19
## 5 african-amer male 24
## 6 african-amer female 34
## 7 white male 197
## 8 white female 240# the means of reading, writing, math, and social science scores
# mean can be replaced with any summary function:
# mean(), median(), sum(), min(), max(), sd(), var(), ...
HSB_tbl %>%
summarise(readm = mean(read), writem = mean(write), mathm = mean(math), ssm = mean(ss))## # A tibble: 1 x 4
## readm writem mathm ssm
## <dbl> <dbl> <dbl> <dbl>
## 1 51.9 52.4 51.8 52.0# the means of reading, writing, math, and social science scores by gender
HSB_tbl %>%
group_by(gender) %>%
summarise(readm = mean(read), writem = mean(write), mathm = mean(math), ssm = mean(ss))## `summarise()` ungrouping output (override with `.groups` argument)## # A tibble: 2 x 5
## gender readm writem mathm ssm
## <fct> <dbl> <dbl> <dbl> <dbl>
## 1 male 52.4 49.8 52.3 51.4
## 2 female 51.5 54.6 51.4 52.6# the means of reading, writing, math, and social science scores by gender and race
HSB_tbl %>%
group_by(gender, race) %>%
summarise(readm = mean(read), writem = mean(write), mathm = mean(math), ssm = mean(ss))## `summarise()` regrouping output by 'gender' (override with `.groups` argument)## # A tibble: 8 x 6
## # Groups: gender [2]
## gender race readm writem mathm ssm
## <fct> <fct> <dbl> <dbl> <dbl> <dbl>
## 1 male hispanic 47.7 45.1 47.7 45.9
## 2 male asian 55.7 54.7 58.9 50.3
## 3 male african-amer 46.6 44.7 44.2 47.2
## 4 male white 53.7 50.9 53.7 53.0
## 5 female hispanic 44.0 48.5 43.6 49.0
## 6 female asian 52.7 56.5 56.7 53.8
## 7 female african-amer 47.4 47.5 46.8 49.2
## 8 female white 53.1 56.3 52.8 53.5# the means of all numeric variables by gender
HSB_tbl %>%
group_by(gender) %>%
summarise(across(where(is.numeric), mean, na.rm = TRUE))## `summarise()` ungrouping output (override with `.groups` argument)## # A tibble: 2 x 10
## gender id locus concept mot read write math sci ss
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 male 305. 0.0134 0.102 0.624 52.4 49.8 52.3 53.2 51.4
## 2 female 297. 0.166 -0.0762 0.692 51.5 54.6 51.4 50.5 52.6# the means of all numeric variables by ses for only asian and hispanic
HSB_tbl %>%
filter(race %in% c("asian", "hispanic")) %>%
group_by(ses) %>%
summarise(across(where(is.numeric), mean, na.rm = TRUE))## `summarise()` ungrouping output (override with `.groups` argument)## # A tibble: 3 x 10
## ses id locus concept mot read write math sci ss
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 low 44.8 -0.142 -0.189 0.537 44.7 45.9 45.8 43.0 45.5
## 2 middle 50.8 -0.122 0.0525 0.584 47.3 48.6 48.3 47.5 47.9
## 3 high 66.9 0.118 0.0619 0.705 55.6 55.9 56.6 54.0 55.09.12 Mutating joins
- During data analysis, you often want to combine two (or more) data frames (or tibbles) to answer your questions.
- A join is a way of matching each row in x to zero, one, or more rows in y.
- The variables used to match two data frames are called key variables (or keys).
- The mutating joins add columns from one data frame y to another data frame x by matching rows based the key variable.
- The functions for mutating joins include (see the cheatsheet to see graphical representation of the functions):
inner_join()keeps observations that appear in both data framesleft_join()keeps all observations in xright_join()keeps all observations in yfull_join()keeps all observations in x and y
- The following examples come from the Relational data chapter in R for data science.
# tribble() creates a tibble
x <- tribble(
~key, ~val_x,
1, "x1",
2, "x2",
3, "x3"
)
y <- tribble(
~key, ~val_y,
1, "y1",
2, "y2",
4, "y3"
)## # A tibble: 3 x 2
## key val_x
## <dbl> <chr>
## 1 1 x1
## 2 2 x2
## 3 3 x3## # A tibble: 3 x 2
## key val_y
## <dbl> <chr>
## 1 1 y1
## 2 2 y2
## 3 4 y3## # A tibble: 2 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2## # A tibble: 2 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 4 <NA> y3## # A tibble: 3 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 4 <NA> y3## # A tibble: 4 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>
## 4 4 <NA> y3## # A tibble: 4 x 3
## key val_x val_y
## <dbl> <chr> <chr>
## 1 1 x1 y1
## 2 2 x2 y2
## 3 3 x3 <NA>
## 4 4 <NA> y3