Chapter 8 Generalized Linear Models
8.1 Problem Setup
In the Linear Models Chapter 7, we assumed the generative process to be linear in the effects of the predictors \(x\). We now write that same linear model, slightly differently: \[ y|x \sim \mathcal{N}(x'\beta, \sigma^2). \]
This model not allow for the non-linear relations of Example 8.1, nor does it allow for the distrbituion of \(\varepsilon\) to change with \(x\), as in Example 8.2. Generalize linear models (GLM), as the name suggests, are a generalization of the linear models in Chapter 7 that allow that13.
For Example 8.1, we would like something in the lines of \[ y|x \sim Binom(1,p(x)) \]
For Example 8.2, we would like something in the lines of \[ y|x \sim \mathcal{N}(x'\beta,\sigma^2(x)), \] or more generally \[ y|x \sim \mathcal{N}(\mu(x),\sigma^2(x)), \] or maybe not Gaussian \[ y|x \sim Pois(\lambda(x)). \]
Even more generally, for some distribution \(F(\theta)\), with a parameter \(\theta\), we would like to assume that the data is generated via \[\begin{align} \tag{8.1} y|x \sim F(\theta(x)) \end{align}\]
Possible examples include \[\begin{align} y|x &\sim Poisson(\lambda(x)) \\ y|x &\sim Exp(\lambda(x)) \\ y|x &\sim \mathcal{N}(\mu(x),\sigma^2(x)) \end{align}\]
GLMs allow models of the type of Eq.(8.1), while imposing some constraints on \(F\) and on the relation \(\theta(x)\). GLMs assume the data distribution \(F\) to be in a “well-behaved” family known as the Natural Exponential Family of distributions. This family includes the Gaussian, Gamma, Binomial, Poisson, and Negative Binomial distributions. These five include as special cases the exponential, chi-squared, Rayleigh, Weibull, Bernoulli, and geometric distributions.
GLMs also assume that the distribution’s parameter, \(\theta\), is some simple function of a linear combination of the effects. In our cigarettes example this amounts to assuming that each cigarette has an additive effect, but not on the probability of cancer, but rather, on some simple function of it. Formally \[g(\theta(x))=x'\beta,\] and we recall that \[x'\beta=\beta_0 + \sum_j x_j \beta_j.\] The linear predictor \(x'\beta\) is usually denoted by \(\eta\), i.e., \(\eta = x'\beta\) The function \(g\) is called the link function, its inverse, \(g^{-1}\) is the mean function. It is called the mean function as \[E(y|x) = \mu = g^{-1}(\eta)\].
We say that “the effects of each cigarette is linear in link scale”. This terminology will later be required to understand R’s output.
8.2 Logistic Regression
The best known of the GLM class of models is the logistic regression that deals with Binomial, or more precisely, Bernoulli-distributed data. The link function in the logistic regression is the logit function \[\begin{align} g(t)=log\left( \frac{t}{(1-t)} \right) \tag{8.2} \end{align}\] implying that under the logistic model assumptions \[\begin{align} y|x \sim Binom \left( 1, p=\frac{e^{x'\beta}}{1+e^{x'\beta}} \right). \tag{8.3} \end{align}\]
Before we fit such a model, we try to justify this construction, in particular, the enigmatic link function in Eq.(8.2). Let’s look at the simplest possible case: the comparison of two groups indexed by \(x\): \(x=0\) for the first, and \(x=1\) for the second. We start with some definitions.
Odds are the same as probabilities, but instead of telling me there is a \(66\%\) of success, they tell me the odds of success are “2 to 1”. If you ever placed a bet, the language of “odds” should not be unfamiliar to you.
Odds ratios (OR) compare between the probabilities of two groups, only that it does not compare them in probability scale, but rather in odds scale. You can also think of ORs as a measure of distance between two Brenoulli distributions. ORs have better mathematical properties than other candidate distance measures, such as \(P(y_1=1)-P(y_2=1)\).
Under the logit link assumption formalized in Eq.(8.3), the OR between two conditions indexed by \(y|x=1\) and \(y|x=0\), returns: \[\begin{align} OR(y|x=1,y|x=0) = \frac{P(y=1|x=1)/P(y=0|x=1)}{P(y=1|x=0)/P(y=0|x=0)} = e^{\beta_1}. \end{align}\]
The last equality demystifies the choice of the link function in the logistic regression: it allows us to interpret \(\beta\) of the logistic regression as a measure of change of binary random variables, namely, as the (log) odds-ratios due to a unit increase in \(x\).
8.2.1 Logistic Regression with R
Let’s get us some data.
The PlantGrowth data records the weight of plants under three conditions: control, treatment1, and treatment2.
## weight group
## 1 4.17 ctrl
## 2 5.58 ctrl
## 3 5.18 ctrl
## 4 6.11 ctrl
## 5 4.50 ctrl
## 6 4.61 ctrlWe will now attach the data so that its contents is available in the workspace (don’t forget to detach afterwards, or you can expect some conflicting object names).
We will also use the cut function to create a binary response variable for Light, and Heavy plants (we are doing logistic regression, so we need a two-class response).
As a general rule of thumb, when we discretize continuous variables, we lose information.
For pedagogical reasons, however, we will proceed with this bad practice.
Look at the following output and think: how many group effects do we expect? What should be the sign of each effect?
attach(PlantGrowth)
weight.factor<- cut(weight, 2, labels=c('Light', 'Heavy')) # binarize weights
plot(table(group, weight.factor))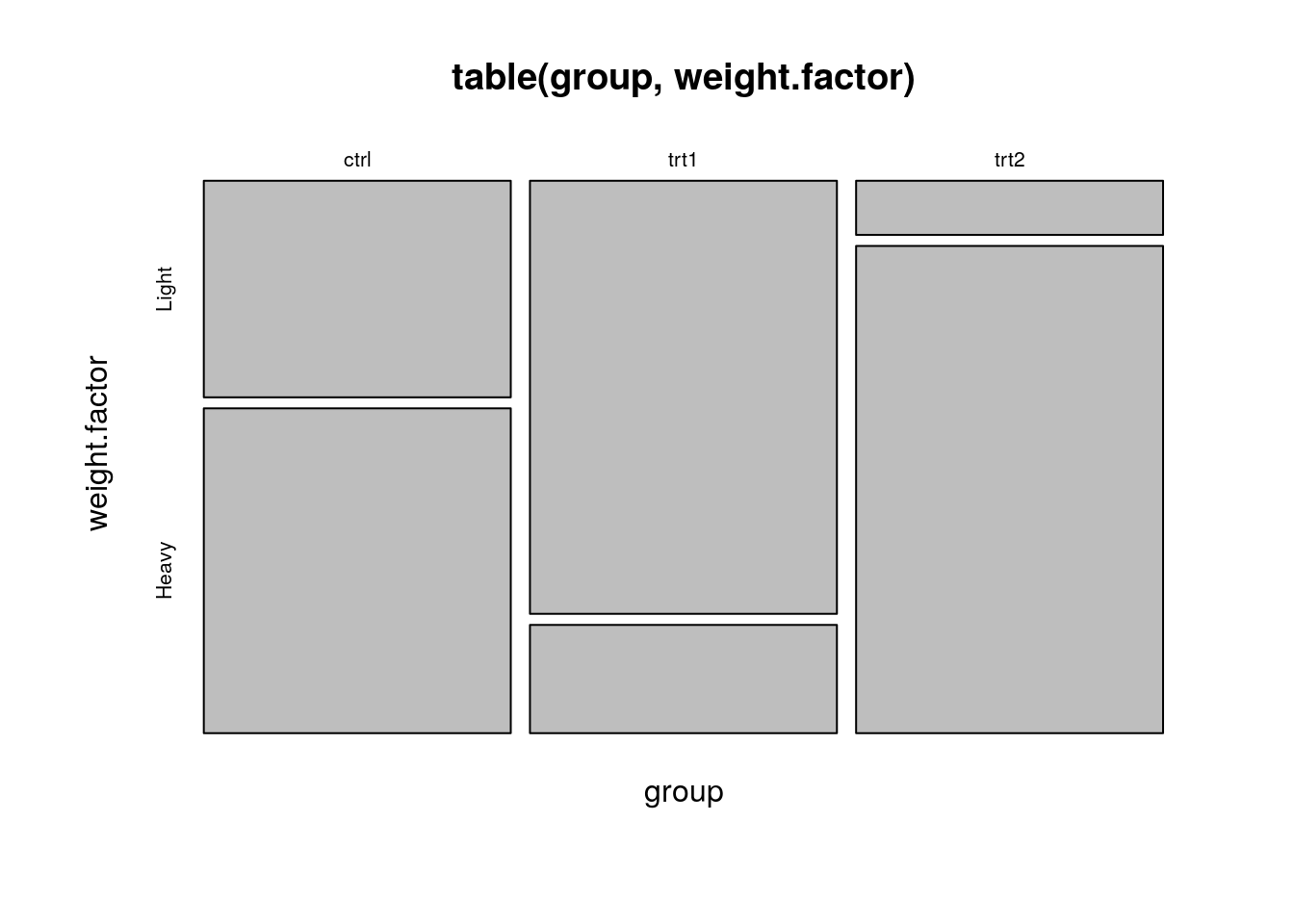
Let’s fit a logistic regression, and inspect the output.
##
## Call:
## glm(formula = weight.factor ~ group, family = binomial)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.1460 -0.6681 0.4590 0.8728 1.7941
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 0.4055 0.6455 0.628 0.5299
## grouptrt1 -1.7918 1.0206 -1.756 0.0792 .
## grouptrt2 1.7918 1.2360 1.450 0.1471
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 41.054 on 29 degrees of freedom
## Residual deviance: 29.970 on 27 degrees of freedom
## AIC: 35.97
##
## Number of Fisher Scoring iterations: 4Things to note:
The
glmfunction is our workhorse for all GLM models.The
familyargument ofglmtells R the respose variable is brenoulli, thus, performing a logistic regression.The
summaryfunction is content aware. It gives a different output forglmclass objects than for other objects, such as thelmwe saw in Chapter 7. In fact, whatsummarydoes is merely callsummary.glm.As usual, we get the coefficients table, but recall that they are to be interpreted as (log) odd-ratios, i.e., in “link scale”. To return to probabilities (“response scale”), we will need to undo the logistic transformation.
As usual, we get the significance for the test of no-effect, versus a two-sided alternative. P-values are asymptotic, thus, only approximate (and can be very bad approximations in small samples).
The residuals of
glmare slightly different than thelmresiduals, and called Deviance Residuals.AIC (Akaike Information Criteria) is the equivalent of R2 in logistic regression. It measures the fit when a penalty is applied to the number of parameters (see AIC definition here). Smaller AIC values indicate the model is closer to the truth.
Null deviance: Fits the model only with the intercept. The degree of freedom is n-1. We can interpret it as a Chi-square value (fitted value different from the actual value hypothesis testing).
Residual Deviance: Model with all the variables. It is also interpreted as a Chi-square hypothesis testing.
Number of Fisher Scoring iterations: Number of iterations before converging ().
For help see
?glm,?family, and?summary.glm.
Like in the linear models, we can use an ANOVA table to check if treatments have any effect, and not one treatment at a time. In the case of GLMs, this is called an analysis of deviance table.
## Analysis of Deviance Table
##
## Model: binomial, link: logit
##
## Response: weight.factor
##
## Terms added sequentially (first to last)
##
##
## Df Deviance Resid. Df Resid. Dev Pr(>Chi)
## NULL 29 41.054
## group 2 11.084 27 29.970 0.003919 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Things to note:
- The
anovafunction, like thesummaryfunction, are content-aware and produce a different output for theglmclass than for thelmclass. All thatanovadoes is callanova.glm. - In GLMs there is no canonical test (like the F test for
lm).LRTimplies we want an approximate Likelihood Ratio Test. We thus specify the type of test desired with thetestargument. - The distribution of the weights of the plants does vary with the treatment given, as we may see from the significance of the
groupfactor. - Readers familiar with ANOVA tables, should know that we computed the GLM equivalent of a type I sum- of-squares.
Run
drop1(glm.1, test='Chisq')for a GLM equivalent of a type III sum-of-squares. - For help see
?anova.glm.
Let’s predict the probability of a heavy plant for each treatment.
## 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
## 0.6 0.6 0.6 0.6 0.6 0.6 0.6 0.6 0.6 0.6 0.2 0.2 0.2 0.2 0.2 0.2 0.2 0.2
## 19 20 21 22 23 24 25 26 27 28 29 30
## 0.2 0.2 0.9 0.9 0.9 0.9 0.9 0.9 0.9 0.9 0.9 0.9Things to note:
- Like the
summaryandanovafunctions, thepredictfunction is aware that its input is ofglmclass. All thatpredictdoes is callpredict.glm. - In GLMs there are many types of predictions. The
typeargument controls which type is returned. Usetype=responsefor predictions in probability scale; use `type=link’ for predictions in log-odds scale. - How do I know we are predicting the probability of a heavy plant, and not a light plant? Just run
contrasts(weight.factor)to see which of the categories of the factorweight.factoris encoded as 1, and which as 0. - For help see
?predict.glm.
Let’s detach the data so it is no longer in our workspace, and object names do not collide.
We gave an example with a factorial (i.e. discrete) predictor. We can do the same with multiple continuous predictors.
## npreg glu bp skin bmi ped age type
## 1 6 148 72 35 33.6 0.627 50 Yes
## 2 1 85 66 29 26.6 0.351 31 No
## 3 1 89 66 23 28.1 0.167 21 No
## 4 3 78 50 32 31.0 0.248 26 Yes
## 5 2 197 70 45 30.5 0.158 53 Yes
## 6 5 166 72 19 25.8 0.587 51 Yes## Start: AIC=301.79
## type ~ npreg + glu + bp + skin + bmi + ped + age
##
## Df Deviance AIC
## - skin 1 286.22 300.22
## - bp 1 286.26 300.26
## - age 1 286.76 300.76
## <none> 285.79 301.79
## - npreg 1 291.60 305.60
## - ped 1 292.15 306.15
## - bmi 1 293.83 307.83
## - glu 1 343.68 357.68
##
## Step: AIC=300.22
## type ~ npreg + glu + bp + bmi + ped + age
##
## Df Deviance AIC
## - bp 1 286.73 298.73
## - age 1 287.23 299.23
## <none> 286.22 300.22
## - npreg 1 292.35 304.35
## - ped 1 292.70 304.70
## - bmi 1 302.55 314.55
## - glu 1 344.60 356.60
##
## Step: AIC=298.73
## type ~ npreg + glu + bmi + ped + age
##
## Df Deviance AIC
## - age 1 287.44 297.44
## <none> 286.73 298.73
## - npreg 1 293.00 303.00
## - ped 1 293.35 303.35
## - bmi 1 303.27 313.27
## - glu 1 344.67 354.67
##
## Step: AIC=297.44
## type ~ npreg + glu + bmi + ped
##
## Df Deviance AIC
## <none> 287.44 297.44
## - ped 1 294.54 302.54
## - bmi 1 303.72 311.72
## - npreg 1 304.01 312.01
## - glu 1 349.80 357.80##
## Call:
## glm(formula = type ~ npreg + glu + bmi + ped, family = binomial,
## data = Pima.te)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.9845 -0.6462 -0.3661 0.5977 2.5304
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -9.552177 1.096207 -8.714 < 2e-16 ***
## npreg 0.178066 0.045343 3.927 8.6e-05 ***
## glu 0.037971 0.005442 6.978 3.0e-12 ***
## bmi 0.084107 0.021950 3.832 0.000127 ***
## ped 1.165658 0.444054 2.625 0.008664 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 420.30 on 331 degrees of freedom
## Residual deviance: 287.44 on 327 degrees of freedom
## AIC: 297.44
##
## Number of Fisher Scoring iterations: 5Things to note:
- We used the
~.syntax to tell R to fit a model with all the available predictors. - Since we want to focus on significant predictors, we used the
stepfunction to perform a step-wise regression, i.e. sequentially remove non-significant predictors. The function reports each model it has checked, and the variable it has decided to remove at each step. - The output of
stepis a single model, with the subset of selected predictors.
8.2.2 Another Logistic regression example and model evaluation
Let’s model the relationship between the probability that an iris flower is of type “setosa”, and the length of its Sepal.
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species is_setosa
## 1 5.1 3.5 1.4 0.2 setosa 1
## 2 4.9 3.0 1.4 0.2 setosa 1
## 3 4.7 3.2 1.3 0.2 setosa 1
## 4 4.6 3.1 1.5 0.2 setosa 1
## 5 5.0 3.6 1.4 0.2 setosa 1
## 6 5.4 3.9 1.7 0.4 setosa 1
Again, we do logistic regression. Let’s check the fitted probabilities:
m_glm <- glm(d$is_setosa~d$Sepal.Length, family = binomial)
yhat_glm <- predict(m_glm, type = "response")
hist(yhat_glm)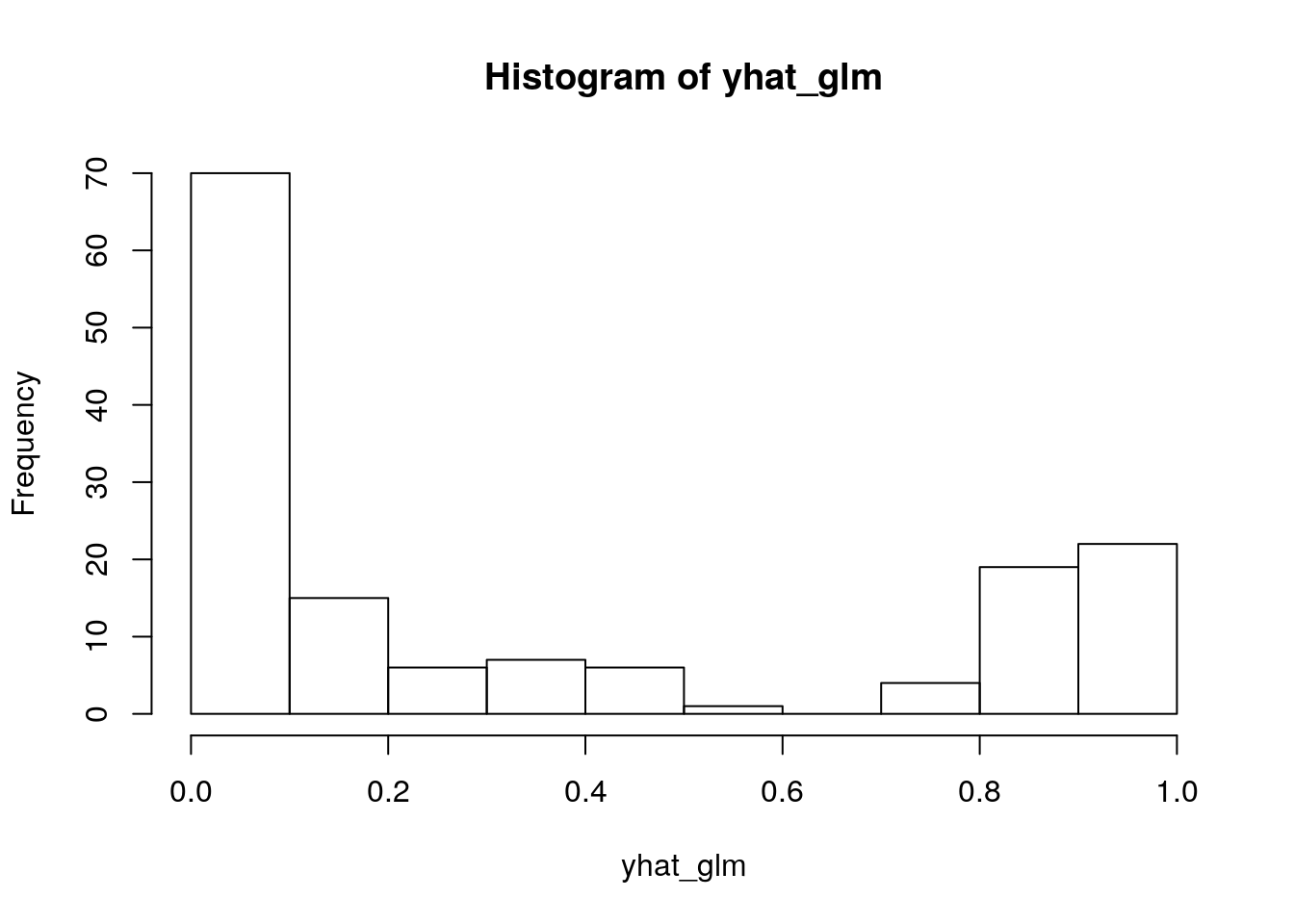
If we are interested in a prediction whether a flower is “setosa” or not, we need a decision rule. A trivial rule is: classify as “1” (’is_setosa`=1) if \(\hat{P}(\text{is_setosa}=1|\text{Sepal.Length})>0.5\)
We can plot the data and distinguish between samples we classed to 1 and 0. We also add the predicted probabilities and the 0.5 threshold:
plot(d$is_setosa~d$Sepal.Length, col = as.factor(yhat_glm_binar))
lines(yhat_glm[order(d$Sepal.Length)]~d$Sepal.Length[order(d$Sepal.Length)])
abline(h=0.5, lty = 2)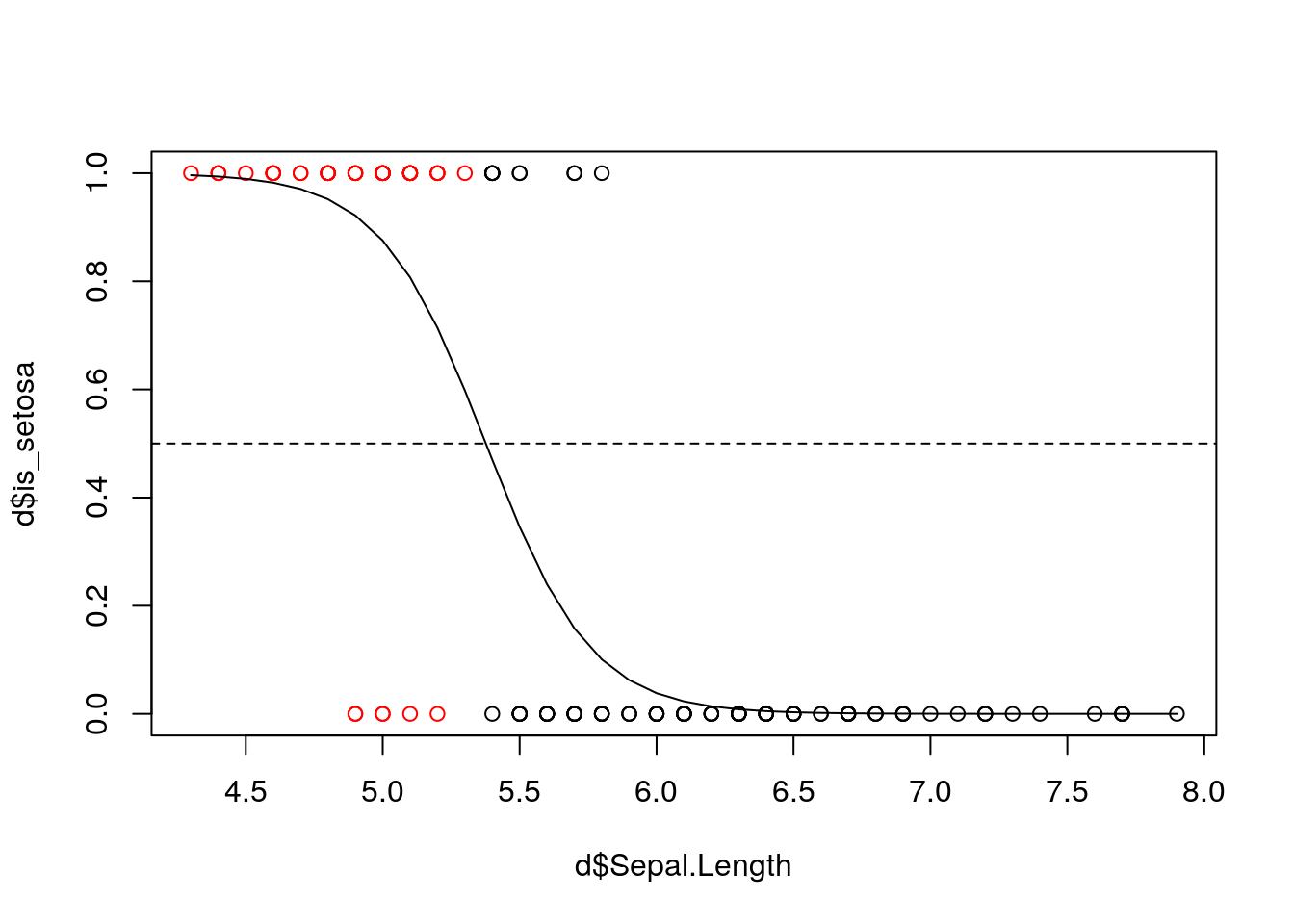
Note that other thresholds are also possible (this is a question of False positives and false negatives)
Let’s compare a logistic and a linear models in a prediction problem. We use the spam dataset from the ElemStatLearn package (see ?ElemStatLearn::spam):
## A.1 A.2 A.3 A.4 A.5 A.6 A.7 A.8 A.9 A.10 A.11 A.12 A.13 A.14
## 1 0.00 0.64 0.64 0 0.32 0.00 0.00 0.00 0.00 0.00 0.00 0.64 0.00 0.00
## 2 0.21 0.28 0.50 0 0.14 0.28 0.21 0.07 0.00 0.94 0.21 0.79 0.65 0.21
## 3 0.06 0.00 0.71 0 1.23 0.19 0.19 0.12 0.64 0.25 0.38 0.45 0.12 0.00
## 4 0.00 0.00 0.00 0 0.63 0.00 0.31 0.63 0.31 0.63 0.31 0.31 0.31 0.00
## 5 0.00 0.00 0.00 0 0.63 0.00 0.31 0.63 0.31 0.63 0.31 0.31 0.31 0.00
## 6 0.00 0.00 0.00 0 1.85 0.00 0.00 1.85 0.00 0.00 0.00 0.00 0.00 0.00
## A.15 A.16 A.17 A.18 A.19 A.20 A.21 A.22 A.23 A.24 A.25 A.26 A.27 A.28
## 1 0.00 0.32 0.00 1.29 1.93 0.00 0.96 0 0.00 0.00 0 0 0 0
## 2 0.14 0.14 0.07 0.28 3.47 0.00 1.59 0 0.43 0.43 0 0 0 0
## 3 1.75 0.06 0.06 1.03 1.36 0.32 0.51 0 1.16 0.06 0 0 0 0
## 4 0.00 0.31 0.00 0.00 3.18 0.00 0.31 0 0.00 0.00 0 0 0 0
## 5 0.00 0.31 0.00 0.00 3.18 0.00 0.31 0 0.00 0.00 0 0 0 0
## 6 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0 0.00 0.00 0 0 0 0
## A.29 A.30 A.31 A.32 A.33 A.34 A.35 A.36 A.37 A.38 A.39 A.40 A.41 A.42
## 1 0 0 0 0 0 0 0 0 0.00 0 0 0.00 0 0
## 2 0 0 0 0 0 0 0 0 0.07 0 0 0.00 0 0
## 3 0 0 0 0 0 0 0 0 0.00 0 0 0.06 0 0
## 4 0 0 0 0 0 0 0 0 0.00 0 0 0.00 0 0
## 5 0 0 0 0 0 0 0 0 0.00 0 0 0.00 0 0
## 6 0 0 0 0 0 0 0 0 0.00 0 0 0.00 0 0
## A.43 A.44 A.45 A.46 A.47 A.48 A.49 A.50 A.51 A.52 A.53 A.54 A.55
## 1 0.00 0 0.00 0.00 0 0 0.00 0.000 0 0.778 0.000 0.000 3.756
## 2 0.00 0 0.00 0.00 0 0 0.00 0.132 0 0.372 0.180 0.048 5.114
## 3 0.12 0 0.06 0.06 0 0 0.01 0.143 0 0.276 0.184 0.010 9.821
## 4 0.00 0 0.00 0.00 0 0 0.00 0.137 0 0.137 0.000 0.000 3.537
## 5 0.00 0 0.00 0.00 0 0 0.00 0.135 0 0.135 0.000 0.000 3.537
## 6 0.00 0 0.00 0.00 0 0 0.00 0.223 0 0.000 0.000 0.000 3.000
## A.56 A.57 spam
## 1 61 278 spam
## 2 101 1028 spam
## 3 485 2259 spam
## 4 40 191 spam
## 5 40 191 spam
## 6 15 54 spam##
## 0 1
## 2788 1813Creating a training and test sets:
set.seed(256)
in_train <- sample(1:nrow(db), 0.5*nrow(db))
db_train <- db[in_train, ]
db_test <- db[-in_train, ]
table(db_train$spam)##
## 0 1
## 1396 904##
## 0 1
## 1392 909Comparing the prediction accuracy of the models over a “fresh” test set:
## Warning: glm.fit: fitted probabilities numerically 0 or 1 occurredy_hat_lm <- predict(m_lm, db_test)
y_hat_glm <- predict(m_glm, db_test, type = "response")
y_hat_lm_binar <- (y_hat_lm>0.5)*1
y_hat_glm_binar <- (y_hat_glm>0.5)*1
mean(db_test$spam==y_hat_lm_binar)## [1] 0.8813559## [1] 0.9269883It seems that the logistic regression does a better job
8.2.3 Measuring Accuracy, Precision and Recall:
Confusion Matrix
In the field of machine learning and specifically the problem of statistical classification, a confusion matrix, also known as an error matrix, is a specific table layout that allows visualization of the performance of a prediction algorithm (as logistic regression), typically a supervised learning one. Each row of the matrix represents the instances in a predicted class while each column represents the instances in an actual class (or vice versa). The name stems from the fact that it makes it easy to see if the system is confusing two classes (i.e. commonly mislabeling one as another).
The confusion matrix is a better choice to evaluate the classification performance compared with the different metrics as AIC which you saw before - it compute the performance on new set, which we did not use at model fitting. The general idea is to count the number of times True/False instances are classified as True/False:
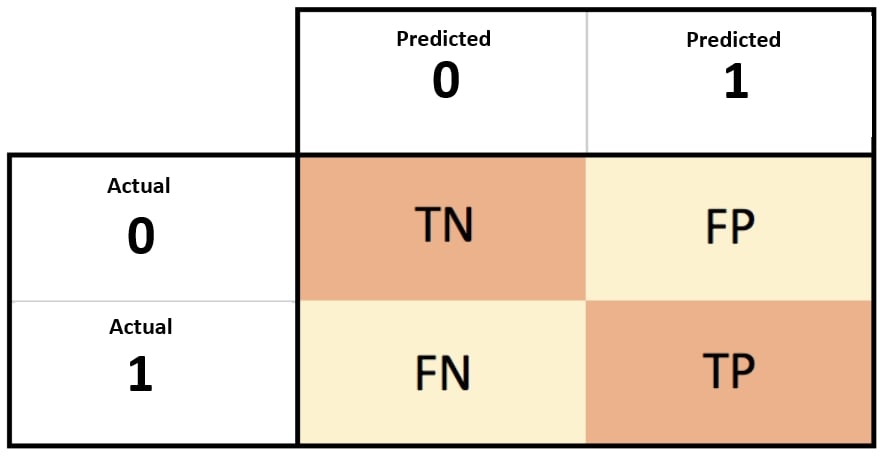
confusion matrix
It can be obtained by:
## predicted
## true 0 1
## 0 1330 62
## 1 106 803The Accuracy is defined as: \[Accuracy = \frac{TP + TN}{TP + TN + FP + FN}\] and can be obtained with:
## [1] 0.9269883But accuracy can sometimes be a bad metric for predictive classification models (see The Accuracy Paradox. For example, if the incidence of category A is dominant, being found in 99% of cases, then predicting that every case is category A will have an accuracy of 99%. In that case an unuseful model may have a high level of accuracy.
Precision and recall are better measures in such cases. \[Precision = \frac{TP}{TP + FP}\] \[Recall = \frac{TP}{TP + FN}\] Precision looks at the accuracy of the positive prediction. Recall is the ratio of positive instances that are correctly detected by the classifier. While recall expresses the ability to find all relevant instances in a dataset, precision expresses the proportion of the data points our model says was relevant actually were relevant.
We can get them with:
## [1] 0.9283237## [1] 0.8833883Note that we can not have have high precision and high recall at the same time, and there is a tradeoff between the two. Precision is more important than Recall when you would like to have less False Positives in trade off to have more False Negatives. Meaning, getting a False Positive is very costly, and a False Negative is not as much. In some situations, we might know that we want to maximize either recall or precision at the expense of the other metric. For example, in preliminary disease screening of patients for follow-up examinations, we would probably want a recall near 1.0, as we want to find all patients who actually have the disease, and we can accept a low precision if the cost of the follow-up examination is not significant. You may know this tradoff as Type I and type II errors in statistical hypothesis testing.
In cases where we want to find an optimal blend of precision and recall we can combine the two metrics using what is called the F1 score: \[ F_1 = 2 \frac{precision*recall}{precision+recall}\].
There is a concave relationship between precision and recall. The ROC curve is a graphical plot that illustrates the diagnostic ability of a binary classification model (such as the logistic regression) as its discrimination threshold is varied. The ROC curve is created by plotting the True Positive Rate (Recall) against the False Positive Rate (\(FP/(FP+TN)\)) at various threshold settings.
here is an example of how to compute it:
alphas <- seq(0,1,0.01)
TPRs <- numeric(length(alphas))
FPRs <- numeric(length(alphas))
for (i in seq_along(alphas)) {
pr_i <- ifelse(y_hat_glm>alphas[i],1,0)
CM_i <- table(db_test$spam, pr_i)
TPRs[i] <- CM_i[4] / sum(CM_i[2,])
FPRs[i] <- CM_i[3] / sum(CM_i[1,])
}
plot(TPRs~FPRs, type = "l")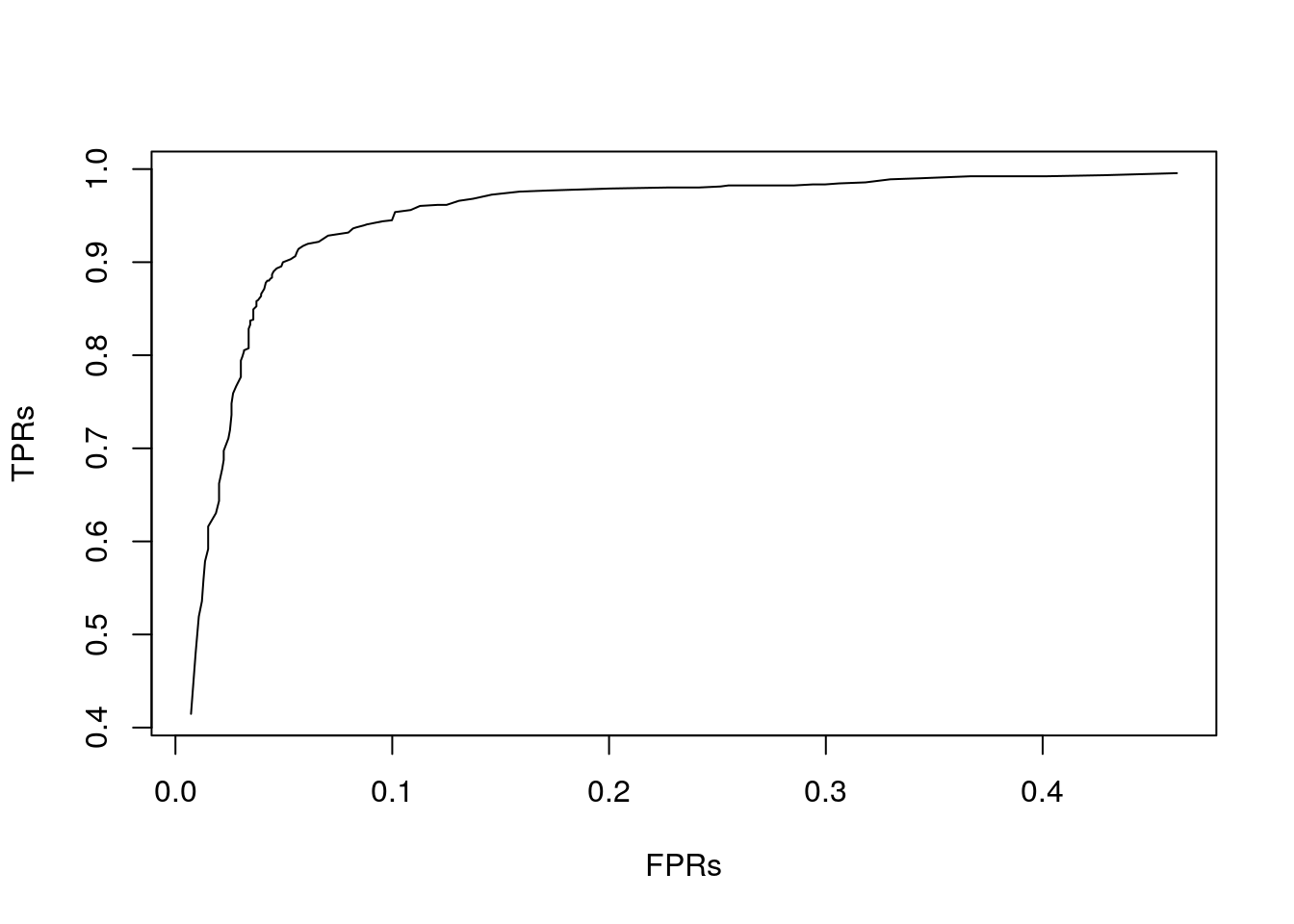 And see here fancier ROC libraries, e.g., with the
And see here fancier ROC libraries, e.g., with the ROCit package:
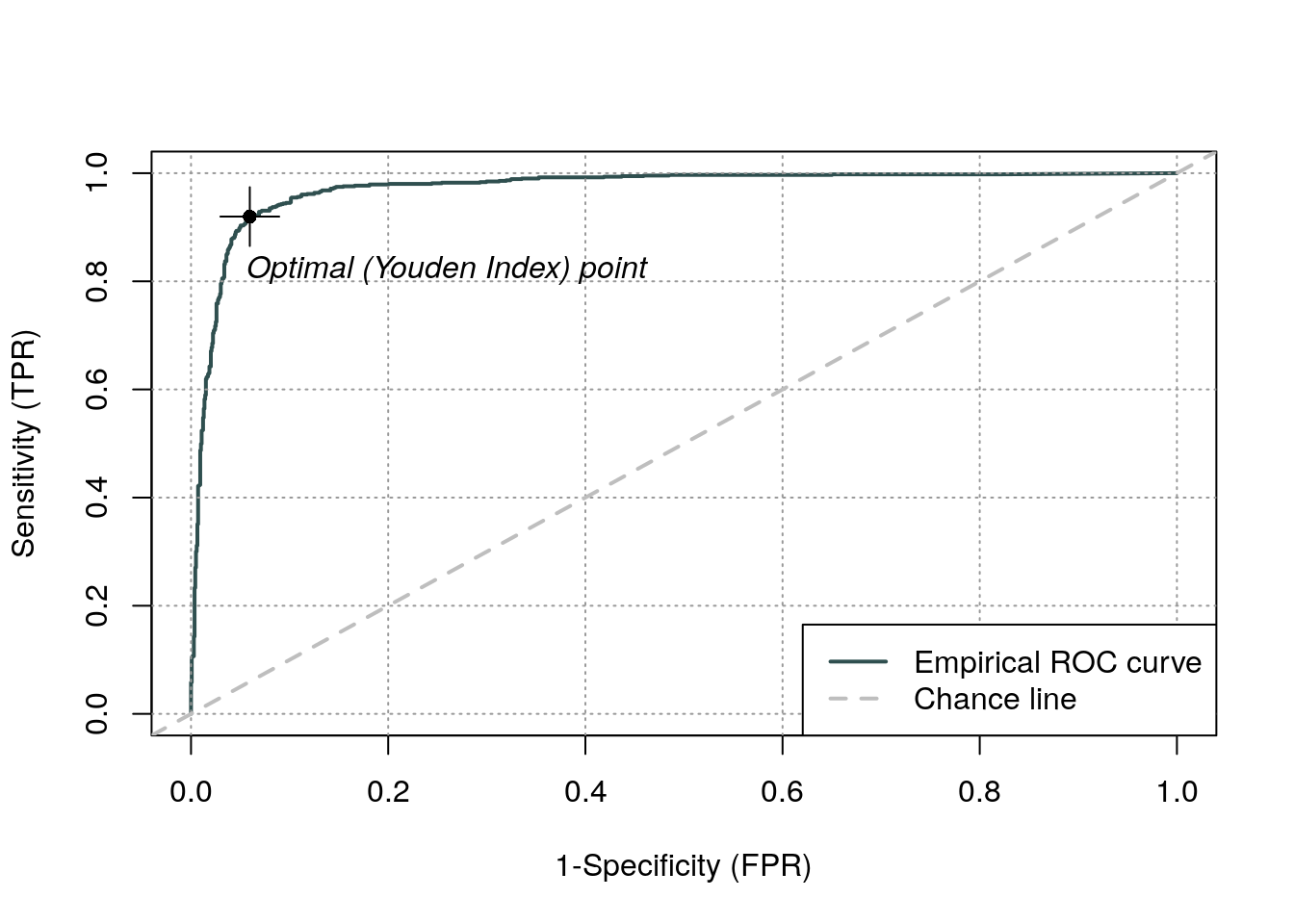
ROC analysis provides tools to select possibly optimal models and to discard suboptimal ones independently from (and prior to specifying) the cost context or the class distribution.

The ROC space for a “better” and “worse” classfier. Source: Wikipedia
8.3 Poisson Regression
Poisson regression means we fit a model assuming \(y|x \sim Poisson(\lambda(x))\). Put differently, we assume that for each treatment, encoded as a combinations of predictors \(x\), the response is Poisson distributed with a rate that depends on the predictors.
The typical link function for Poisson regression is the logarithm: \(g(t)=log(t)\). This means that we assume \(y|x \sim Poisson(\lambda(x) = e^{x'\beta})\). Why is this a good choice? We again resort to the two-group case, encoded by \(x=1\) and \(x=0\), to understand this model: \(\lambda(x=1)=e^{\beta_0+\beta_1}=e^{\beta_0} \; e^{\beta_1}= \lambda(x=0) \; e^{\beta_1}\). We thus see that this link function implies that a change in \(x\) multiples the rate of events by \(e^{\beta_1}\).
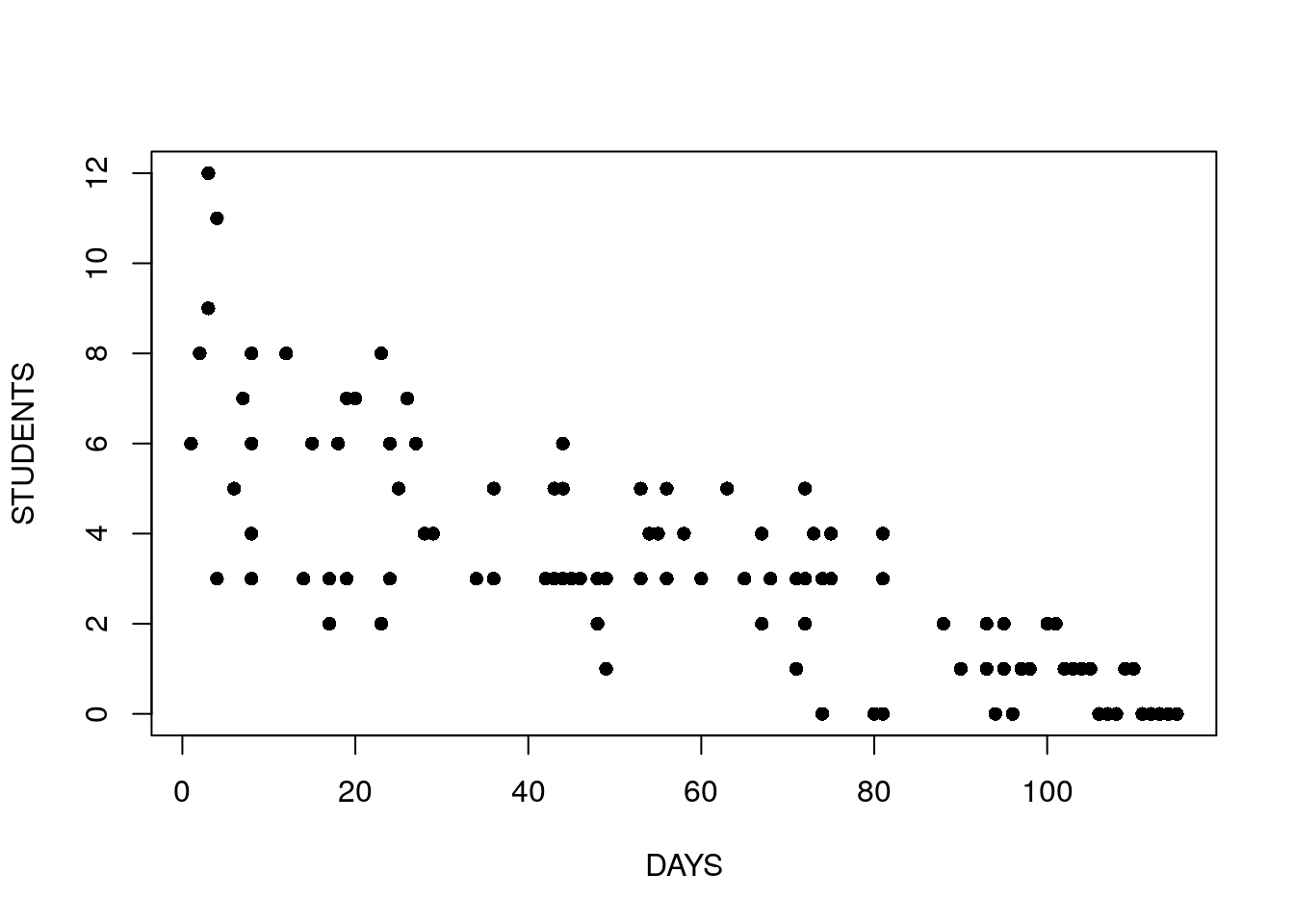
For our example14 we inspect the number of infected high-school kids, as a function of the days since an outbreak.
cases <-
structure(list(Days = c(1L, 2L, 3L, 3L, 4L, 4L, 4L, 6L, 7L, 8L,
8L, 8L, 8L, 12L, 14L, 15L, 17L, 17L, 17L, 18L, 19L, 19L, 20L,
23L, 23L, 23L, 24L, 24L, 25L, 26L, 27L, 28L, 29L, 34L, 36L, 36L,
42L, 42L, 43L, 43L, 44L, 44L, 44L, 44L, 45L, 46L, 48L, 48L, 49L,
49L, 53L, 53L, 53L, 54L, 55L, 56L, 56L, 58L, 60L, 63L, 65L, 67L,
67L, 68L, 71L, 71L, 72L, 72L, 72L, 73L, 74L, 74L, 74L, 75L, 75L,
80L, 81L, 81L, 81L, 81L, 88L, 88L, 90L, 93L, 93L, 94L, 95L, 95L,
95L, 96L, 96L, 97L, 98L, 100L, 101L, 102L, 103L, 104L, 105L,
106L, 107L, 108L, 109L, 110L, 111L, 112L, 113L, 114L, 115L),
Students = c(6L, 8L, 12L, 9L, 3L, 3L, 11L, 5L, 7L, 3L, 8L,
4L, 6L, 8L, 3L, 6L, 3L, 2L, 2L, 6L, 3L, 7L, 7L, 2L, 2L, 8L,
3L, 6L, 5L, 7L, 6L, 4L, 4L, 3L, 3L, 5L, 3L, 3L, 3L, 5L, 3L,
5L, 6L, 3L, 3L, 3L, 3L, 2L, 3L, 1L, 3L, 3L, 5L, 4L, 4L, 3L,
5L, 4L, 3L, 5L, 3L, 4L, 2L, 3L, 3L, 1L, 3L, 2L, 5L, 4L, 3L,
0L, 3L, 3L, 4L, 0L, 3L, 3L, 4L, 0L, 2L, 2L, 1L, 1L, 2L, 0L,
2L, 1L, 1L, 0L, 0L, 1L, 1L, 2L, 2L, 1L, 1L, 1L, 1L, 0L, 0L,
0L, 1L, 1L, 0L, 0L, 0L, 0L, 0L)), .Names = c("Days", "Students"
), class = "data.frame", row.names = c(NA, -109L))
attach(cases)
head(cases) ## Days Students
## 1 1 6
## 2 2 8
## 3 3 12
## 4 3 9
## 5 4 3
## 6 4 3Look at the following plot and think:
- Can we assume that the errors have constant variace?
- What is the sign of the effect of time on the number of sick students?
- Can we assume a linear effect of time?
We now fit a model to check for the change in the rate of events as a function of the days since the outbreak.
##
## Call:
## glm(formula = Students ~ Days, family = poisson)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.00482 -0.85719 -0.09331 0.63969 1.73696
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) 1.990235 0.083935 23.71 <2e-16 ***
## Days -0.017463 0.001727 -10.11 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for poisson family taken to be 1)
##
## Null deviance: 215.36 on 108 degrees of freedom
## Residual deviance: 101.17 on 107 degrees of freedom
## AIC: 393.11
##
## Number of Fisher Scoring iterations: 5Things to note:
- We used
family=poissonin theglmfunction to tell R that we assume a Poisson distribution. - The coefficients table is there as usual. When interpreting the table, we need to recall that the effect, i.e. the \(\hat \beta\), are multiplicative due to the assumed link function.
- Each day decreases the rate of events by a factor of about \(e^{\beta_1}=\) 0.983.
- For more information see
?glmand?family.
8.4 Extensions
As we already implied, GLMs are a very wide class of models.
We do not need to use the default link function,but more importantly, we are not constrained to Binomial, or Poisson distributed response.
For exponential, gamma, and other response distributions, see ?glm or the references in the Bibliographic Notes section.
8.5 Bibliographic Notes
The ultimate reference on GLMs is McCullagh (1984). For a less technical exposition, we refer to the usual Venables and Ripley (2013).
8.6 Practice Yourself
- Try using
lmfor analyzing the plant growth data inweight.factoras a function ofgroupin thePlantGrowthdata. - Generate some synthetic data for a logistic regression:
- Generate two predictor variables of length \(100\). They can be random from your favorite distribution.
- Fix
beta<- c(-1,2), and generate the response with:rbinom(n=100,size=1,prob=exp(x %*% beta)/(1+exp(x %*% beta))). Think: why is this the model implied by the logistic regression? - Fit a Logistic regression to your synthetic data using
glm. - Are the estimated coefficients similar to the true ones you used?
- What is the estimated probability of an event at
x=1,1? Usepredict.glmbut make sure to read the documentation on thetypeargument.
- Read about the
epildataset using? MASS::epil. Inspect the dependency of the number of seizures (\(y\)) in the age of the patient (age) and the treatment (trt).- Fit a Poisson regression with
glmandfamily = "poisson". - Are the coefficients significant?
- Does the treatment reduce the frequency of the seizures?
- According to this model, what would be the number of seizures for 20 years old patient with progabide treatment?
- Fit a Poisson regression with
References
McCullagh, Peter. 1984. “Generalized Linear Models.” European Journal of Operational Research 16 (3): 285–92.
Venables, William N, and Brian D Ripley. 2013. Modern Applied Statistics with S-Plus. Springer Science & Business Media.