Module 3 Additional tools
3.1 Identifier tools
Supported identifier types:
- UniProtKB
- Entrez
- Gene Symbol
- Ensembl Gene ID
- Ensembl Protein ID
If you have a list of proteins with a supported identifier type, you can convert it to any of the other supported identifier types. Paste the identifiers in the box and click Convert IDs. A table with all matches appears. Because some identifier types match one-to-many, you can select the option “Single” to only show the first match for an inserted identifier, or “All matches” to show all matching identifiers for that idenfitier.
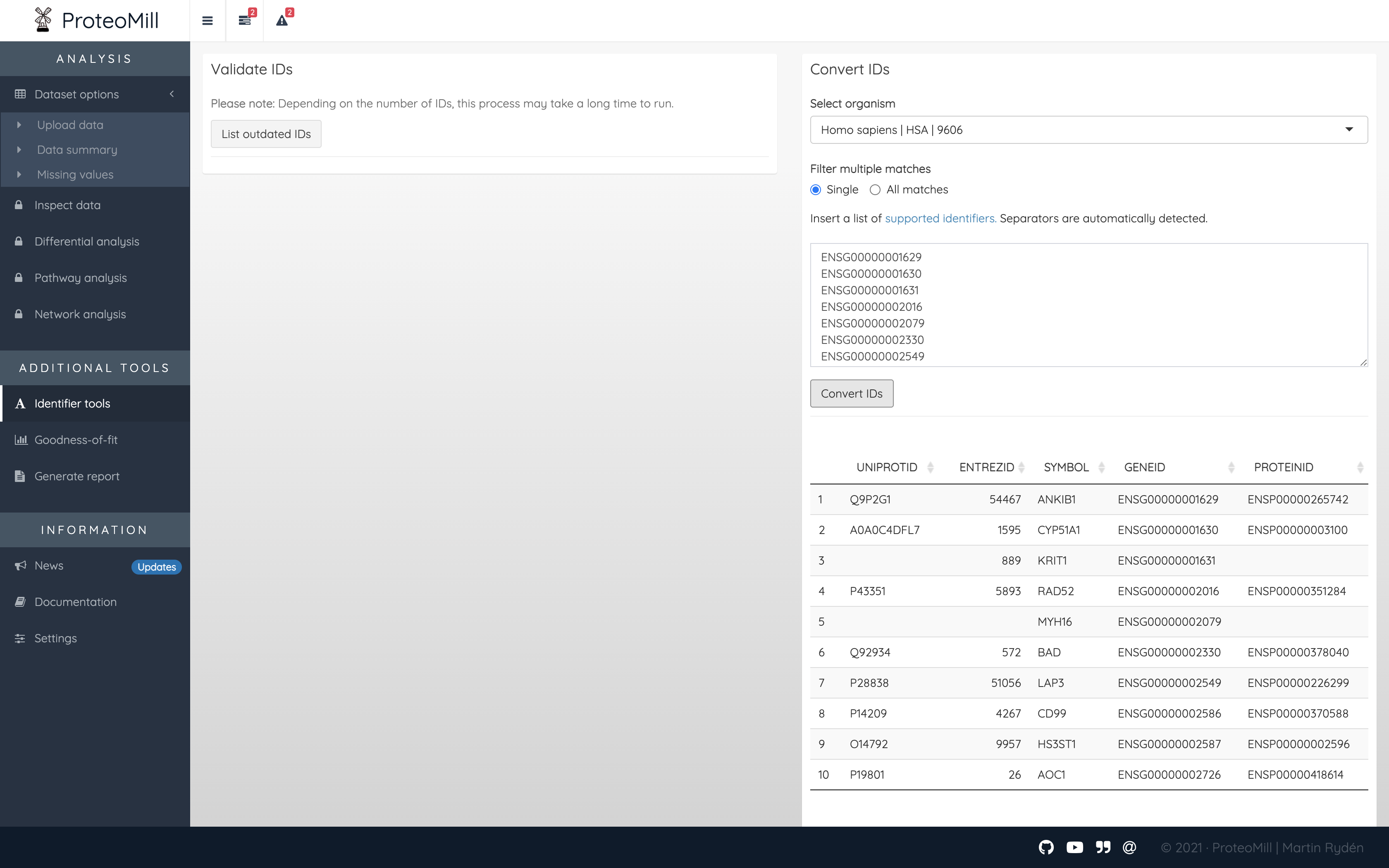
Figure 3.1: Convert IDs. A list of identifiers has been converted to all supported identifier types.
3.2 Goodness-of-fit
This information will be updated soon.
3.3 Generate report
All the figures produced in ProteoMill can be automatically distilled into a report. The only requirement is that you have uploaded a dataset. In the Generate report box, click on the drop-down menu and click Select all. Then click Generate report. The report will be saved to your computer as a HTML file.
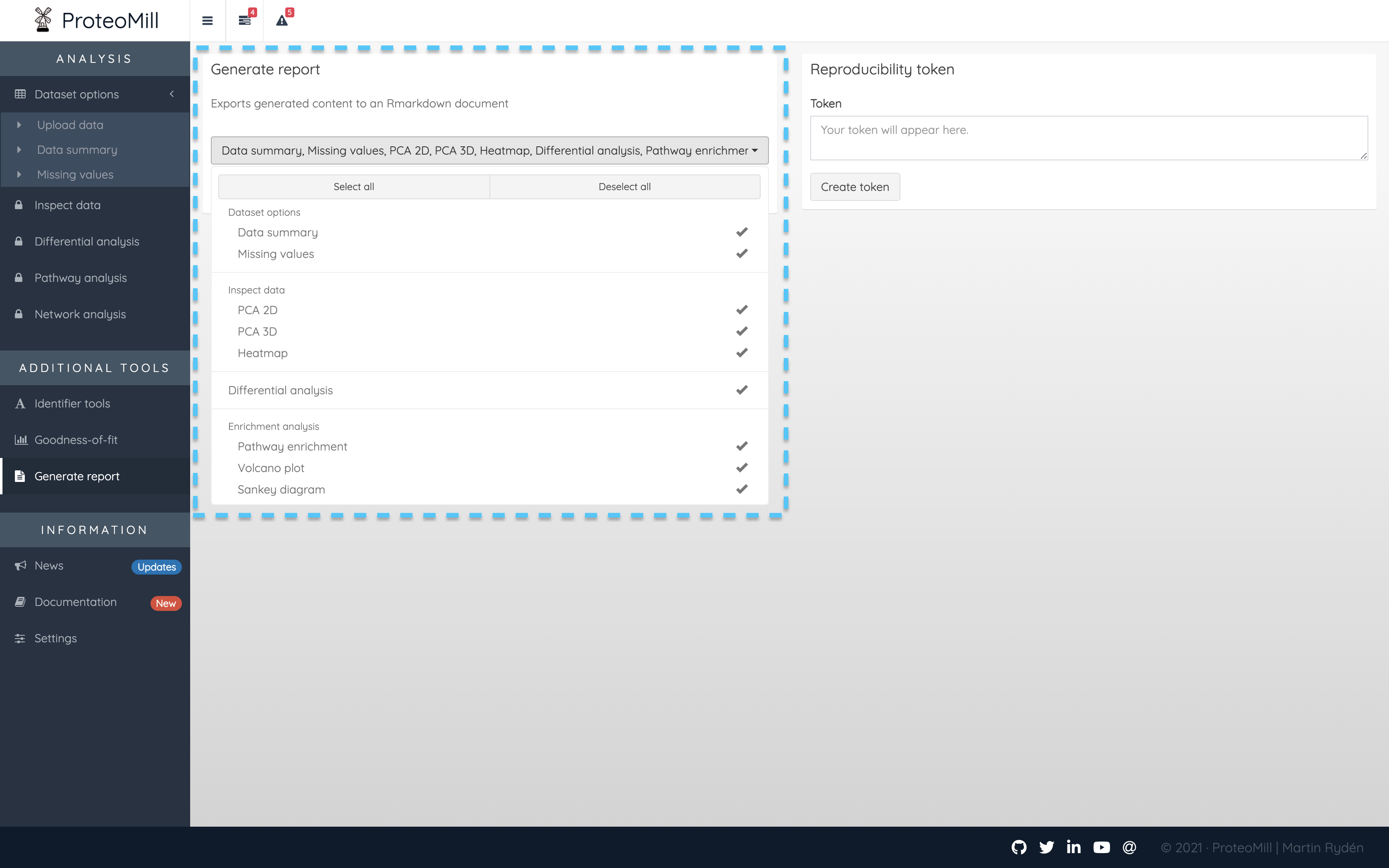
Figure 3.2: Generate a report
3.4 Reproduce experiments
ProteoMill now supports reproducibility tokens as a simple way to load the settings and database versions used in a previous experiment. This feature is found in the Generate report tab.
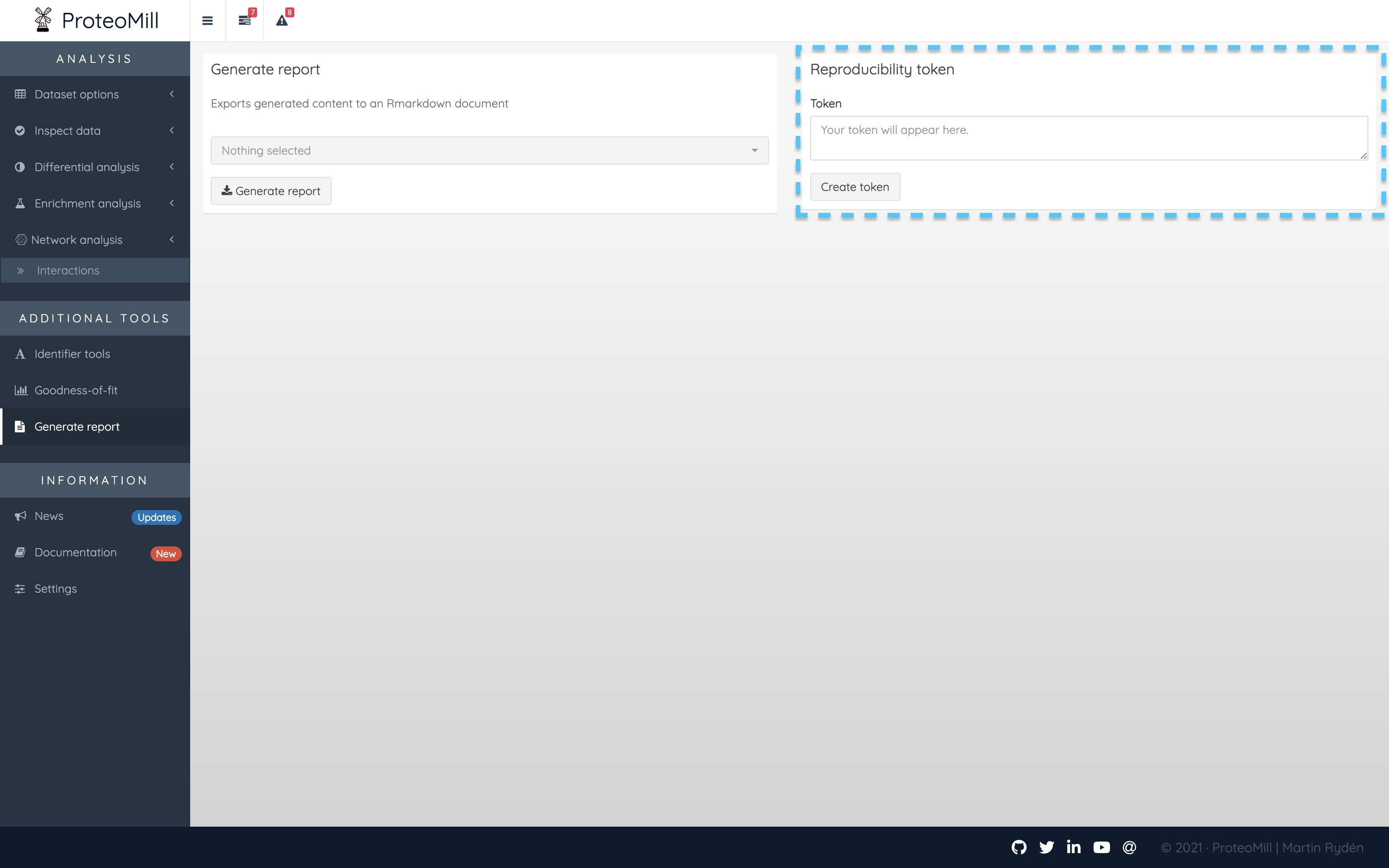
Figure 3.3: Generate a token
When you have finished your analysis, and want to save the settings used so that you or other researchers (who have access to the input file used) can reproduce the experiment, click on “Create token”. This will generate a key that you need to save exactly as it is. Any modifications made to the token OR the input files means the token is not valid.
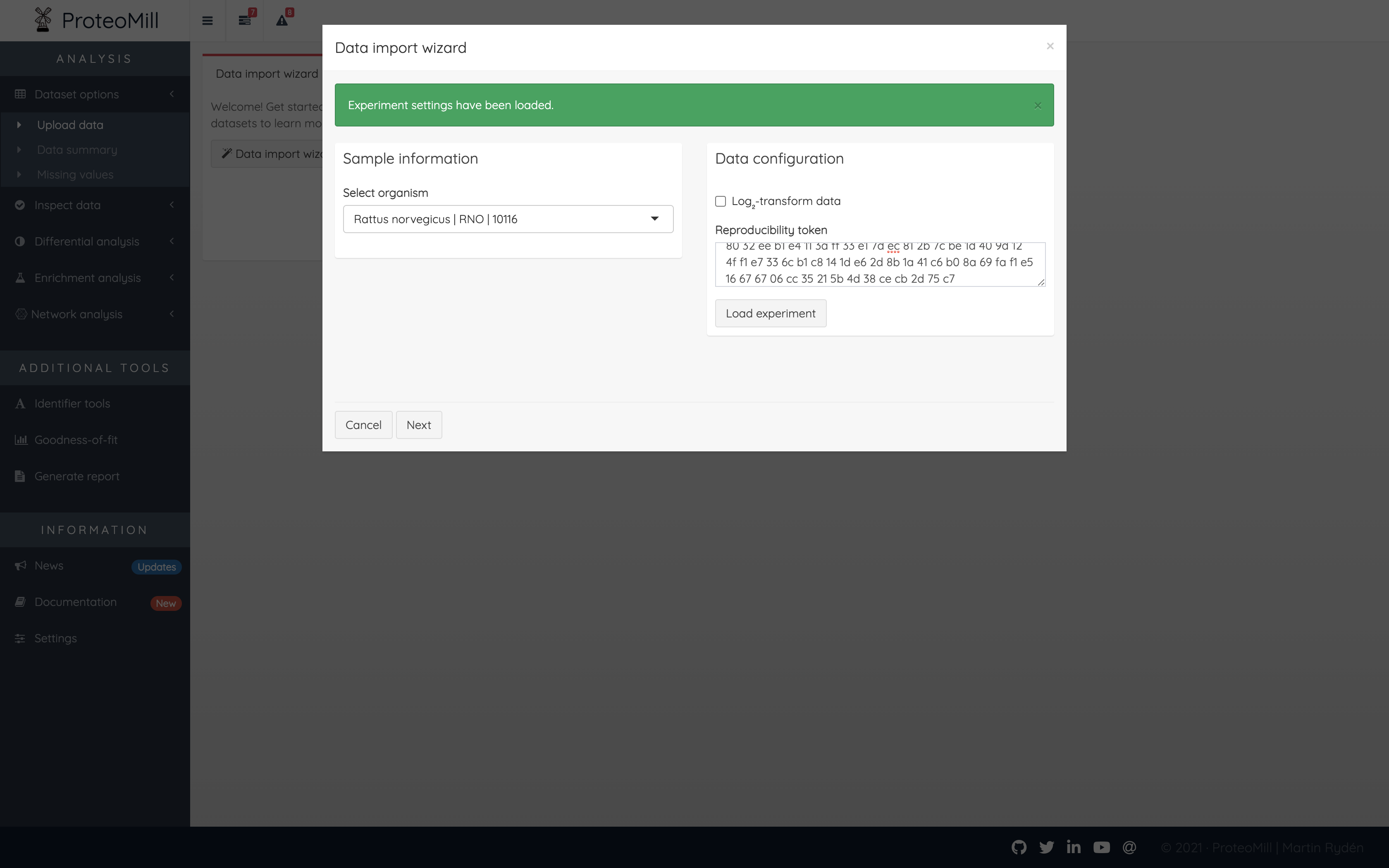
Figure 3.4: Use a token
If you want to load experiment settings using a token, open the Data import wizard and insert the token in the Reproducibility token field, then click Load experiment.
Figure 3.5: ProteoMill verifies that the previously used file is identical to the currently uploaded file using MD5 hashes.