Note 3 Simulation Example on Structural Equation Modeling (SEM)
# Load required packages
library(tidyverse)
theme_set(theme_classic() +
theme(panel.grid.major.y = element_line(color = "grey92")))3.1 Simulate Multivariate Data
In SEM, when multivariate normality is assumed, one can either generate data directly using matrix algebra, or generate the latent variables first before generating the observed variables. The first method is faster, but the second method is more general and can be applied to situations like categorical data or multilevel data. Therefore, in this note we’ll use the second method.
Let’s do a latent growth model (LGM) similar to the one in the note “Simulating Multilevel Data.” Here is the model in a latent growth model representation: \[\begin{bmatrix} y_{0i} \\ y_{1i} \\ y_{2i} \\ y_{3i} \end{bmatrix} = \boldsymbol{\mathbf{\Lambda }} \begin{bmatrix} \eta_{1i} \\ \eta_{2i} \end{bmatrix} + \begin{bmatrix} e_{0i} \\ e_{1i} \\ e_{2i} \\ e_{3i} \end{bmatrix}\] where \(y_{0i}, \ldots, y_{3i}\) are the outcome values for person \(i\) from time 0 to time 3, \(\eta_{1i}\) is the specific intercept for person \(i\), \(\eta_{2i}\) is the specific slope for person \(i\), and \(e_{0i}, \ldots, e_{3i}\) are the within-person level error term. The distributional assumptions are
\[\begin{align*} \begin{bmatrix} \eta_{1i} \\ \eta_{2i} \end{bmatrix} & \sim \mathcal{N}\left(\begin{bmatrix} \alpha_1 \\ \alpha_2 \end{bmatrix}, \begin{bmatrix} \phi_{11} & \phi_{21} \\ \phi_{21} & \phi_{22} \end{bmatrix}\right) \\ \begin{bmatrix} e_{0i} \\ e_{1i} \\ e_{2i} \\ e_{3i} \end{bmatrix} & \sim \mathcal{N}\left(\begin{bmatrix} 0 \\ 0 \\ 0 \\ 0 \end{bmatrix}, \begin{bmatrix} \theta_{11} & 0 & 0 & 0 \\ 0 & \theta_{22} & 0 & 0 \\ 0 & 0 & \theta_{33} & 0 \\ 0 & 0 & 0 & \theta_{44} \\ \end{bmatrix}\right) \end{align*}\]
A path diagram is shown below:
growth_model <- "i =~ 1*y1 + 1*y2 + 1*y3 + 1*y4
s =~ 0*y1 + 1*y2 + 2*y3 + 3*y4
i ~~ 1 * i
s ~~ 0.2 * s + 0.1 * i
y1 ~~ 0.5 * y1
y2 ~~ 0.5 * y2
y3 ~~ 0.5 * y3
y4 ~~ 0.5 * y4
i ~ 1 * 1
s ~ 0.5 * 1"
library(semPlot)
semPaths(semPlotModel_lavaanModel(growth_model))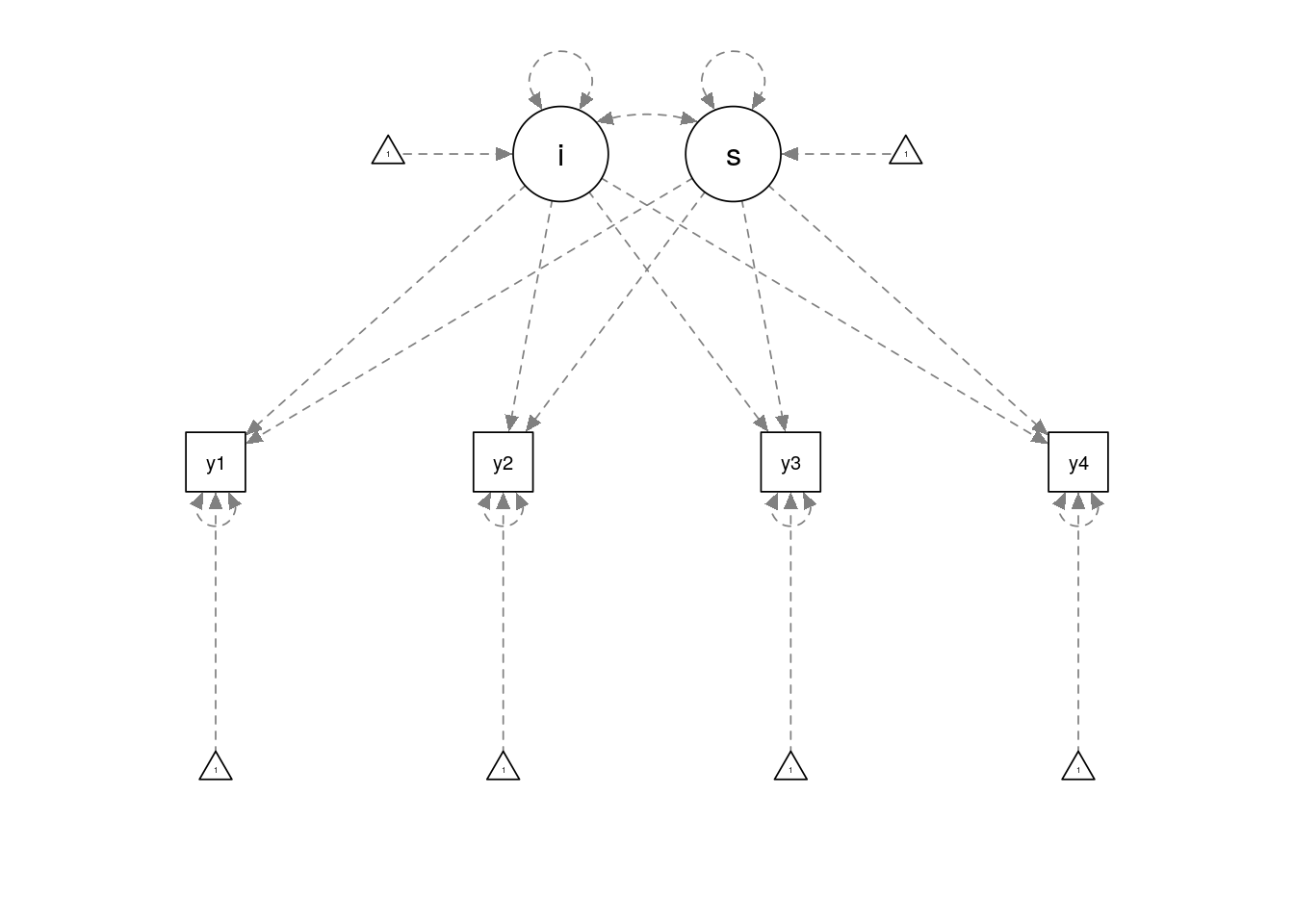
3.1.1 Using R
Here’s the code for generating the data:
library(mnormt)
set.seed(123)
# Define sample size
N <- 100
# Define the Fixed Parameters
alpha <- c(1, 0.5) # latent means
Phi <- matrix(c(1, 0.1,
0.1, 0.2), nrow = 2) # latent variances/covariances
Lambda <- cbind(c(1, 1, 1, 1),
c(0, 1, 2, 3)) # factor loadings
Theta <- diag(0.5, nrow = 4)
# Generate latent factor scores
eta <- rmnorm(N, mean = alpha, varcov = Phi)
# Generate residuals:
e <- rmnorm(N, varcov = Theta)
# Compute outcome scores: y_i = t(Lambda %*% eta_i) + e
y <- tcrossprod(eta, Lambda) + e
# Make it a data frame
colnames(y) <- paste0("y", 1:4)
df <- as.data.frame(y)If you want to check whether the simulated data is correct, generate with a large sample size, and check the means and covariances:
N_test <- 1e5
eta_test <- rmnorm(N_test, mean = alpha, varcov = Phi)
colMeans(eta_test)># [1] 1.0016925 0.5021968cov(eta_test)># [,1] [,2]
># [1,] 1.001489 0.1000130
># [2,] 0.100013 0.2011578e_test <- rmnorm(N_test, varcov = Theta)
colMeans(e_test)># [1] 0.0008598494 0.0025900286 -0.0051191571 0.0011246080cov(e_test)># [,1] [,2] [,3] [,4]
># [1,] 0.4995076410 -0.0007194104 0.0010100393 -0.0007624743
># [2,] -0.0007194104 0.4993700934 -0.0016984945 0.0024070969
># [3,] 0.0010100393 -0.0016984945 0.4965962722 0.0003983313
># [4,] -0.0007624743 0.0024070969 0.0003983313 0.5027934517It’s handy to wrap this as a function:
gen_lgm_data <- function(N, alpha, Phi, Lambda, Theta) {
# Generate latent factor scores
eta <- rmnorm(N, mean = alpha, varcov = Phi)
# Generate residuals:
e <- rmnorm(N, varcov = Theta)
# Compute outcome scores
y <- tcrossprod(eta, Lambda) + e
colnames(y) <- paste0("y", 1:4)
# Make it a data frame
as.data.frame(y)
}
# Test it:
set.seed(123)
gen_lgm_data(100,
alpha = c(1, 0.5),
Phi = matrix(c(1, 0.1,
0.1, 0.2), nrow = 2),
Lambda = cbind(c(1, 1, 1, 1),
c(0, 1, 2, 3)),
Theta = diag(0.5, nrow = 4)) %>%
head() # shows only the first six cases># y1 y2 y3 y4
># 1 1.994318062 1.71116086 0.93927927 1.8544817
># 2 2.265725727 2.90855563 3.37429135 4.1980646
># 3 2.296655600 2.35159235 3.73462339 5.0831256
># 4 2.332408257 1.09066818 0.74843609 2.6298055
># 5 0.001197646 0.03891746 -0.08691963 0.1158814
># 6 1.818221258 3.44031115 4.56735772 5.0621645Indeed, you get the same data!
3.1.2 Other methods for generating SEM data
Many SEM software or packages have capability in generating data with input of
an SEM model. For example, in R, you can call Mplus using the
MplusAutomation package and use their MONTECARLO routine. In R, you can
generate SEM data using the lavaan package with the simulateData()
function, like the following example:
# Using a previously defined SEM model:
lavaan::simulateData(growth_model) %>%
head() # shows only the first six cases># y1 y2 y3 y4
># 1 0.7164342 0.5568492 0.4726109 -0.1122304
># 2 1.8000324 1.8018441 1.5462532 2.4277768
># 3 0.6034256 0.6713423 2.2884345 3.1191797
># 4 1.1521190 2.7229104 4.7980347 5.8445554
># 5 0.6229306 -0.1175073 1.7054730 0.5659889
># 6 0.5230306 2.1871287 2.6141488 2.6783935Personally, however, I prefer directly simulating data in R because
- it forces you to specify everything in the model in the way you want. Especially in Mplus there are a lot of hidden default settings that may mess up with your simulation;
- it makes the process of generating data more transparent;
- it helps you learn the math behind the model;
- it is more flexible as you can specify any distributional assumptions or models not supported by the SEM packages.
3.2 Analyzing the Simulated Data
In R, for running SEM models, the most common options are lavaan, OpenMx,
and Mplus (via MplusAutomation). When possible, I’ll stick to lavaan to
avoid jumping between programs, so let’s analyze the simulated data twice, first
with the true model and second with a misspecified model where the random slope
term is omitted (i.e., the variance of s is constrained to zero).
library(lavaan)># This is lavaan 0.6-3># lavaan is BETA software! Please report any bugs.# True model
m1 <- 'i =~ 1*y1 + 1*y2 + 1*y3 + 1*y4
s =~ 0*y1 + 1*y2 + 2*y3 + 3*y4
i ~~ s'
# Model without random slopes
m2 <- 'i =~ 1*y1 + 1*y2 + 1*y3 + 1*y4
s =~ 0*y1 + 1*y2 + 2*y3 + 3*y4
s ~~ 0*i + 0*s'
m1_fit <- growth(m1, data = df)
m2_fit <- growth(m2, data = df)Here’s some examples of things you can extract (without showing all the results)
fitMeasures(m1_fit) # return more than 30 fit indices
# Specific fit indices
fitMeasures(m1_fit, c("chisq", "df", "pvalue", "cfi", "rmsea"))
coef(m1_fit) # parameter estimates
vcov(m1_fit) # asymptotic covariance matrix of parameters
# Asymptotic standard errors of latent mean of s
sqrt(diag(vcov(m1_fit))["s~1"])
parameterEstimates(m1_fit, standardized = FALSE) # coefficients, SE, CI
modificationIndices(m1_fit, minimum.value = 3.84) # modification indices
# Bootstrapped fit index
bootstrapLavaan(m1_fit, R = 1000L, type = "bollen.stine",
FUN = fitMeasures, fit.measures = c("cfi"))3.3 Full Example of a Small Scale Simulation
In a methodological experiment with Monte Carlo simulation, one usually generates millions of data sets across tens or hundreds of carefully chosen conditions. As an example, here is a small scale simulation study on LGM. The two goals are: (a) to understand the bias on the mean of slopes and its standard error estimates, and (b) to illustrate the difference between estimated and empirical standard error.
For simplicity, I’ll only choose three designed factors (i.e., manipulated independent variables), namely sample size, variance of the random slope in the data generating model, and the mean of the slopes. The design factors are summarized here:
- Sample size (N): 50, 100, 200
- Variance of slopes (\(\phi_{22}\)): 0.1, 0.5 (i.e., 1/10 and 1/2 of the intercept variance)
- Mean of slopes (\(\alpha_2\)): 1, 0.5
Therefore, it’s a 3 \(\times\) 2 \(\times\) 2 factorial design. Now let’s walk through each component.
3.3.1 Fixed Values for the Study
In the study I decided to run 500 replications for each condition. I can estimate the simulation error to see whether it’s enough.
set.seed(515) # set the seed for reproducibility
NREP <- 500 # number of replications
# Fixed parameters (all caps)
ALPHA1 <- 1 # latent mean of intercepts
PHI11 <- 1 # intercept variance
LAMBDA <- cbind(c(1, 1, 1, 1),
c(0, 1, 2, 3)) # factor loadings
THETA <- diag(0.5, nrow = 4) # residual variancesWe should also define everything that will not change across replications and across conditions, including syntax for running growth models, functions to generate data, etc.
# Function for generating data:
gen_lgm_data <- function(N, alpha, Phi, Lambda, Theta) {
# Generate latent factor scores
eta <- rmnorm(N, mean = alpha, varcov = Phi)
# Generate residuals:
e <- rmnorm(N, varcov = Theta)
# Compute outcome scores
y <- tcrossprod(eta, Lambda) + e
colnames(y) <- paste0("y", 1:4)
# Make it a data frame
as.data.frame(y)
}
# lavaan syntax
# True model
m1 <- 'i =~ 1*y1 + 1*y2 + 1*y3 + 1*y4
s =~ 0*y1 + 1*y2 + 2*y3 + 3*y4
i ~~ s'
# Model without random slopes
m2 <- 'i =~ 1*y1 + 1*y2 + 1*y3 + 1*y4
s =~ 0*y1 + 1*y2 + 2*y3 + 3*y4
s ~~ 0*i + 0*s'3.3.2 Keep track of simulation conditions
I’ll create a data frame to store the design factors:
# Design factors:
DESIGNFACTOR <- expand.grid(
N = c(50, 100, 200),
phi22 = c(0.1, 0.5),
alpha2 = c(1, 0.5)
)
# Add condition number:
DESIGNFACTOR <- rowid_to_column(DESIGNFACTOR, "cond")
DESIGNFACTOR># cond N phi22 alpha2
># 1 1 50 0.1 1.0
># 2 2 100 0.1 1.0
># 3 3 200 0.1 1.0
># 4 4 50 0.5 1.0
># 5 5 100 0.5 1.0
># 6 6 200 0.5 1.0
># 7 7 50 0.1 0.5
># 8 8 100 0.1 0.5
># 9 9 200 0.1 0.5
># 10 10 50 0.5 0.5
># 11 11 100 0.5 0.5
># 12 12 200 0.5 0.5An additional benefit is that I only need to loop over one dimension (condition number) instead of writing three loops, one for each factor.
3.3.3 Writing a function to conduct the simulation
While this step is not necessary, it’s recommended to wrap your steps of
generating data and extracting results into one big function, so that the code
is easier to read, like the runsim() function below:
runsim <- function(to_run, # conditions to run
nrep, # number of replications
alpha1 = ALPHA1, # latent mean of intercepts
phi11 = PHI11, # intercept variance
Lambda = LAMBDA, # factor loadings
Theta = THETA, # residual variances
designfactors = DESIGNFACTOR) {
# Extract design parameters for the given condition
N <- designfactors[to_run, "N"]
phi22 <- designfactors[to_run, "phi22"]
alpha2 <- designfactors[to_run, "alpha2"]
# Put the values back to the matrix
alpha <- c(alpha1, alpha2)
Phi <- matrix(c(phi11, phi22 / 2,
phi22 / 2, phi22), nrow = 2)
# Initialize place holders for the results
rep_result <- vector("list", nrep)
for (i in seq_len(nrep)) {
# Generate data
df <- gen_lgm_data(N, alpha, Phi, Lambda, Theta)
# Run model 1
m1_fit <- growth(m1, data = df)
# Run model 2
m2_fit <- growth(m2, data = df)
# Save results
rep_result[[i]] <- list(m1_fit = m1_fit,
m2_fit = m2_fit)
}
# Return results
return(rep_result)
}We can do a trial run:
# Run condition 1 with 2 replications
sim_results <- runsim(1, 2)># Warning in lav_object_post_check(object): lavaan WARNING: covariance matrix of latent variables
># is not positive definite;
># use lavInspect(fit, "cov.lv") to investigate.# Examine the results:
sim_results[[1]]># $m1_fit
># lavaan 0.6-3 ended normally after 31 iterations
>#
># Optimization method NLMINB
># Number of free parameters 9
>#
># Number of observations 50
>#
># Estimator ML
># Model Fit Test Statistic 2.093
># Degrees of freedom 5
># P-value (Chi-square) 0.836
>#
># $m2_fit
># lavaan 0.6-3 ended normally after 21 iterations
>#
># Optimization method NLMINB
># Number of free parameters 7
>#
># Number of observations 50
>#
># Estimator ML
># Model Fit Test Statistic 17.169
># Degrees of freedom 7
># P-value (Chi-square) 0.0163.3.4 Running the simulation
This step is easy. You can use a for loop, or the purrr::map() function
sim_results <-
map(seq_len(nrow(DESIGNFACTOR)),
~ runsim(.x, nrep = NREP))3.3.5 Extract the results
Right now I’m saving everything for each step. I’ll need a function to extract the relevant output, which are
- Estimated coefficients of \(\alpha_2\)
- Estimated standard errors of \(\alpha_2\)
It’s usually helpful to start extract results on one replication first:
# Try on one replication
with(sim_results[[1]][[1]],
tibble(coef(m1_fit)["s~1"],
sqrt(vcov(m1_fit)["s~1", "s~1"]),
coef(m2_fit)["s~1"],
sqrt(vcov(m2_fit)["s~1", "s~1"])))
# Now, wrap it as a function:
extract_coef <- function(res) {
out <- with(res,
tibble(coef(m1_fit)["s~1"],
sqrt(vcov(m1_fit)["s~1", "s~1"]),
coef(m2_fit)["s~1"],
sqrt(vcov(m2_fit)["s~1", "s~1"])))
# Add names:
names(out) <- c("m1_est", "m1_se", "m2_est", "m2_se")
out
}
extract_coef(sim_results[[1]][[1]])Now, we can do it on all replications and all conditions:
library(tidyverse)
sim_outdata <- sim_results %>%
# Iterate across conditions
map_dfr(
# Iterate across replications
~ map_dfr(.x, extract_coef, .id = "rep"),
.id = "cond") %>%
# Make `cond` a integer variable
mutate(rep = as.integer(rep),
cond = as.integer(cond)) %>%
# Add design factors
left_join(DESIGNFACTOR)
# Save the results
write_csv(sim_outdata, "example_sem_results.csv")3.3.6 Summarize the Results
Finally, let’s summarize the results in terms of the standardized bias and the relative standard error bias.
sim_outdata <- sim_outdata %>%
# Make it long format
gather("var", "val", m1_est:m2_se) %>%
separate(var, c("model", "var")) %>%
spread(var, val) %>%
# (Optional) Rename the levels of design factors to make it better looking in
# the plots
mutate(model = factor(model, labels = c("Correctly specified",
"Misspecified")),
alpha2_lab = as_factor(paste0("alpha[2] == ", alpha2)),
phi22_lab = as_factor(paste0("phi[22] == ", phi22)),
N_lab = as_factor(paste0("italic(N) == ", N)))
sim_sum <- sim_outdata %>%
# Summarize results by conditions
group_by(model, alpha2, phi22, N, alpha2_lab, phi22_lab, N_lab) %>%
summarise(ave_est = mean(est),
emp_sd = sd(est),
ave_se = mean(se)) %>%
# Compute standardized bias and relative SE bias
mutate(bias = ave_est - alpha2,
bias_mcse = emp_sd / sqrt(NREP),
std_bias = bias / emp_sd,
rse_bias = (ave_se - emp_sd) / emp_sd) %>%
ungroup()With relatively small number of conditions, one can present the results in a table (and it’s handy in R):
sim_sum %>%
select(model:N,
Bias = bias, `Monte Carlo Error` = bias_mcse,
`Standardized Bias` = std_bias,
`Relative SE Error` = rse_bias) %>%
knitr::kable(digits = 3L)| model | alpha2 | phi22 | N | Bias | Monte Carlo Error | Standardized Bias | Relative SE Error |
|---|---|---|---|---|---|---|---|
| Correctly specified | 0.5 | 0.1 | 50 | 0.006 | 0.003 | 0.101 | -0.054 |
| Correctly specified | 0.5 | 0.1 | 100 | -0.001 | 0.002 | -0.024 | -0.009 |
| Correctly specified | 0.5 | 0.1 | 200 | 0.002 | 0.001 | 0.059 | 0.017 |
| Correctly specified | 0.5 | 0.5 | 50 | 0.002 | 0.005 | 0.015 | -0.014 |
| Correctly specified | 0.5 | 0.5 | 100 | -0.001 | 0.003 | -0.012 | 0.020 |
| Correctly specified | 0.5 | 0.5 | 200 | -0.002 | 0.002 | -0.045 | 0.008 |
| Correctly specified | 1.0 | 0.1 | 50 | -0.001 | 0.003 | -0.019 | -0.076 |
| Correctly specified | 1.0 | 0.1 | 100 | 0.002 | 0.002 | 0.038 | -0.031 |
| Correctly specified | 1.0 | 0.1 | 200 | -0.001 | 0.001 | -0.045 | -0.016 |
| Correctly specified | 1.0 | 0.5 | 50 | 0.001 | 0.005 | 0.013 | -0.031 |
| Correctly specified | 1.0 | 0.5 | 100 | 0.002 | 0.004 | 0.023 | -0.082 |
| Correctly specified | 1.0 | 0.5 | 200 | 0.002 | 0.003 | 0.043 | -0.039 |
| Misspecified | 0.5 | 0.1 | 50 | 0.007 | 0.003 | 0.113 | -0.161 |
| Misspecified | 0.5 | 0.1 | 100 | -0.002 | 0.002 | -0.040 | -0.132 |
| Misspecified | 0.5 | 0.1 | 200 | 0.002 | 0.001 | 0.056 | -0.112 |
| Misspecified | 0.5 | 0.5 | 50 | 0.001 | 0.005 | 0.013 | -0.249 |
| Misspecified | 0.5 | 0.5 | 100 | -0.002 | 0.003 | -0.030 | -0.238 |
| Misspecified | 0.5 | 0.5 | 200 | -0.002 | 0.002 | -0.033 | -0.251 |
| Misspecified | 1.0 | 0.1 | 50 | -0.002 | 0.003 | -0.025 | -0.186 |
| Misspecified | 1.0 | 0.1 | 100 | 0.002 | 0.002 | 0.037 | -0.155 |
| Misspecified | 1.0 | 0.1 | 200 | -0.001 | 0.001 | -0.043 | -0.136 |
| Misspecified | 1.0 | 0.5 | 50 | 0.002 | 0.005 | 0.018 | -0.275 |
| Misspecified | 1.0 | 0.5 | 100 | 0.002 | 0.004 | 0.028 | -0.307 |
| Misspecified | 1.0 | 0.5 | 200 | 0.002 | 0.003 | 0.035 | -0.286 |
It is, however, recommended you try to plot the results, both for exploratory purpose and for better presentation of the results.
# Summarize estimates
sim_outdata %>%
ggplot(aes(x = factor(N), y = est, color = model)) +
geom_boxplot() +
geom_hline(aes(yintercept = alpha2)) +
facet_grid(alpha2_lab ~ phi22_lab, labeller = label_parsed) +
labs(x = "Sample Size (N)", y = "Estimates of Mean Slope")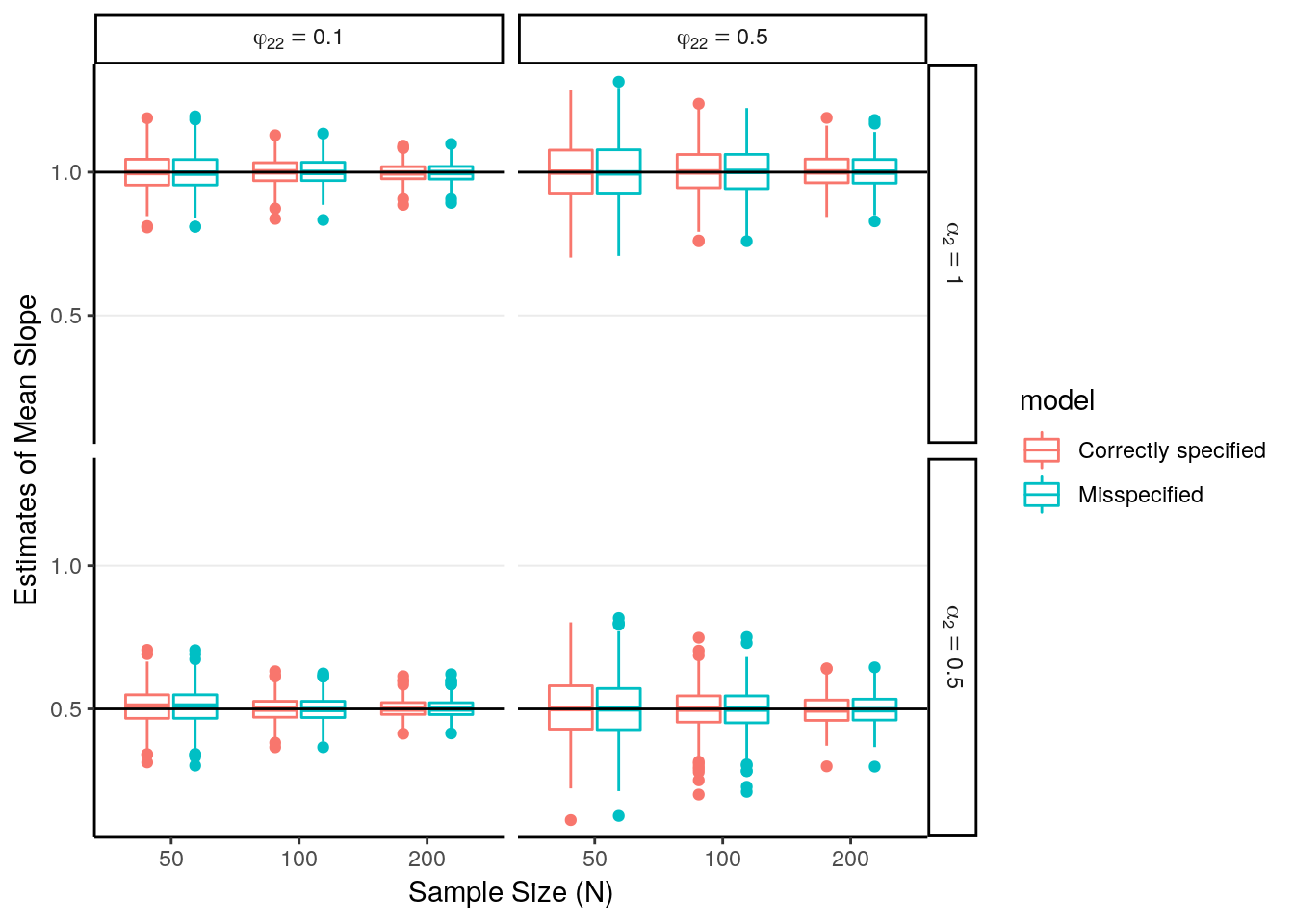
# Standardized bias
sim_sum %>%
ggplot(aes(x = factor(N), y = std_bias, color = model)) +
geom_line(aes(group = model)) +
geom_hline(aes(yintercept = 0)) +
facet_grid(alpha2_lab ~ phi22_lab, labeller = label_parsed) +
labs(x = "Sample Size (N)", y = "Standardized Bias")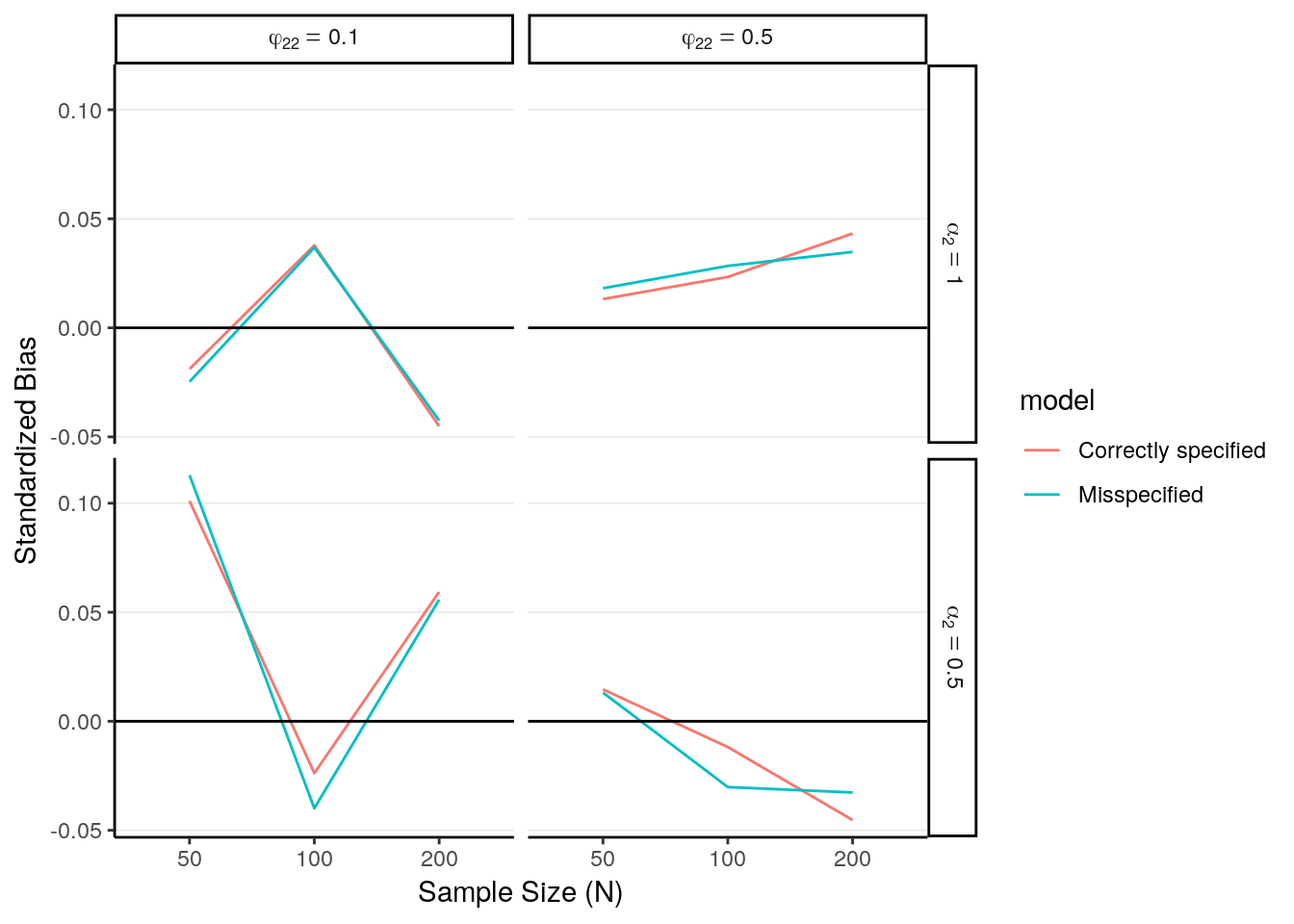
# Relative SE bias
sim_sum %>%
ggplot(aes(x = factor(N), y = rse_bias, color = model)) +
geom_line(aes(group = model)) +
geom_hline(aes(yintercept = 0)) +
facet_grid(alpha2_lab ~ phi22_lab, labeller = label_parsed) +
labs(x = "Sample Size (N)", y = "Relative Standard Error Bias")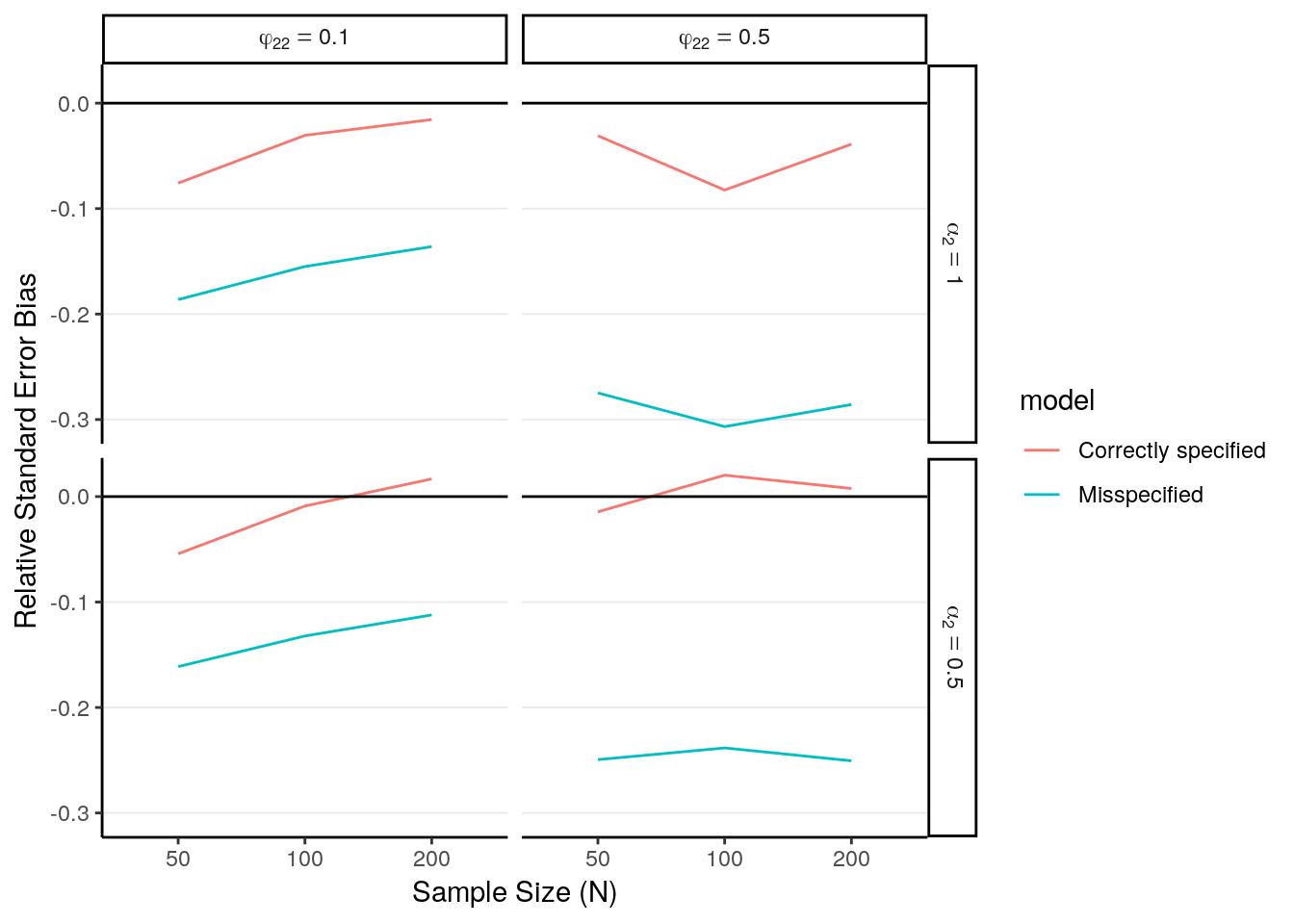
3.4 Using the SimDesign package
Recently, the SimDesign package was developed so that designing and running
simulation studies can be more structured and organized. The package also
provides some great features such as parallel computing, fail-safe stopping,
gathering of error or warning messages, among others. Check out the paper by
Sigal & Chalmers (2016) as well as the package vignettes (https://cran.r-project.org/web/packages/SimDesign/index.html) for more
information. Below is the R code for running the latent growth example with
their package. Try it yourself!
library(tidyverse)
library(SimDesign)
library(mnormt)
library(lavaan)
# Design factors:
DESIGNFACTOR <- expand.grid(
N = c(50, 100, 200),
phi22 = c(0.1, 0.5),
alpha2 = c(0, 0.5)
)
# Add condition number:
DESIGNFACTOR <- rowid_to_column(DESIGNFACTOR, "cond")
DESIGNFACTOR
# Function for generating data:
gen_lgm_data <- function(condition, fixed_objects = NULL) {
N <- condition$N
phi22 <- condition$phi22
alpha2 <- condition$alpha2
alpha <- c(1, alpha2)
Phi <- matrix(c(1, phi22 / 2,
phi22 / 2, phi22), nrow = 2)
Lambda <- cbind(c(1, 1, 1, 1),
c(0, 1, 2, 3))
Theta <- diag(0.5, nrow = 4)
# Generate latent factor scores
eta <- rmnorm(N, mean = alpha, varcov = Phi)
# Generate residuals:
e <- rmnorm(N, varcov = Theta)
# Compute outcome scores
y <- tcrossprod(eta, Lambda) + e
colnames(y) <- paste0("y", 1:4)
# Make it a data frame
as.data.frame(y)
}
# Test: generate data
# test_df <- gen_lgm_data(DESIGNFACTOR[1, ])
# lavaan syntax (to be passed through `fixed_objects`)
# True model
M1 <- 'i =~ 1*y1 + 1*y2 + 1*y3 + 1*y4
s =~ 0*y1 + 1*y2 + 2*y3 + 3*y4
i ~~ s'
# Model without random slopes
M2 <- 'i =~ 1*y1 + 1*y2 + 1*y3 + 1*y4
s =~ 0*y1 + 1*y2 + 2*y3 + 3*y4
s ~~ 0*i + 0*s'
# Analysis function
run_lgm <- function(condition, dat, fixed_objects = NULL) {
# Run model 1
m1_fit <- growth(fixed_objects$M1, data = dat)
# Run model 2
m2_fit <- growth(fixed_objects$M2, data = dat)
# Extract parameter estimates and standard errors
ret <- c(coef(m1_fit)["s~1"],
sqrt(vcov(m1_fit)["s~1", "s~1"]),
coef(m2_fit)["s~1"],
sqrt(vcov(m2_fit)["s~1", "s~1"]))
names(ret) <- c("m1_est", "m1_se", "m2_est", "m2_se")
ret
}
# Test: run analyses for simulated data
# run_lgm(dat = test_df)
# Helper function for computing relative SE bias
rse_bias <- function(est_se, est) {
est_se <- as.matrix(est_se)
est <- as.matrix(est)
est_se <- colMeans(est_se)
emp_sd <- apply(est, 2L, sd)
est_se / emp_sd - 1
}
# Evaluative function
evaluate_lgm <- function(condition, results, fixed_objects = NULL) {
alpha2 <- condition$alpha2
c(bias = bias(results[ , c(1, 3)], parameter = alpha2),
std_bias = bias(results[ , c(1, 3)], parameter = alpha2,
type = "standardized"),
rmse = RMSE(results[ , c(1, 3)], parameter = alpha2),
rse_bias = rse_bias(results[ , c(2, 4)], results[ , c(1, 3)])
)
}
# Put all together
sim_results <- runSimulation(DESIGNFACTOR,
replications = 500,
generate = gen_lgm_data,
analyse = run_lgm,
summarise = evaluate_lgm,
fixed_objects =
list(M1 = M1, M2 = M2))
# With parallel processing (4 cores in this example; uncomment the following to
# run)
# sim_results <- runSimulation(DESIGNFACTOR,
# replications = 500,
# generate = gen_lgm_data,
# analyse = run_lgm,
# summarise = evaluate_lgm,
# packages = c("mnormt", "lavaan"),
# parallel = TRUE,
# ncores = 4L,
# fixed_objects =
# list(M1 = M1, M2 = M2))3.5 Exercise
- From the simulation results, evaluate the relative efficiency of the estimated average slope (i.e., \(\alpha_2\)) under model 2 relative to that under model 1.