1 Genes, Genomes and Genome Browsers
1.1 What is a gene?
A gene is a segment of DNA that directs the production of a functional product (i.e. protein, see Figure 1.1). A significant number of eukaroytic genes are protein coding. Each protein-coding gene is first used as a template to create an RNA transcript through a process called transcription. The RNA transcript is then processed into a messenger RNA (mRNA). The most notable change that occurs during RNA processing is that intron sequences are removed (dark gray segments in Figure 1.1). The mRNA is then exported from the nucleus and used as a template to make a protein in a process called translation. Most translation occurs in the cytoplasm. Translation of secreted and transmembrane proteins occur at the surface of the endoplasmic reticulum. Proteins do most of the work necessary to maintain cell structure, function and behavior.
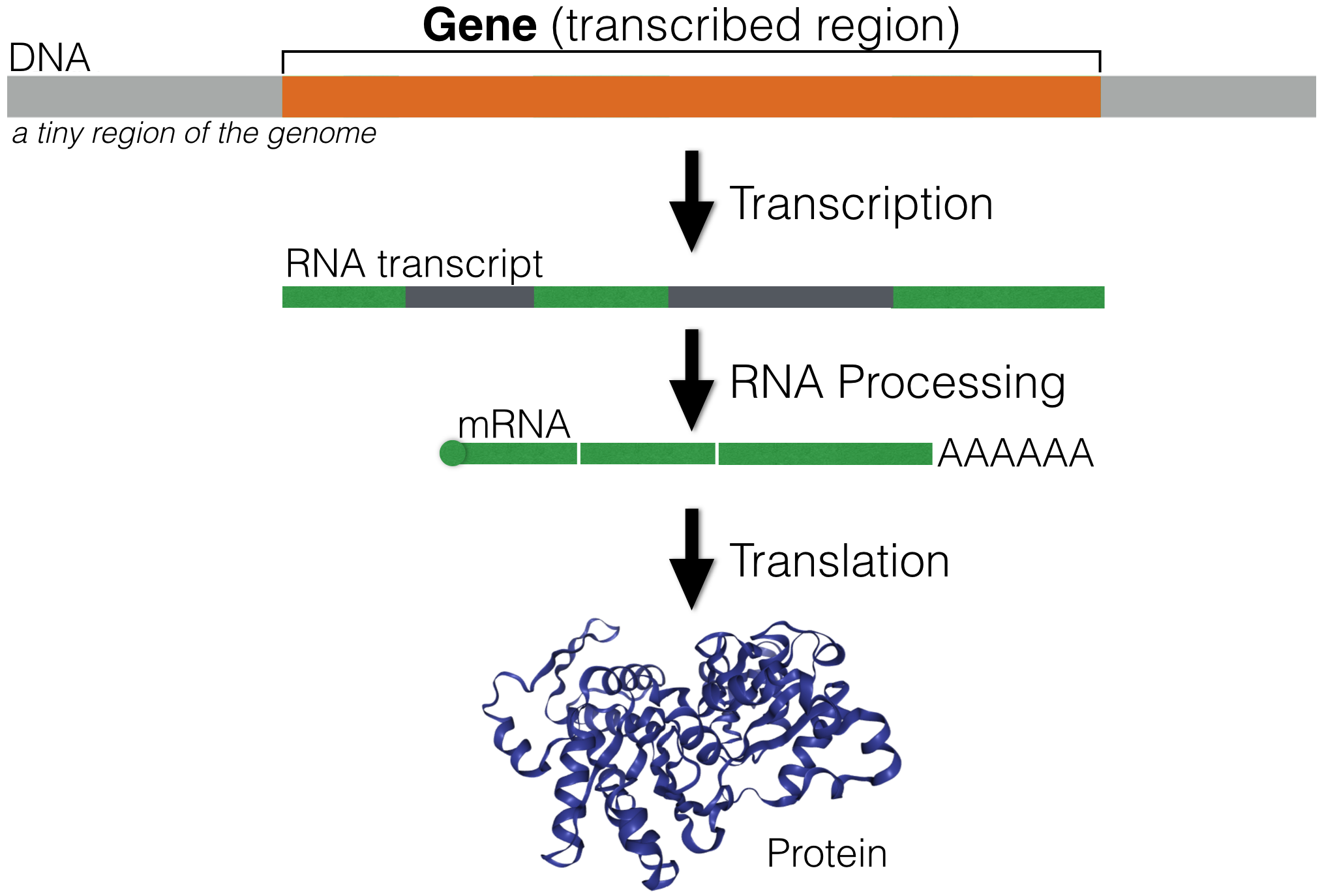
Figure 1.1: An overview of Gene Expression. A small segment of DNA (light grey) containing a single gene (orange) is shown. The RNA transcript is highlighted in green (exons) and dark gray (introns).
While the transcribed region a gene requires adjacent sequence to become transcribed at the right time and place (i.e. promoter, see Chapter 5), for simplicity, the size of a gene (number of base pairs) is typically defined by where transcription begins and ends. Thus, the sites of transcription initiation and transcription termination define both gene size and RNA transcript size (but not mRNA size which is typically shorter as most eukaryotic genes have introns).
1.2 What is a genome?
Genes are distributed linearly along chromosomes2 (Figure 1.2). One complete set of chromosomes for a given specifies makes up what we call a genome (sometimes called the “haploid genome”)3.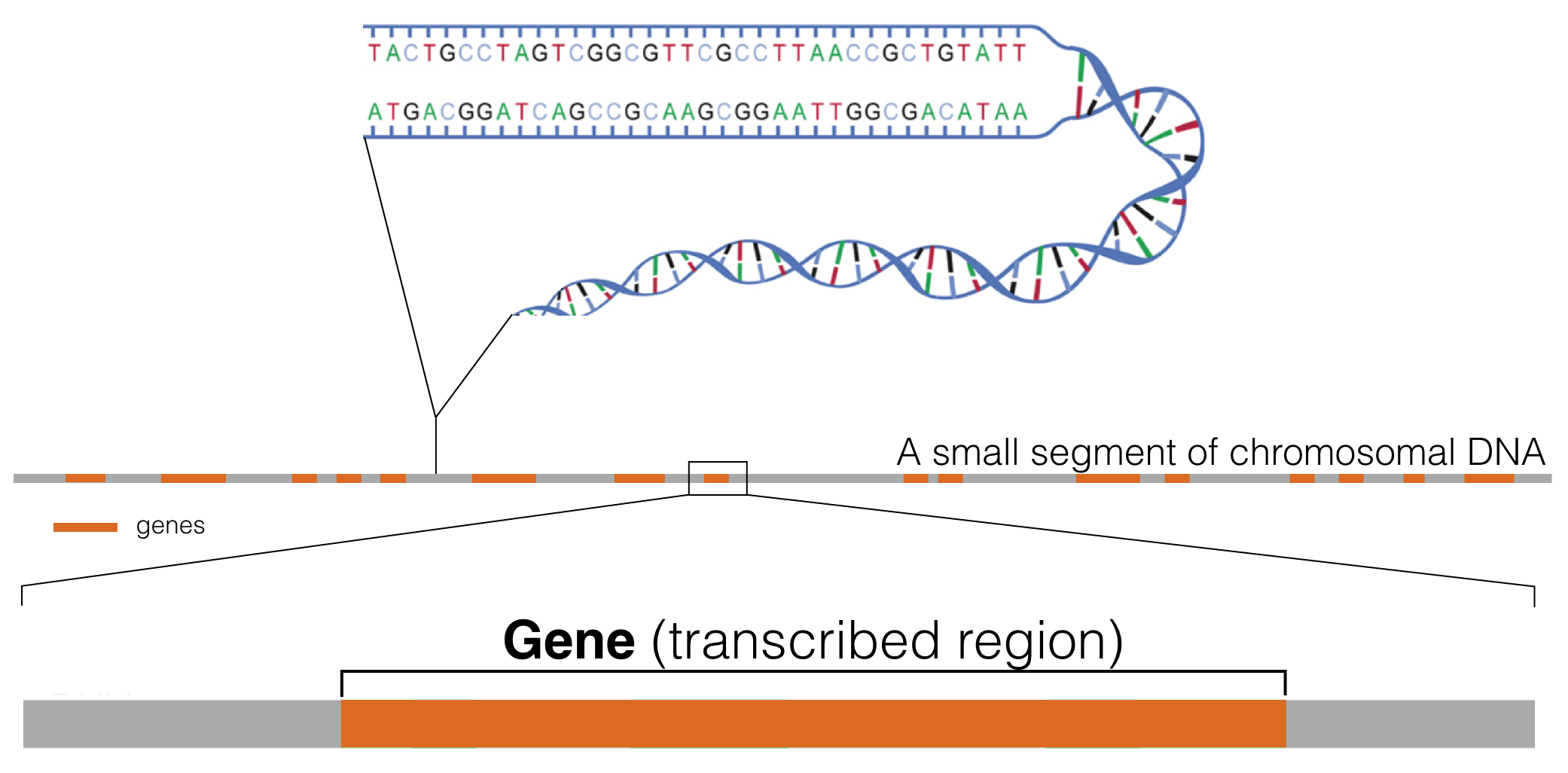
Figure 1.2: This schematic represents a hypothetical segment of a chromosome with orange rectangles representing genes distributed linearly along the DNA segment shown.
The haploid genome for humans consists of 24 linear chromosomes and one circular mitochondrial chromosome. Each chromosome varies in length. For example, chromosome one is the longest at 248,956,422 base pairs (bp). Given that 10 bp of a double helix measures 34 angstroms in length, chromosome one is calculated to be 82.7 mm!4 (Figure 1.3).
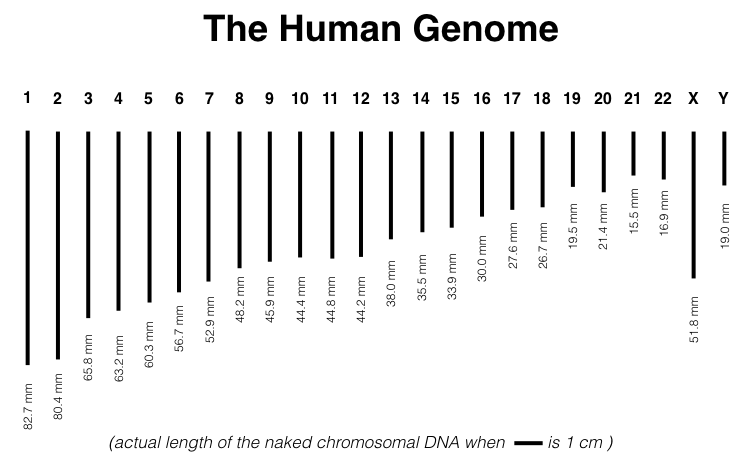
Figure 1.3: Actual length of the human genome when scale bar equals one centimeter. Naken mitochondrial DNA is too small to be shown. It is a small circular DNA molecule, with a circumference of 55 microns.
Like all animals, humans are diploid. Thus, their somatic cells5 harbor 22 pairs of autosomes6 and one pair of sex chromosomes (either XX or XY). Thus, over two meters of DNA is crammed into each somatic nucleus! But the diameter of the average mammalian nucleus is only 6 microns or 0.006 of a millimeter. How does the diploid genome fit? First, the diameter of the DNA double helix is only two angstroms7 but also, each chromosome is carefully packaged by proteins in a systematic and stereotypical manner (Figure 1.4).
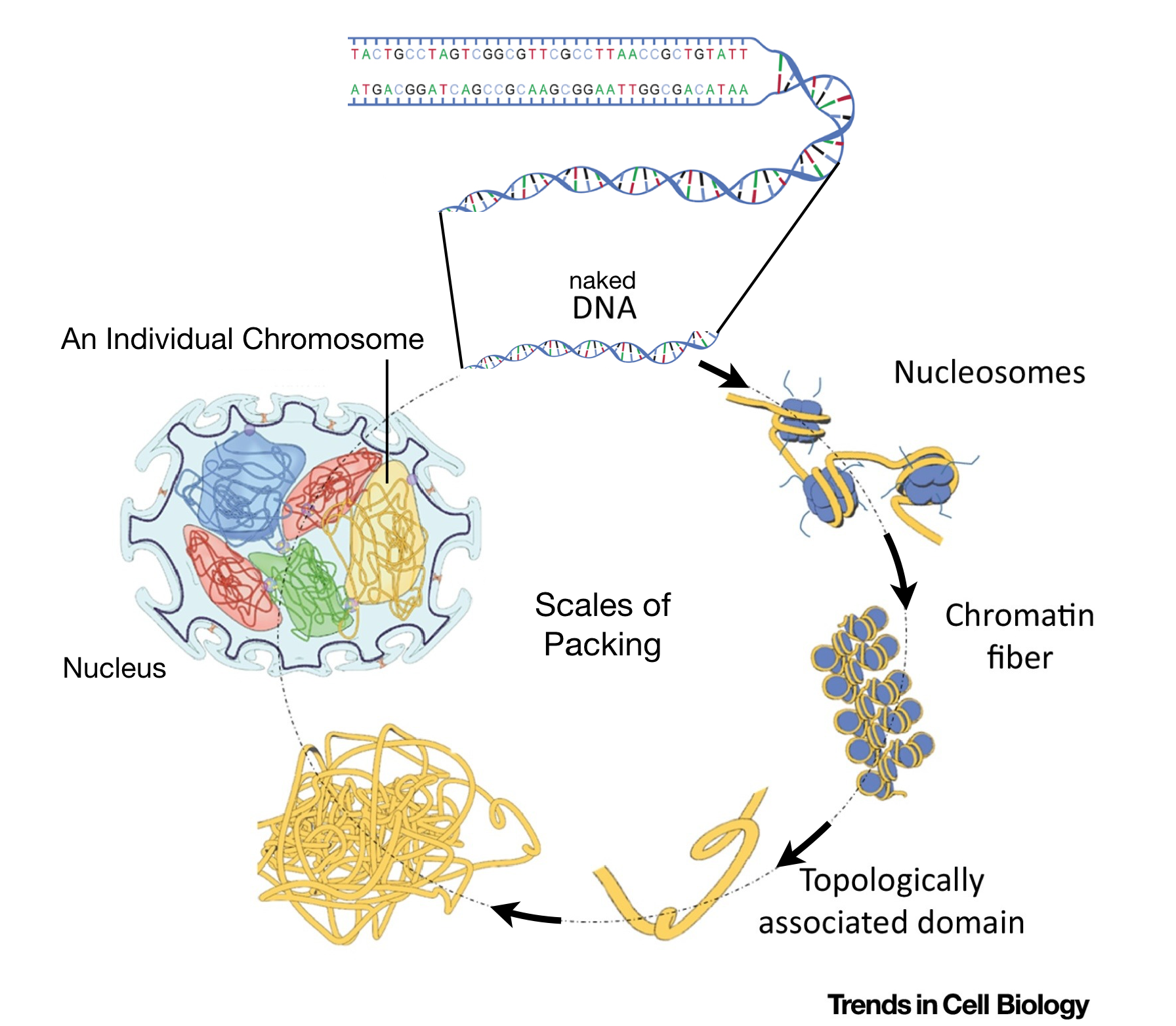
Figure 1.4: This image is modified from Uhler and Shivashankar, 2017. It shows the various levels of chromosome packing found in nuclei during interphase of the cell cycle. The first level of packing requires an octamer of histone proteins that assemble into a ball-like structure called a nucleosome. Naked DNA wraps around these octamers forming ‘beads on a string’. Additional levels of packing into chromatin fibers and topological associated domains requires additional proteins. Individual chromosomes (23 pairs in humans) are then carefully orgnanized within the cell nucleus in a cell type-dependent manner.
1.2.1 Test Your Understanding
- Place each gene expression step in the correct order (RNA processing,transcription, translation)
- Where does RNA processing occur?
- Where does transcription occur?
- Where does translation occur?
- TRUE or FALSE. If I told you the size of a mature mRNA transcript you would know the size of the gene in the genome?
- Fill in the blank. The first level of chromosomal packing requires an octamer of histone proteins that assemble into a ball-like structure called a _______
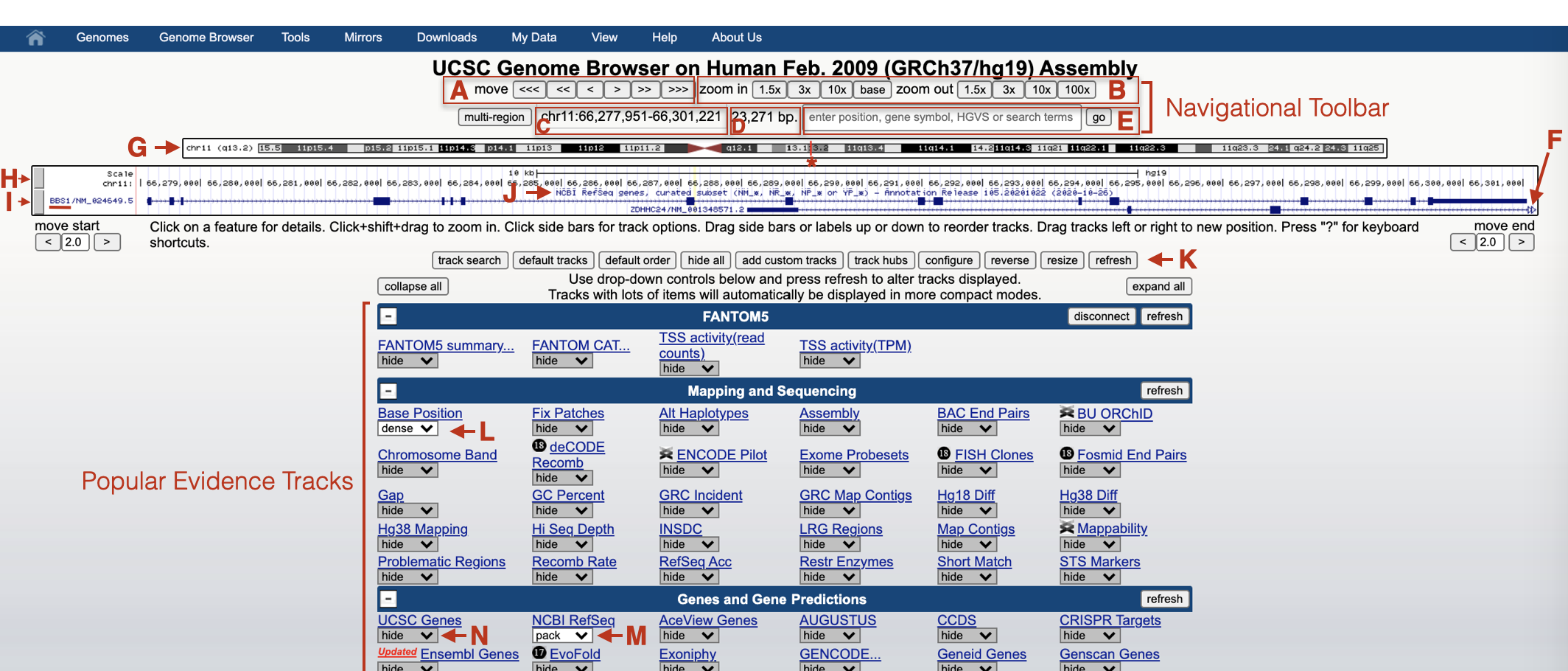
1.3 What is a Genome Browser?
According to Wikipedia, “Genome browsers enable researchers to visualize and browse entire genomes with annotated data including gene structure, protein structure, expression, variation, etc. They differ from ordinary biological databases in that they display data in a graphical format”. Genome coordinates are displayed along the X-axis. The annotations and graphics that describe gene structure, function, expression etc. are stacked along the Y-axis. Finally, the data used to create the graphics are contributed by multiple sources.
There are numerous Genome Browsers available. We will be using the UCSC Genome Browser. The UCSC Genome Browser harbors the sequence of sequenced genomes called “reference genomes”8 from a variety of species (24 linear chromosomes and the mitochondrial genome). Our focus will be on the human genome. Initially, we will focus our Genome Browser window on a single human gene, BBS1. In this chapter, you will learn how the UCSC Genome Browser is organized, how to configure so-called “evidence tracks”9 and how to navigate through Genome Browser window (scrolling left and right, zooming in and out).