1 Getting Started with R
R is a popular programming language within the Data Scientist’s arsenal of tools. R has become popular over the past several years due to its flexibility, various data science uses and it is cost effective. It is widely used due to its array of various uses such as: data wrangling, statistics, machine learning and data visualization. In addition, there is no cost to using R, however there is also no support unless you use a third-party service. Over the course of this text, we will start to get more familiar with R and show how it might be used to complete complicated tasks. Will will be using RStudio as the front-end user interface with R.
1.1 Launch RStudio
Downloading R and RStudio is free. You can download R from https://cran.r-project.org/ and RStudio from https://www.rstudio.com/products/rstudio/ and download the desktop version.
In the first chapter, we introduce you to some base functionality within R and showcase how to perform some basic statistical tasks.
For starters, you can interface directly with the R terminal and enter in basic calculations. Below we show a simple mathematical equation and how R appropriately returns the correct answer:
(2*3-1)^2 - 5
#> [1] 20Often we will want to save a value, to do this we use one of the assignment operators: <- , ->, or =, as you can see below the example below:
1.2 Basic Data Types in R
R has 6 basic data types.
- numeric (real or decimal)
- integer
- character
- logical
- complex
The class function can let you know the data type:
class(3.14)
#> [1] "numeric"
class(pi) # pi = R approximation for pi
#> [1] "numeric"
class(45)
#> [1] "numeric"
class(45L) # add L if you want R to think of this as an integer
#> [1] "integer"
class("integer")
#> [1] "character"
class(TRUE) # logical TRUE
#> [1] "logical"
cx <- complex(length.out = 1, # 1 + 1*sqrt(-1)
real = 1,
imaginary = 1)
cx
#> [1] 1+1i
class(cx)
#> [1] "complex"R works with each of these can test if a statement is TRUE or FALSE, for example:
TRUE == FALSE
#> [1] FALSE
1.3 Summary of basic R Data Types
| Example | Type |
| “male,” “Diabetes” | Character / String |
| 3, 20.6, 100.222 | Numeric |
| 26L (add an ‘L’ to denote integer) | Integer |
| TRUE, FALSE | Logical |
| 1+1i | Complex |
\(~\)
\(~\)
1.4 Lists and Vectors
Lists and vectors are similar in R but a few differences separate them.
| List | Vector |
|---|---|
| A list holds different data such as Numeric, Character, logical, etc. | Vector stores elements of the same type or converts implicitly. |
| list is a multidimensional object | The vector is one-dimensional |
| Lists are recursive | vector is not recursive |
We can make lists or vectors within R by using the c() function, in the example below we make a few example lists:
list_of_ints <- c(2L,4L,6L,8L)
list_of_strings <- c("Data", "Science", "is a", 'blast!')
list_of_logicals <- c(TRUE, FALSE, TRUE, FALSE)
list_of_mixed_type <- c(3.14, 1L, "cat", TRUE)
list_of_numbers <- c(22/7, 18, 42, 65.2)
list_of_sexes <- c("Male","Female","Female","Male")R provides many functions to examine features of vectors and other objects, for example
-
class()- what kind of object is it (high-level)? -
typeof()- what is the object’s data type (low-level)? -
length()- how long is it? What about two dimensional objects? -
attributes()- does it have any metadata?
class(list_of_strings)
#> [1] "character"
length(list_of_strings)
#> [1] 4
typeof(list_of_strings)
#> [1] "character"
attributes(list_of_strings)
#> NULL
class(list_of_ints)
#> [1] "integer"
length(list_of_ints)
#> [1] 4
typeof(list_of_ints)
#> [1] "integer"
attributes(list_of_ints)
#> NULL
class(list_of_numbers)
#> [1] "numeric"
length(list_of_numbers)
#> [1] 4
typeof(list_of_numbers)
#> [1] "double"
attributes(list_of_numbers)
#> NULL
class(list_of_mixed_type)
#> [1] "character"
length(list_of_mixed_type)
#> [1] 4
typeof(list_of_mixed_type)
#> [1] "character"
attributes(list_of_mixed_type)
#> NULL1.4.1 Accessing elements in a list
We can access certain elements from within the lists by passing the location of the element of interest within the list:
list_of_mixed_type[1]
#> [1] "3.14"
list_of_mixed_type[2]
#> [1] "1"
list_of_mixed_type[3]
#> [1] "cat"
list_of_mixed_type[4]
#> [1] "TRUE"
list_of_lists <- list(list_of_ints,
list_of_strings,
list_of_logicals,
list_of_mixed_type,
list_of_numbers,
list_of_sexes)
list_of_lists
#> [[1]]
#> [1] 2 4 6 8
#>
#> [[2]]
#> [1] "Data" "Science" "is a" "blast!"
#>
#> [[3]]
#> [1] TRUE FALSE TRUE FALSE
#>
#> [[4]]
#> [1] "3.14" "1" "cat" "TRUE"
#>
#> [[5]]
#> [1] 3.142857 18.000000 42.000000 65.200000
#>
#> [[6]]
#> [1] "Male" "Female" "Female" "Male"
list_of_lists[[2]][[4]]
#> [1] "blast!"
names(list_of_lists) <- list('list_of_ints',
'list_of_strings',
'list_of_logicals',
'list_of_mixed_type',
'list_of_numbers',
'list_of_sexes')
list_of_lists
#> $list_of_ints
#> [1] 2 4 6 8
#>
#> $list_of_strings
#> [1] "Data" "Science" "is a" "blast!"
#>
#> $list_of_logicals
#> [1] TRUE FALSE TRUE FALSE
#>
#> $list_of_mixed_type
#> [1] "3.14" "1" "cat" "TRUE"
#>
#> $list_of_numbers
#> [1] 3.142857 18.000000 42.000000 65.200000
#>
#> $list_of_sexes
#> [1] "Male" "Female" "Female" "Male"
list_of_lists[['list_of_strings']][[4]]
#> [1] "blast!"1.4.2 Factors
Factors are the data objects which are used to categorize the data and store it as levels. They can store both strings and integers. They are useful in the columns which have a limited number of unique values. If we use the factor function on list_of_sexes it returns back the list as a factored list with the levels as the unique set of distinct values in the list:
list_of_sexes_factor <- factor(list_of_sexes)
list_of_sexes_factor
#> [1] Male Female Female Male
#> Levels: Female Male1.4.2.1 levels and ordered levels
We can access the levels of a factor with the levels function:
levels(list_of_sexes_factor)
#> [1] "Female" "Male"The order of the levels in a factor can be changed by applying the factor function again with new order of the levels:
list_of_sexes_factor_relevel <- factor(list_of_sexes, levels = c("Male","Female"))
list_of_sexes_factor_relevel
#> [1] Male Female Female Male
#> Levels: Male FemaleWe can also place an ordering on the factors with the ordered function :
list_of_costs <- c('$0 - $100',
'$100 - $200',
'$200 - $300',
'$300 - $400')
ordered_list_of_costs <- ordered(list_of_costs, levels = list_of_costs)
ordered_list_of_costs
#> [1] $0 - $100 $100 - $200 $200 - $300 $300 - $400
#> Levels: $0 - $100 < $100 - $200 < $200 - $300 < $300 - $400\(~\)
\(~\)
1.5 Matriceis
A matrix (plural matrices) is a rectangular array or table of numbers, symbols, or expressions, arranged in rows and columns, which is used to represent a mathematical object or a property of such an object. In R a matrix \(M\) is an \(n \times k\) array with \(n\) rows with \(k\) columns, we say the dimension of \(M\) is \(n \times k\).
M <- matrix(data = c(1,3,5,2,4,6), nrow = 2, byrow = TRUE)
M
#> [,1] [,2] [,3]
#> [1,] 1 3 5
#> [2,] 2 4 6
dim(M)
#> [1] 2 3
nrow(M)
#> [1] 2
ncol(M)
#> [1] 3
class(M)
#> [1] "matrix" "array"
length(M)
#> [1] 6
typeof(M)
#> [1] "double"
attributes(M)
#> $dim
#> [1] 2 3We can also add on additional attributes to the matrix such as dimensional names:
rownames = c("odds", "evens")
colnames = c("col1", "col2", "col3")
M <- matrix(data = c(1,3,5,2,4,6), nrow = 2, byrow = TRUE, dimnames = list(rownames, colnames))
M
#> col1 col2 col3
#> odds 1 3 5
#> evens 2 4 6
class(M)
#> [1] "matrix" "array"
length(M)
#> [1] 6
typeof(M)
#> [1] "double"
attributes(M)
#> $dim
#> [1] 2 3
#>
#> $dimnames
#> $dimnames[[1]]
#> [1] "odds" "evens"
#>
#> $dimnames[[2]]
#> [1] "col1" "col2" "col3"\(~\)
\(~\)
1.6 The Data Frame
A Data Frame is a two-dimensional array-like structure in which each column contains values of one variable and each row contains one set of values from each column.
my_dataframe <- data.frame(list_of_ints,
list_of_strings,
list_of_logicals,
list_of_mixed_type,
list_of_numbers,
list_of_sexes_factor,
ordered_list_of_costs)
my_dataframe
#> list_of_ints list_of_strings list_of_logicals list_of_mixed_type
#> 1 2 Data TRUE 3.14
#> 2 4 Science FALSE 1
#> 3 6 is a TRUE cat
#> 4 8 blast! FALSE TRUE
#> list_of_numbers list_of_sexes_factor ordered_list_of_costs
#> 1 3.142857 Male $0 - $100
#> 2 18.000000 Female $100 - $200
#> 3 42.000000 Female $200 - $300
#> 4 65.200000 Male $300 - $400
class(my_dataframe)
#> [1] "data.frame"
length(my_dataframe)
#> [1] 7
typeof(my_dataframe)
#> [1] "list"
attributes(my_dataframe)
#> $names
#> [1] "list_of_ints" "list_of_strings" "list_of_logicals"
#> [4] "list_of_mixed_type" "list_of_numbers" "list_of_sexes_factor"
#> [7] "ordered_list_of_costs"
#>
#> $class
#> [1] "data.frame"
#>
#> $row.names
#> [1] 1 2 3 4The primary difference between dataframes and matrices is that dataframes allow for columns to be of different datatypes.
We can see the first few rows of a dataframe with the head function:
head(my_dataframe)
#> list_of_ints list_of_strings list_of_logicals list_of_mixed_type
#> 1 2 Data TRUE 3.14
#> 2 4 Science FALSE 1
#> 3 6 is a TRUE cat
#> 4 8 blast! FALSE TRUE
#> list_of_numbers list_of_sexes_factor ordered_list_of_costs
#> 1 3.142857 Male $0 - $100
#> 2 18.000000 Female $100 - $200
#> 3 42.000000 Female $200 - $300
#> 4 65.200000 Male $300 - $400
1.6.0.1 The dim function
We can determine the dimension by using the dim function:
dim(my_dataframe)
#> [1] 4 7so we can see that this data-frame has 4 rows and 7 columns. We can also get those values by using nrow and ncol:
1.6.1 Matrix Notation
We can use familiar matrix notation to select specific elements from the data frame:
my_dataframe[3,5]
#> [1] 42We can use similar notation to select a row:
my_dataframe[3,]
#> list_of_ints list_of_strings list_of_logicals list_of_mixed_type
#> 3 6 is a TRUE cat
#> list_of_numbers list_of_sexes_factor ordered_list_of_costs
#> 3 42 Female $200 - $300Or a column:
my_dataframe[,5]
#> [1] 3.142857 18.000000 42.000000 65.200000
1.6.2 Data Frames have colnames
It is important to have meaningful column names in our dataframe. This is useful for quickly understanding what variables you are viewing and logical for programming purposes. You can review the column names of a dataframe with the colnames functions:
colnames(my_dataframe)
#> [1] "list_of_ints" "list_of_strings" "list_of_logicals"
#> [4] "list_of_mixed_type" "list_of_numbers" "list_of_sexes_factor"
#> [7] "ordered_list_of_costs"It is also helpful to be able to pass in these column names to select a column rather than recall which column number the information is associated with:
my_dataframe[,'list_of_ints']
#> [1] 2 4 6 8We may also use the following code to access a column in a dataframe:
my_dataframe$list_of_strings
#> [1] "Data" "Science" "is a" "blast!"\(~\)
\(~\)
1.7 Structure
The R Structure function will compactly display the internal structure of an R object.
To see help page for a function use ? before the function name, for example try: ?str
For example, if we use str on my_dataframe it will show the user the number of observations, variables and data type of each list:
str(my_dataframe)
#> 'data.frame': 4 obs. of 7 variables:
#> $ list_of_ints : int 2 4 6 8
#> $ list_of_strings : chr "Data" "Science" "is a" "blast!"
#> $ list_of_logicals : logi TRUE FALSE TRUE FALSE
#> $ list_of_mixed_type : chr "3.14" "1" "cat" "TRUE"
#> $ list_of_numbers : num 3.14 18 42 65.2
#> $ list_of_sexes_factor : Factor w/ 2 levels "Female","Male": 2 1 1 2
#> $ ordered_list_of_costs: Ord.factor w/ 4 levels "$0 - $100"<"$100 - $200"<..: 1 2 3 4When we call str on list_of_costs we see that it is a character vector of length 4 and are shown the values
str(list_of_costs)
#> chr [1:4] "$0 - $100" "$100 - $200" "$200 - $300" "$300 - $400"\(~\)
\(~\)
1.8 summary
The summary is a generic function used to produce summaries of the results of various model fitting functions.
See ?summary for more information.
In the case of a dataframe the summary function will give summary level information on the dataframe, for continuous variables it will display the minimum, first quartile, median, mean, third quartile and max; for categorical data counts of each of the classes
summary(my_dataframe)
#> list_of_ints list_of_strings list_of_logicals list_of_mixed_type
#> Min. :2.0 Length:4 Mode :logical Length:4
#> 1st Qu.:3.5 Class :character FALSE:2 Class :character
#> Median :5.0 Mode :character TRUE :2 Mode :character
#> Mean :5.0
#> 3rd Qu.:6.5
#> Max. :8.0
#> list_of_numbers list_of_sexes_factor ordered_list_of_costs
#> Min. : 3.143 Female:2 $0 - $100 :1
#> 1st Qu.:14.286 Male :2 $100 - $200:1
#> Median :30.000 $200 - $300:1
#> Mean :32.086 $300 - $400:1
#> 3rd Qu.:47.800
#> Max. :65.200We can also get these statistics by class, here we use the information in the dataframe column list_of_sexes
by(my_dataframe, my_dataframe$list_of_sexes, summary)
#> my_dataframe$list_of_sexes: Female
#> list_of_ints list_of_strings list_of_logicals list_of_mixed_type
#> Min. :4.0 Length:2 Mode :logical Length:2
#> 1st Qu.:4.5 Class :character FALSE:1 Class :character
#> Median :5.0 Mode :character TRUE :1 Mode :character
#> Mean :5.0
#> 3rd Qu.:5.5
#> Max. :6.0
#> list_of_numbers list_of_sexes_factor ordered_list_of_costs
#> Min. :18 Female:2 $0 - $100 :0
#> 1st Qu.:24 Male :0 $100 - $200:1
#> Median :30 $200 - $300:1
#> Mean :30 $300 - $400:0
#> 3rd Qu.:36
#> Max. :42
#> ------------------------------------------------------------
#> my_dataframe$list_of_sexes: Male
#> list_of_ints list_of_strings list_of_logicals list_of_mixed_type
#> Min. :2.0 Length:2 Mode :logical Length:2
#> 1st Qu.:3.5 Class :character FALSE:1 Class :character
#> Median :5.0 Mode :character TRUE :1 Mode :character
#> Mean :5.0
#> 3rd Qu.:6.5
#> Max. :8.0
#> list_of_numbers list_of_sexes_factor ordered_list_of_costs
#> Min. : 3.143 Female:0 $0 - $100 :1
#> 1st Qu.:18.657 Male :2 $100 - $200:0
#> Median :34.171 $200 - $300:0
#> Mean :34.171 $300 - $400:1
#> 3rd Qu.:49.686
#> Max. :65.200\(~\)
\(~\)
1.9 Save and Read RDS files
You can save important data, variables, or models as R data files using saveRDS:
saveRDS(my_dataframe,'my_dataframe.RDS')by default this saves to your working directory.
1.10 getwd()
You can see your current working directory path by using getwd():
getwd()
#> [1] "C:/Users/jkyle/Documents/GitHub/Jeff_Data_Wrangling"You can alter the paths in a number of different ways.
In this example, we will want to save results to a sub-folder called y_data.
First, I will tell R to create a directory:
new_path <- file.path(getwd(),'y_data')
dir.create(new_path)Now I can save y within this folder:
1.12 Remove Items from your enviroment
WARNING once something is removed from your environment it is GONE! SAVE WHAT YOU NEED with saveRDS otherwise you will need to rerun portions of code.
You can remove items in your R environment by using the rm function.
rm(my_dataframe2)
#> Warning in rm(my_dataframe2): object 'my_dataframe2' not found1.12.1 Remove all but a list
Let’s say that we want to remove everything except for my_dataframe and y, then we might do something like:
Above we are using multiple functions in conjunction with one-another:
- We make a variable called
keepto contain the items we wish to maintain -
lsis returning a list of items within the environment -
setdiffis taking the set-difference betweenlswhich returns all the items within the environment and the items that we have specified withinkeep.
1.12.2 Delete Files / Folders
unlink deletes a file, we can use recursive = TRUE to delete directories:
unlink(new_path, recursive = TRUE)\(~\)
\(~\)
1.13 Functions
A function is an R object that takes in arguments or parameters and follows the collection of statements to provide the requested return.
1.13.1 Example R functions.
The R function set.seed is designed to assist with pseudo-random number generation.
The R function rnorm will produce a given number of randomly generated numbers with a provided mean and standard-deviation, Below we define 3 distributions each of 1000 numbers:
-
X,Y, andZwill be drawn from a distribution with mean of 0 and standard deviation of 1. -
XandZwill be drawn with the same seed where asYwill be drawn with a different seed. -
Ywill be drawn from a distribution with mean of 5 and standard deviation of 2.
{
set.seed(12345)
X <- rnorm(1000, mean = 0, sd = 1)
}
{
set.seed(5)
Y <- rnorm(1000, mean = 0, sd = 1)
W <- rnorm(1000, mean = 5, sd = 2)
}
{
set.seed(12345)
Z <- rnorm(1000, mean = 0, sd = 1)
}Here are the first 10 from each of the distributions:
cat("X \n")
#> X
X[1:10]
#> [1] 0.5855288 0.7094660 -0.1093033 -0.4534972 0.6058875 -1.8179560
#> [7] 0.6300986 -0.2761841 -0.2841597 -0.9193220
cat("Y \n")
#> Y
Y[1:10]
#> [1] -0.84085548 1.38435934 -1.25549186 0.07014277 1.71144087 -0.60290798
#> [7] -0.47216639 -0.63537131 -0.28577363 0.13810822
cat("W \n")
#> W
W[1:10]
#> [1] 2.089891 7.489126 4.136106 5.013739 5.249102 4.180743 6.126827 8.213916
#> [9] 2.996097 6.018652
cat("Z \n")
#> Z
Z[1:10]
#> [1] 0.5855288 0.7094660 -0.1093033 -0.4534972 0.6058875 -1.8179560
#> [7] 0.6300986 -0.2761841 -0.2841597 -0.9193220Note that X and Z will be exactly the same sets since we used the same seed to generate the two sets of numbers whereas Y will be different, even though we used the same parameters, for example
X[546] == Z[546]
#> [1] TRUE
X[546] == Y[546]
#> [1] FALSEEven though X and Y are not the same, they are sampled from the same distribution therefore the p-value on the t-test should be well above .05:
t.test(X,Y)
#>
#> Welch Two Sample t-test
#>
#> data: X and Y
#> t = 0.6405, df = 1997.7, p-value = 0.5219
#> alternative hypothesis: true difference in means is not equal to 0
#> 95 percent confidence interval:
#> -0.05938059 0.11697799
#> sample estimates:
#> mean of x mean of y
#> 0.04619816 0.01739946Comparatively, X and W were not sampled from the same distribution, therefore the p-value should be closer to 0:
t.test(X,W)
#>
#> Welch Two Sample t-test
#>
#> data: X and W
#> t = -72.569, df = 1474.2, p-value < 2.2e-16
#> alternative hypothesis: true difference in means is not equal to 0
#> 95 percent confidence interval:
#> -5.237795 -4.962086
#> sample estimates:
#> mean of x mean of y
#> 0.04619816 5.14613865We can see the difference between the distributions by using the hist function:
hist(X, col=rgb(1,0,0,0.5), main="Overlapping Histogram") # red
hist(Y, col=rgb(0,1,0,0.5) , add=T) # green
hist(W, col=rgb(0,0,1,0.5) , add=T) # blue 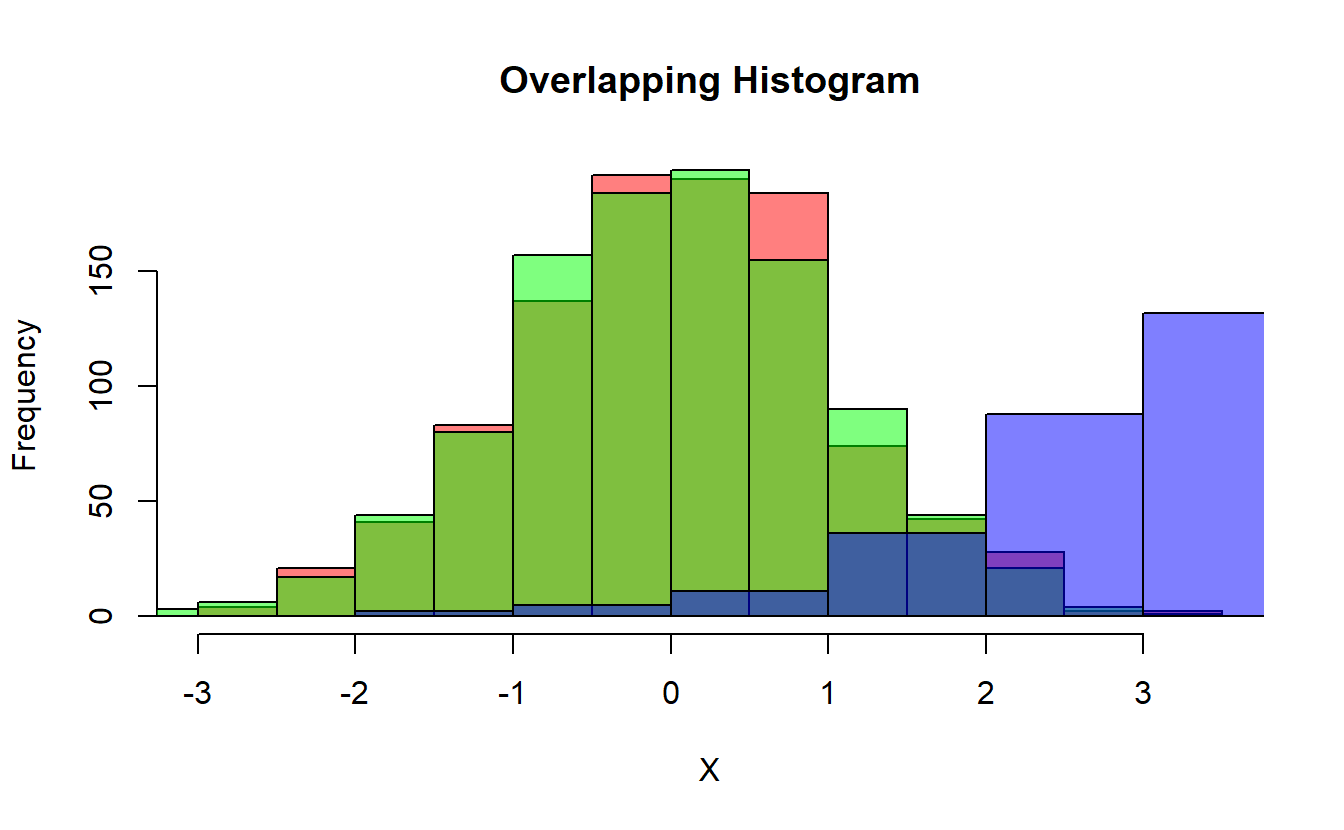
(#fig:hist1 )Overlapping Histogram of X, Y, & W
1.14 User defined functions
Many of you may recall the quadratic formula from algebra; given a general quadratic equation of the form:
\[ax^2 + bx + c = 0 \]
with \(a\), \(b\), and \(c\) representing constants with \(a \neq 0\) then
\[x = -b \pm \frac{\sqrt{b^2 - 4ac}}{2a}\]
are the two solutions or roots of the quadratic equation.
We can program an R function that takes in as inputs the coefficients from a quadratic equation and returns the two solutions:
quadratic_forumla <- function(a,b,c){
x_1 <- (-b + sqrt(b^2 - 4*a*c))/(2*a)
x_2 <- (-b - sqrt(b^2 - 4*a*c))/(2*a)
return(c(x_2,x_1))
}
quadratic_forumla(1,5,6)
#> [1] -3 -2
quadratic_forumla(5,3,-10)
#> [1] -1.745683 1.145683
quadratic_forumla(2,5,3)
#> [1] -1.5 -1.0\(~\)
\(~\)
1.15 Libraries & Packages
You may want to use newer or more specialized packages or libraries to read in or handle data. First you will need to make sure the library is installed with install.packages('package_name') then you can load the library with library('package_name') , and you will have access to all of the functionality withing the package.
Additionally, we can access a function from a library without loading the entire library, this can be done by using a command such as package::function, for instance, readr::read_csv tells R to look in the readr package for the function read_csv. This can be useful in programming functions and packages as well as if multiple packages contain functions with the same name.
1.15.1 installed packages
To see which packages are installed on your machine you can use:
# which packages are installed
library()$results[,1]
sample(library()$results[,1], 20, replace = FALSE)
#> [1] "bitops" "clock" "htmlwidgets" "crayon" "inum"
#> [6] "rematch" "askpass" "waldo" "RColorBrewer" "tools"
#> [11] "reprex" "modelr" "gtools" "sourcetools" "labeling"
#> [16] "lattice" "backports" "dbplyr" "sandwich" "mgcv"The following function will check if a list of packages are already installed, if not then it will install them:
# install package if missing
install_if_not <- function( list.of.packages ) {
new.packages <- list.of.packages[!(list.of.packages %in% installed.packages()[,"Package"])]
if(length(new.packages)) { install.packages(new.packages, repos = "http://cran.us.r-project.org") } else { print(paste0("the package '", list.of.packages , "' is already installed")) }
}You can use the function like this:
# test function
install_if_not(c("tidyverse"))
#> [1] "the package 'tidyverse' is already installed"some additional information on the installed packages including the version can be found now:
tibble::as_tibble(installed.packages())
#> # A tibble: 290 × 16
#> Package LibPath Version Prior…¹ Depends Imports Linki…² Sugge…³ Enhan…⁴
#> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr> <chr>
#> 1 abind C:/Users/j… 1.4-5 <NA> R (>= … method… <NA> <NA> <NA>
#> 2 Amelia C:/Users/j… 1.8.1 <NA> R (>= … foreig… Rcpp (… "tcltk… <NA>
#> 3 AMR C:/Users/j… 1.8.2 <NA> R (>= … <NA> <NA> "curl,… cleane…
#> 4 arsenal C:/Users/j… 3.6.3 <NA> R (>= … knitr … <NA> "broom… <NA>
#> 5 askpass C:/Users/j… 1.1 <NA> <NA> sys (>… <NA> "testt… <NA>
#> 6 assertthat C:/Users/j… 0.2.1 <NA> <NA> tools <NA> "testt… <NA>
#> # … with 284 more rows, 7 more variables: License <chr>, License_is_FOSS <chr>,
#> # License_restricts_use <chr>, OS_type <chr>, MD5sum <chr>,
#> # NeedsCompilation <chr>, Built <chr>, and abbreviated variable names
#> # ¹Priority, ²LinkingTo, ³Suggests, ⁴Enhances1.15.2 loaded packages
To check which packages are loaded you can use:
# loaded packages
(.packages())
#> [1] "stats" "graphics" "grDevices" "utils" "datasets" "methods"
#> [7] "base"Make sure that the tidyverse and dplyr packages are installed. You can run install.packages(c('tidyverse','dplyr')) to install both.
1.15.3 read sample data
my_dataframe <- readRDS('my_dataframe.RDS' )We can use the head command to see the first few rows:
head(my_dataframe)
#> list_of_ints list_of_strings list_of_logicals list_of_mixed_type
#> 1 2 Data TRUE 3.14
#> 2 4 Science FALSE 1
#> 3 6 is a TRUE cat
#> 4 8 blast! FALSE TRUE
#> list_of_numbers list_of_sexes_factor ordered_list_of_costs
#> 1 3.142857 Male $0 - $100
#> 2 18.000000 Female $100 - $200
#> 3 42.000000 Female $200 - $300
#> 4 65.200000 Male $300 - $400\(~\)
\(~\)
1.16 Using Packages
We make sections of code accessible to installed packages by using library command:
loaded_package_before <- (.packages())
# everthing below here can call functions in dplyr package
library('dplyr')
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
glimpse(my_dataframe)
#> Rows: 4
#> Columns: 7
#> $ list_of_ints <int> 2, 4, 6, 8
#> $ list_of_strings <chr> "Data", "Science", "is a", "blast!"
#> $ list_of_logicals <lgl> TRUE, FALSE, TRUE, FALSE
#> $ list_of_mixed_type <chr> "3.14", "1", "cat", "TRUE"
#> $ list_of_numbers <dbl> 3.142857, 18.000000, 42.000000, 65.200000
#> $ list_of_sexes_factor <fct> Male, Female, Female, Male
#> $ ordered_list_of_costs <ord> $0 - $100, $100 - $200, $200 - $300, $300 - $400
(.packages())
#> [1] "dplyr" "stats" "graphics" "grDevices" "utils" "datasets"
#> [7] "methods" "base"
loaded_package_after <- (.packages())
setdiff(loaded_package_after, loaded_package_before)
#> [1] "dplyr"
detach(package:dplyr)
#the dplyr package has now been detached. calls to functions may have errors
packages_cur <- (.packages())
setdiff(loaded_package_after,packages_cur)
#> [1] "dplyr"Additionally, we can access a function from a library without loading the entire library, this can be done by using a command such as package::function. This notation is needed in any instance where two or more loaded packages have at least one function with the same name. This notation is also useful in development of functions and packages.
For instance, the glimpse function from the dplyr package can also be accessed by using the following command
# glimpse the data
dplyr::glimpse(my_dataframe)
#> Rows: 4
#> Columns: 7
#> $ list_of_ints <int> 2, 4, 6, 8
#> $ list_of_strings <chr> "Data", "Science", "is a", "blast!"
#> $ list_of_logicals <lgl> TRUE, FALSE, TRUE, FALSE
#> $ list_of_mixed_type <chr> "3.14", "1", "cat", "TRUE"
#> $ list_of_numbers <dbl> 3.142857, 18.000000, 42.000000, 65.200000
#> $ list_of_sexes_factor <fct> Male, Female, Female, Male
#> $ ordered_list_of_costs <ord> $0 - $100, $100 - $200, $200 - $300, $300 - $400And just notice that without the library loaded or the dplyr:: in front we can error:
# error
glimpse(my_dataframe)
#> Rows: 4
#> Columns: 7
#> $ list_of_ints <int> 2, 4, 6, 8
#> $ list_of_strings <chr> "Data", "Science", "is a", "blast!"
#> $ list_of_logicals <lgl> TRUE, FALSE, TRUE, FALSE
#> $ list_of_mixed_type <chr> "3.14", "1", "cat", "TRUE"
#> $ list_of_numbers <dbl> 3.142857, 18.000000, 42.000000, 65.200000
#> $ list_of_sexes_factor <fct> Male, Female, Female, Male
#> $ ordered_list_of_costs <ord> $0 - $100, $100 - $200, $200 - $300, $300 - $400After first detaching a package with detach(package:package.name.here) we can check for an update from the console with install.packages(c("package.name.here"))
install.packages(c('dplyr'), repos = "http://cran.us.r-project.org")If a package has a lock you can override the lock with
install.packages(c('dplyr'), repos = "http://cran.us.r-project.org" , INSTALL_opts = '--no-lock')\(~\)
\(~\)
1.17 Welcome to the tidyverse
The tidyverse is a collection of R packages that have been grouped together in order to make “data wrangling” more efficient:
library('tidyverse')
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
#> ✔ ggplot2 3.4.0 ✔ purrr 1.0.0
#> ✔ tibble 3.1.8 ✔ stringr 1.5.0
#> ✔ tidyr 1.2.1 ✔ forcats 0.5.2
#> ✔ readr 2.1.3
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()tibbleis an more modernRversion of a data framereadrread in data-files such as CSV as tibbles or data-frames, write data-frames or tibbles to CSV or supported data-file type.dplyris a popular package to manage many common data manipulation tasks and data summariestidyrreshape your data but keep it ‘tidy’stringrfunctions to support working with text strings.purrrfunctional programming for Rggplot2is an interface for R to create numerous types of graphics of dataforcatstools for dealing with categorical data
Figure 1.1: https://www.tidyverse.org/
1.17.1 and friends
-
broommake outputs tidy -
lubridateworking with dates -
readxlthereadxlpackage contains theread_excelto read in.xlsor.xlsxfiles -
knitrproduce outputs such as HTML, PDF, Docs, PowerPoint, with Rmarkdown -
shinybuild interactive web apps straight from R -
flexdashboardbuild dashboards with R -
furrrparallel mapping -
yardstickfor model evaluation metrics
tidyverse_friends <- c('broom','lubridate','readxl','knitr','shiny','furrr','flexdashboard','yardstick')
install.packages(tidyverse_friends)1.17.2 pipe opperator
The pipe %>% operator originates from the magrittr package. The pipe takes the information on the left and passes it to the information on the right:
f(x)is the same asx %>% f()
Notice how x gets piped into a function f
glimpse(my_dataframe)
#> Rows: 4
#> Columns: 7
#> $ list_of_ints <int> 2, 4, 6, 8
#> $ list_of_strings <chr> "Data", "Science", "is a", "blast!"
#> $ list_of_logicals <lgl> TRUE, FALSE, TRUE, FALSE
#> $ list_of_mixed_type <chr> "3.14", "1", "cat", "TRUE"
#> $ list_of_numbers <dbl> 3.142857, 18.000000, 42.000000, 65.200000
#> $ list_of_sexes_factor <fct> Male, Female, Female, Male
#> $ ordered_list_of_costs <ord> $0 - $100, $100 - $200, $200 - $300, $300 - $400is the same as
my_dataframe %>%
glimpse()
#> Rows: 4
#> Columns: 7
#> $ list_of_ints <int> 2, 4, 6, 8
#> $ list_of_strings <chr> "Data", "Science", "is a", "blast!"
#> $ list_of_logicals <lgl> TRUE, FALSE, TRUE, FALSE
#> $ list_of_mixed_type <chr> "3.14", "1", "cat", "TRUE"
#> $ list_of_numbers <dbl> 3.142857, 18.000000, 42.000000, 65.200000
#> $ list_of_sexes_factor <fct> Male, Female, Female, Male
#> $ ordered_list_of_costs <ord> $0 - $100, $100 - $200, $200 - $300, $300 - $400Note that this pipe operation has become so popular in R version 4.1.0 now comes equipped with a pipe operator of it’s own:
1:10 |> mean()
#> [1] 5.5\(~\)
\(~\)
1.17.3 Other Packages
Other packages we that will will make use of throughout the course of the text:
-
devtoolsdevelopRpackages- Note that
devtoolson Windows also requiresRtoolshttps://cran.r-project.org/bin/windows/Rtools/
- Note that
arsenalcompare dataframes ; create summary tablesskimrautomate Exploratory Data AnalysisDataExplorerautomate Exploratory Data Analysisrsqcomputes Adjusted R2 for various model typesRSQLiteR package for interfacing with SQLite databasedbplyrdatabase back-end for dplyrplotlyinteractive HTML plotsDTcontainsdatatablefunction for interactive HTMLdatatableGGallycontainsggcorrfor correlation plots andggpairsfor other data-plotscorrrcorrelation matrix as a data-frameAMRPrincipal Component Plots
randomForestfit a Random Forest modelcaret(Classification And Regression Training) is a set of functions that attempt to streamline the process for creating predictive models.shinyShiny is an R package that makes it easy to build interactive web apps straight from R.usethisusethis is a workflow package; it automates repetitive tasks that arise during project setup and development, both for R packages and non-package projects.testthattestthat is a package designed to assist with unit testing functions within packages.pkgdownpkgdown is designed to make it quick and easy to build a website for your package.
other_packages <- c('devtools','rsq','arsenal','skimr','DataExplorer',
'RSQLite','dbplyr','plotly','DT','GGally','corrr',
'AMR','caret','randomForest','shiny','usethis','testthat','pkgdown')
install.packages(other_packages)\(~\)
\(~\)
1.18 Package Versions
We already mentioned that install.packages will update the package from CRAN:
install.packages( c( tidyverse_friends , other_packages ), repos = "http://cran.us.r-project.org")We also use devtools to install the most-up-to-date package from github, for example:
devtools::install_github("tidyverse/tidyverse")Here are the versions installed on this system, compare with your own:
as_tibble(installed.packages()) %>%
select(Package, Version, Depends) %>%
filter(Package %in% c( c('tidyverse'),
c('tibble','readr','dplyr','tidyr','stringr','purrr','ggplot2','forcats'),
c( tidyverse_friends , other_packages ) )) %>%
knitr::kable()| Package | Version | Depends |
|---|---|---|
| AMR | 1.8.2 | R (>= 3.0.0) |
| arsenal | 3.6.3 | R (>= 3.4.0), stats (>= 3.4.0) |
| broom | 1.0.2 | R (>= 3.1) |
| caret | 6.0-93 | ggplot2, lattice (>= 0.20), R (>= 3.2.0) |
| corrr | 0.4.4 | R (>= 3.4) |
| DataExplorer | 0.8.2 | R (>= 3.6) |
| dbplyr | 2.2.1 | R (>= 3.1) |
| dplyr | 1.0.10 | R (>= 3.4.0) |
| DT | 0.26 | NA |
| forcats | 0.5.2 | R (>= 3.4) |
| furrr | 0.3.1 | future (>= 1.25.0), R (>= 3.4.0) |
| GGally | 2.1.2 | R (>= 3.1), ggplot2 (>= 3.3.4) |
| ggplot2 | 3.4.0 | R (>= 3.3) |
| knitr | 1.41 | R (>= 3.3.0) |
| lubridate | 1.9.0 | methods, timechange (>= 0.1.1), R (>= 3.2) |
| plotly | 4.10.1.9000 | R (>= 3.2.0), ggplot2 (>= 3.0.0) |
| purrr | 1.0.0 | R (>= 3.2.3) |
| randomForest | 4.7-1.1 | R (>= 4.1.0), stats |
| readr | 2.1.3 | R (>= 3.4) |
| readxl | 1.4.1 | R (>= 3.4) |
| rsq | 2.5 | NA |
| RSQLite | 2.2.20 | R (>= 3.1.0) |
| shiny | 1.7.4 | R (>= 3.0.2), methods |
| skimr | 2.1.5 | R (>= 3.1.2) |
| stringr | 1.5.0 | R (>= 3.3) |
| testthat | 3.1.6 | R (>= 3.1) |
| tibble | 3.1.8 | R (>= 3.1.0) |
| tidyr | 1.2.1 | R (>= 3.1) |
| tidyverse | 1.3.2 | R (>= 3.3) |
| yardstick | 1.1.0 | R (>= 3.4.0) |
You can install specific versions of packages with: devtools::install_version("my.package.name", version = "0.9.1")
1.19 Details on this machine’s version of R
tibble::enframe(Sys.info()) %>%
filter(name %in% c('sysname','release','version','machine')) %>%
knitr::kable()| name | value |
|---|---|
| sysname | Windows |
| release | 10 x64 |
| version | build 22621 |
| machine | x86-64 |
tibble::as_tibble(R.Version()) %>%
pivot_longer(everything()) %>%
knitr::kable()| name | value |
|---|---|
| platform | x86_64-w64-mingw32 |
| arch | x86_64 |
| os | mingw32 |
| crt | ucrt |
| system | x86_64, mingw32 |
| status | |
| major | 4 |
| minor | 2.2 |
| year | 2022 |
| month | 10 |
| day | 31 |
| svn rev | 83211 |
| language | R |
| version.string | R version 4.2.2 (2022-10-31 ucrt) |
| nickname | Innocent and Trusting |
1.19.1 sessionInfo
All the details about the current running session of R:
sessionInfo()
#> R version 4.2.2 (2022-10-31 ucrt)
#> Platform: x86_64-w64-mingw32/x64 (64-bit)
#> Running under: Windows 10 x64 (build 22621)
#>
#> Matrix products: default
#>
#> locale:
#> [1] LC_COLLATE=English_United States.utf8
#> [2] LC_CTYPE=English_United States.utf8
#> [3] LC_MONETARY=English_United States.utf8
#> [4] LC_NUMERIC=C
#> [5] LC_TIME=English_United States.utf8
#>
#> attached base packages:
#> [1] stats graphics grDevices utils datasets methods base
#>
#> other attached packages:
#> [1] forcats_0.5.2 stringr_1.5.0 purrr_1.0.0 readr_2.1.3
#> [5] tidyr_1.2.1 tibble_3.1.8 ggplot2_3.4.0 tidyverse_1.3.2
#> [9] dplyr_1.0.10
#>
#> loaded via a namespace (and not attached):
#> [1] lubridate_1.9.0 assertthat_0.2.1 digest_0.6.31
#> [4] utf8_1.2.2 R6_2.5.1 cellranger_1.1.0
#> [7] backports_1.4.1 reprex_2.0.2 evaluate_0.19
#> [10] httr_1.4.4 highr_0.10 pillar_1.8.1
#> [13] rlang_1.0.6 googlesheets4_1.0.1 readxl_1.4.1
#> [16] rstudioapi_0.14 jquerylib_0.1.4 rmarkdown_2.19
#> [19] googledrive_2.0.0 munsell_0.5.0 broom_1.0.2
#> [22] compiler_4.2.2 modelr_0.1.10 xfun_0.36
#> [25] pkgconfig_2.0.3 htmltools_0.5.4 downlit_0.4.2
#> [28] tidyselect_1.2.0 bookdown_0.31 codetools_0.2-18
#> [31] fansi_1.0.3 crayon_1.5.2 tzdb_0.3.0
#> [34] dbplyr_2.2.1 withr_2.5.0 grid_4.2.2
#> [37] jsonlite_1.8.4 gtable_0.3.1 lifecycle_1.0.3
#> [40] DBI_1.1.3 magrittr_2.0.3 scales_1.2.1
#> [43] cli_3.5.0 stringi_1.7.8 cachem_1.0.6
#> [46] fs_1.5.2 xml2_1.3.3 bslib_0.4.2
#> [49] ellipsis_0.3.2 generics_0.1.3 vctrs_0.5.1
#> [52] tools_4.2.2 glue_1.6.2 hms_1.1.2
#> [55] fastmap_1.1.0 yaml_2.3.6 timechange_0.1.1
#> [58] colorspace_2.0-3 gargle_1.2.1 rvest_1.0.3
#> [61] memoise_2.0.1 knitr_1.41 haven_2.5.1
#> [64] sass_0.4.4