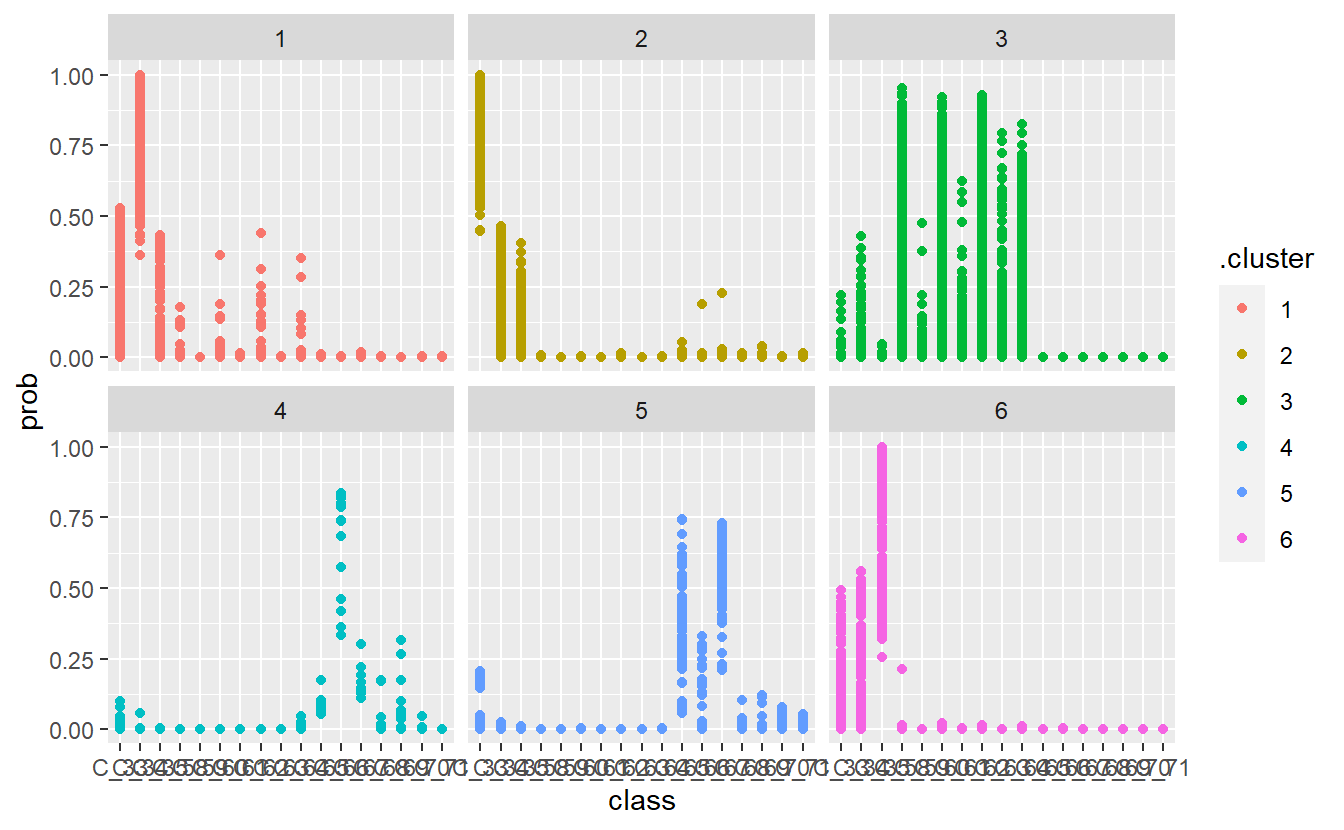
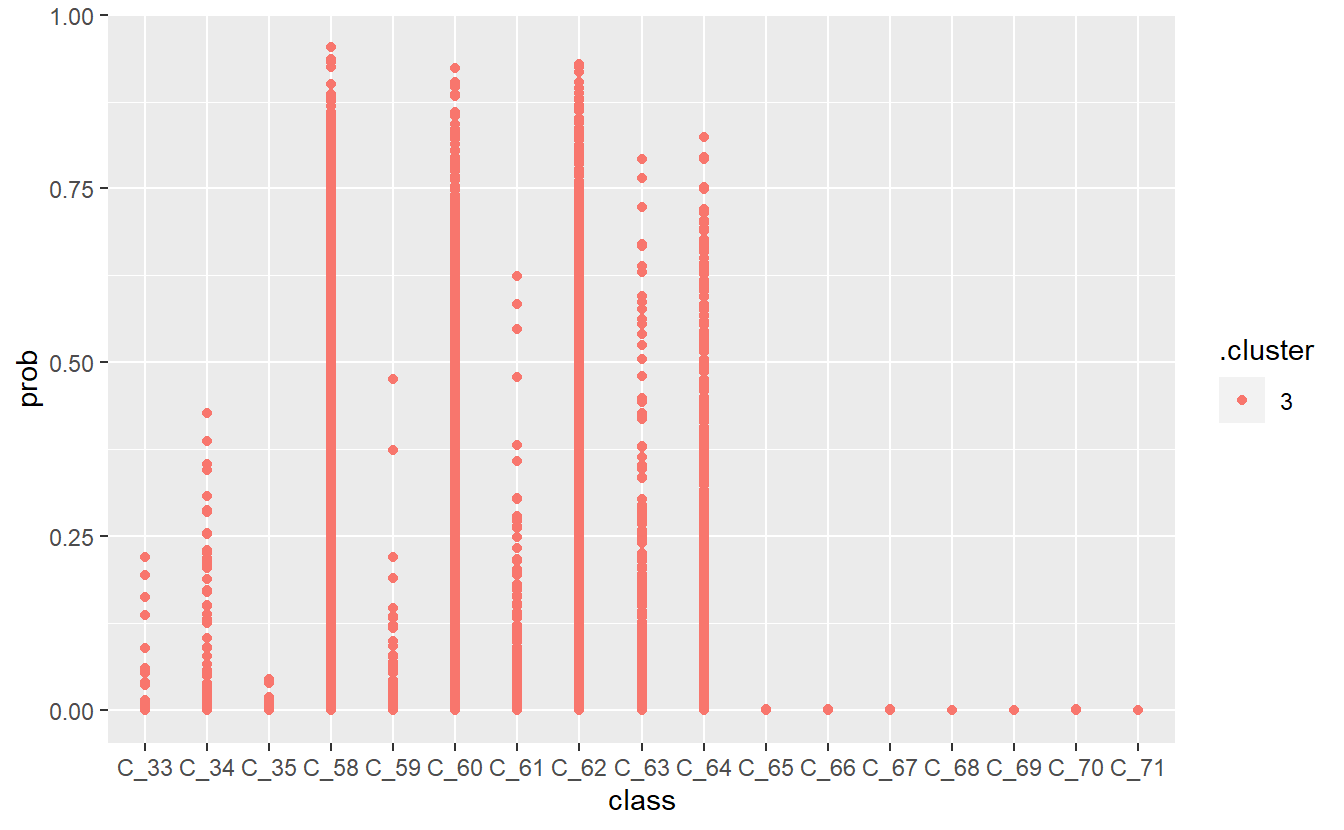
13 Diabetic Monitoring
The following example will show-case one method processing data from strings and processing into dates and factors.
The folder Diabetes-Data is referenced from <https://archive.ics.uci.edu/ml/datasets/Diabetes>. as per the website:
Diabetes Data Set
Diabetes Data Set Download: Data Folder, Data Set Description
Abstract: This diabetes dataset is from AIM ’94
Characteristics of Diabetes Data Set on https://archive.ics.uci.edu/ml/datasets/Diabetes Data Set Characteristics: Multivariate, Time-Series Number of Instances: ? Area: Life Attribute Characteristics: Categorical, Integer Number of Attributes: 20 Date Donated Not Reported Associated Tasks: Missing Values? ? Number of Web Hits: 561559
as reported on of April 2021
Source: Michael Kahn, MD, PhD, Washington University, St. Louis, MO
Data Set Information: Diabetes patient records were obtained from two sources: an automatic electronic recording device and paper records. The automatic device had an internal clock to timestamp events, whereas the paper records only provided “logical time” slots (breakfast, lunch, dinner, bedtime). For paper records, fixed times were assigned to breakfast (08:00), lunch (12:00), dinner (18:00), and bedtime (22:00). Thus paper records have fictitious uniform recording times whereas electronic records have more realistic time stamps.
Diabetes files consist of four fields per record. Each field is separated by a tab and each record is separated by a newline.
File format:
- Date in MM-DD-YYYY format
- Time in XX:YY format
- Code
- Value
The Code field is deciphered as follows:
- 33 = Regular insulin dose
- 34 = NPH insulin dose
- 35 = UltraLente insulin dose
- 48 = Unspecified blood glucose measurement
- 57 = Unspecified blood glucose measurement
- 58 = Pre-breakfast blood glucose measurement
- 59 = Post-breakfast blood glucose measurement
- 60 = Pre-lunch blood glucose measurement
- 61 = Post-lunch blood glucose measurement
- 62 = Pre-supper blood glucose measurement
- 63 = Post-supper blood glucose measurement
- 64 = Pre-snack blood glucose measurement
- 65 = Hypoglycemic symptoms
- 66 = Typical meal ingestion
- 67 = More-than-usual meal ingestion
- 68 = Less-than-usual meal ingestion
- 69 = Typical exercise activity
- 70 = More-than-usual exercise activity
- 71 = Less-than-usual exercise activity
- 72 = Unspecified special event
\(~\)
\(~\)
13.1 Load Data
Week_6_wd <- file.path('Week_6')
Week_6_DATA <- file.path(Week_6_wd,"DATA")
Week_6_Funs <- file.path(Week_6_wd,"FUNCTIONS")
DM2_DATA <- file.path(Week_6_wd,'Diabetes-Data')
DM2_DATA
#> [1] "Week_6/Diabetes-Data"
list_of_file_paths <- list.files(path = DM2_DATA,
pattern = 'data-',
recursive = TRUE,
full.names = TRUE)
list_of_file_paths
#> [1] "Week_6/Diabetes-Data/data-01" "Week_6/Diabetes-Data/data-02"
#> [3] "Week_6/Diabetes-Data/data-03" "Week_6/Diabetes-Data/data-04"
#> [5] "Week_6/Diabetes-Data/data-05" "Week_6/Diabetes-Data/data-06"
#> [7] "Week_6/Diabetes-Data/data-07" "Week_6/Diabetes-Data/data-08"
#> [9] "Week_6/Diabetes-Data/data-09" "Week_6/Diabetes-Data/data-10"
#> [11] "Week_6/Diabetes-Data/data-11" "Week_6/Diabetes-Data/data-12"
#> [13] "Week_6/Diabetes-Data/data-13" "Week_6/Diabetes-Data/data-14"
#> [15] "Week_6/Diabetes-Data/data-15" "Week_6/Diabetes-Data/data-16"
#> [17] "Week_6/Diabetes-Data/data-17" "Week_6/Diabetes-Data/data-18"
#> [19] "Week_6/Diabetes-Data/data-19" "Week_6/Diabetes-Data/data-20"
#> [21] "Week_6/Diabetes-Data/data-21" "Week_6/Diabetes-Data/data-22"
#> [23] "Week_6/Diabetes-Data/data-23" "Week_6/Diabetes-Data/data-24"
#> [25] "Week_6/Diabetes-Data/data-25" "Week_6/Diabetes-Data/data-26"
#> [27] "Week_6/Diabetes-Data/data-27" "Week_6/Diabetes-Data/data-28"
#> [29] "Week_6/Diabetes-Data/data-29" "Week_6/Diabetes-Data/data-30"
#> [31] "Week_6/Diabetes-Data/data-31" "Week_6/Diabetes-Data/data-32"
#> [33] "Week_6/Diabetes-Data/data-33" "Week_6/Diabetes-Data/data-34"
#> [35] "Week_6/Diabetes-Data/data-35" "Week_6/Diabetes-Data/data-36"
#> [37] "Week_6/Diabetes-Data/data-37" "Week_6/Diabetes-Data/data-38"
#> [39] "Week_6/Diabetes-Data/data-39" "Week_6/Diabetes-Data/data-40"
#> [41] "Week_6/Diabetes-Data/data-41" "Week_6/Diabetes-Data/data-42"
#> [43] "Week_6/Diabetes-Data/data-43" "Week_6/Diabetes-Data/data-44"
#> [45] "Week_6/Diabetes-Data/data-45" "Week_6/Diabetes-Data/data-46"
#> [47] "Week_6/Diabetes-Data/data-47" "Week_6/Diabetes-Data/data-48"
#> [49] "Week_6/Diabetes-Data/data-49" "Week_6/Diabetes-Data/data-50"
#> [51] "Week_6/Diabetes-Data/data-51" "Week_6/Diabetes-Data/data-52"
#> [53] "Week_6/Diabetes-Data/data-53" "Week_6/Diabetes-Data/data-54"
#> [55] "Week_6/Diabetes-Data/data-55" "Week_6/Diabetes-Data/data-56"
#> [57] "Week_6/Diabetes-Data/data-57" "Week_6/Diabetes-Data/data-58"
#> [59] "Week_6/Diabetes-Data/data-59" "Week_6/Diabetes-Data/data-60"
#> [61] "Week_6/Diabetes-Data/data-61" "Week_6/Diabetes-Data/data-62"
#> [63] "Week_6/Diabetes-Data/data-63" "Week_6/Diabetes-Data/data-64"
#> [65] "Week_6/Diabetes-Data/data-65" "Week_6/Diabetes-Data/data-66"
#> [67] "Week_6/Diabetes-Data/data-67" "Week_6/Diabetes-Data/data-68"
#> [69] "Week_6/Diabetes-Data/data-69" "Week_6/Diabetes-Data/data-70"
library('tidyverse')
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.2 ──
#> ✔ ggplot2 3.4.0 ✔ purrr 1.0.0
#> ✔ tibble 3.1.8 ✔ dplyr 1.0.10
#> ✔ tidyr 1.2.1 ✔ stringr 1.5.0
#> ✔ readr 2.1.3 ✔ forcats 0.5.2
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
tibble_of_file_paths <- tibble(list_of_file_paths) %>%
mutate(full_path = list_of_file_paths) %>%
mutate(file_no = str_replace(string = full_path,
pattern = paste0(DM2_DATA,'/data-'),
replacement = ''))
tibble_of_file_paths
#> # A tibble: 70 × 3
#> list_of_file_paths full_path file_no
#> <chr> <chr> <chr>
#> 1 Week_6/Diabetes-Data/data-01 Week_6/Diabetes-Data/data-01 01
#> 2 Week_6/Diabetes-Data/data-02 Week_6/Diabetes-Data/data-02 02
#> 3 Week_6/Diabetes-Data/data-03 Week_6/Diabetes-Data/data-03 03
#> 4 Week_6/Diabetes-Data/data-04 Week_6/Diabetes-Data/data-04 04
#> 5 Week_6/Diabetes-Data/data-05 Week_6/Diabetes-Data/data-05 05
#> 6 Week_6/Diabetes-Data/data-06 Week_6/Diabetes-Data/data-06 06
#> # … with 64 more rows
read_in_text_from_file_no <- function(my_file_no){
enquo_by <- enquo(my_file_no)
cur_file <- tibble_of_file_paths %>%
filter(file_no == !!enquo_by)
cur_path <- as.character(cur_file$full_path)
dat <- readr::read_tsv(cur_path,
col_names = c('Date',"Time","Code","Value"),
col_types = cols(col_character(), col_character(), col_character(), col_character())
) %>%
mutate(file_no = as.numeric(cur_file$file_no))
return(dat)
}
library('furrr')
#> Loading required package: future
no_cores <- availableCores() - 1
no_cores
#> system
#> 15
future::plan(multicore, workers = no_cores)
DM2_TIME_RAW <- furrr::future_map_dfr(tibble_of_file_paths$file_no, # list of columns
function(x){ read_in_text_from_file_no(x) })
DM2_TIME_RAW <- readRDS('DATA/DM2_TIME_RAW.RDS')\(~\)
\(~\)
13.2 Decode Table
Code_Table <- tribble(
~ Code, ~ Code_Description,
33 , "Regular insulin dose",
34 , "NPH insulin dose",
35 , "UltraLente insulin dose",
48 , "Unspecified blood glucose measurement", # This decode value is the same as 57
57 , "Unspecified blood glucose measurement", # This decode value is the same as 48
58 , "Pre-breakfast blood glucose measurement",
59 , "Post-breakfast blood glucose measurement",
60 , "Pre-lunch blood glucose measurement",
61 , "Post-lunch blood glucose measurement",
62 , "Pre-supper blood glucose measurement",
63 , "Post-supper blood glucose measurement",
64 , "Pre-snack blood glucose measurement",
65 , "Hypoglycemic symptoms",
66 , "Typical meal ingestion",
67 , "More-than-usual meal ingestion",
68 , "Less-than-usual meal ingestion",
69 , "Typical exercise activity",
70 , "More-than-usual exercise activity",
71 , "Less-than-usual exercise activity",
72 , "Unspecified special event") %>% # What does this mean?
mutate(Code_Invalid = if_else(Code %in% c(48,57,72), TRUE, FALSE)) %>%
mutate(Code_New = if_else(Code_Invalid == TRUE, 999, Code))
Code_Table
#> # A tibble: 20 × 4
#> Code Code_Description Code_Invalid Code_New
#> <dbl> <chr> <lgl> <dbl>
#> 1 33 Regular insulin dose FALSE 33
#> 2 34 NPH insulin dose FALSE 34
#> 3 35 UltraLente insulin dose FALSE 35
#> 4 48 Unspecified blood glucose measurement TRUE 999
#> 5 57 Unspecified blood glucose measurement TRUE 999
#> 6 58 Pre-breakfast blood glucose measurement FALSE 58
#> # … with 14 more rows
Code_Table_Factor <- Code_Table %>%
filter(Code_Invalid == FALSE) %>%
mutate(Code_Factor = factor(paste0("C_",Code_New)))
Code_Table_Factor
#> # A tibble: 17 × 5
#> Code Code_Description Code_Invalid Code_New Code_Fa…¹
#> <dbl> <chr> <lgl> <dbl> <fct>
#> 1 33 Regular insulin dose FALSE 33 C_33
#> 2 34 NPH insulin dose FALSE 34 C_34
#> 3 35 UltraLente insulin dose FALSE 35 C_35
#> 4 58 Pre-breakfast blood glucose measurement FALSE 58 C_58
#> 5 59 Post-breakfast blood glucose measurement FALSE 59 C_59
#> 6 60 Pre-lunch blood glucose measurement FALSE 60 C_60
#> # … with 11 more rows, and abbreviated variable name ¹Code_Factor13.3 Format Data
library('lubridate')
#> Loading required package: timechange
#>
#> Attaching package: 'lubridate'
#> The following objects are masked from 'package:base':
#>
#> date, intersect, setdiff, union
DM2_TIME <- DM2_TIME_RAW %>%
mutate(Date_Time_Char = paste0(Date, ' ', Time)) %>%
mutate(Date_Time = mdy_hm(Date_Time_Char)) %>%
mutate(Value_Num = as.numeric(Value)) %>%
mutate(Code = as.numeric(Code)) %>%
mutate(Record_ID = row_number())
#> Warning: 45 failed to parse.
#> Warning in mask$eval_all_mutate(quo): NAs introduced by coercion
DM2_TIME
#> # A tibble: 29,330 × 9
#> Date Time Code Value file_no Date_…¹ Date_Time Value…² Recor…³
#> <chr> <chr> <dbl> <chr> <dbl> <chr> <dttm> <dbl> <int>
#> 1 04-21-1… 9:09 58 100 1 04-21-… 1991-04-21 09:09:00 100 1
#> 2 04-21-1… 9:09 33 009 1 04-21-… 1991-04-21 09:09:00 9 2
#> 3 04-21-1… 9:09 34 013 1 04-21-… 1991-04-21 09:09:00 13 3
#> 4 04-21-1… 17:08 62 119 1 04-21-… 1991-04-21 17:08:00 119 4
#> 5 04-21-1… 17:08 33 007 1 04-21-… 1991-04-21 17:08:00 7 5
#> 6 04-21-1… 22:51 48 123 1 04-21-… 1991-04-21 22:51:00 123 6
#> # … with 29,324 more rows, and abbreviated variable names ¹Date_Time_Char,
#> # ²Value_Num, ³Record_ID
UnKnown_Measurements <- DM2_TIME %>%
left_join(Code_Table_Factor, by='Code') %>%
filter(is.na(Code_Description) | is.na(Date_Time) | is.na(Value_Num) | Code_Invalid == TRUE | Code_New == 999 )
UnKnown_Measurements
#> # A tibble: 3,171 × 13
#> Date Time Code Value file_no Date_…¹ Date_Time Value…² Recor…³
#> <chr> <chr> <dbl> <chr> <dbl> <chr> <dttm> <dbl> <int>
#> 1 04-21-1… 22:51 48 123 1 04-21-… 1991-04-21 22:51:00 123 6
#> 2 04-24-1… 22:09 48 340 1 04-24-… 1991-04-24 22:09:00 340 23
#> 3 04-25-1… 21:54 48 288 1 04-25-… 1991-04-25 21:54:00 288 31
#> 4 04-28-1… 22:30 48 200 1 04-28-… 1991-04-28 22:30:00 200 49
#> 5 04-29-1… 22:28 48 081 1 04-29-… 1991-04-29 22:28:00 81 58
#> 6 04-30-1… 23:06 48 104 1 04-30-… 1991-04-30 23:06:00 104 67
#> # … with 3,165 more rows, 4 more variables: Code_Description <chr>,
#> # Code_Invalid <lgl>, Code_New <dbl>, Code_Factor <fct>, and abbreviated
#> # variable names ¹Date_Time_Char, ²Value_Num, ³Record_ID
Known_Measurements <- DM2_TIME %>%
anti_join(UnKnown_Measurements %>% select(Record_ID)) %>%
left_join(Code_Table_Factor, by='Code')
#> Joining, by = "Record_ID"
Known_Measurements
#> # A tibble: 26,159 × 13
#> Date Time Code Value file_no Date_…¹ Date_Time Value…² Recor…³
#> <chr> <chr> <dbl> <chr> <dbl> <chr> <dttm> <dbl> <int>
#> 1 04-21-1… 9:09 58 100 1 04-21-… 1991-04-21 09:09:00 100 1
#> 2 04-21-1… 9:09 33 009 1 04-21-… 1991-04-21 09:09:00 9 2
#> 3 04-21-1… 9:09 34 013 1 04-21-… 1991-04-21 09:09:00 13 3
#> 4 04-21-1… 17:08 62 119 1 04-21-… 1991-04-21 17:08:00 119 4
#> 5 04-21-1… 17:08 33 007 1 04-21-… 1991-04-21 17:08:00 7 5
#> 6 04-22-1… 7:35 58 216 1 04-22-… 1991-04-22 07:35:00 216 7
#> # … with 26,153 more rows, 4 more variables: Code_Description <chr>,
#> # Code_Invalid <lgl>, Code_New <dbl>, Code_Factor <fct>, and abbreviated
#> # variable names ¹Date_Time_Char, ²Value_Num, ³Record_ID\(~\)
\(~\)
13.4 View Data
Known_Measurements %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=Code_Factor)) +
geom_point()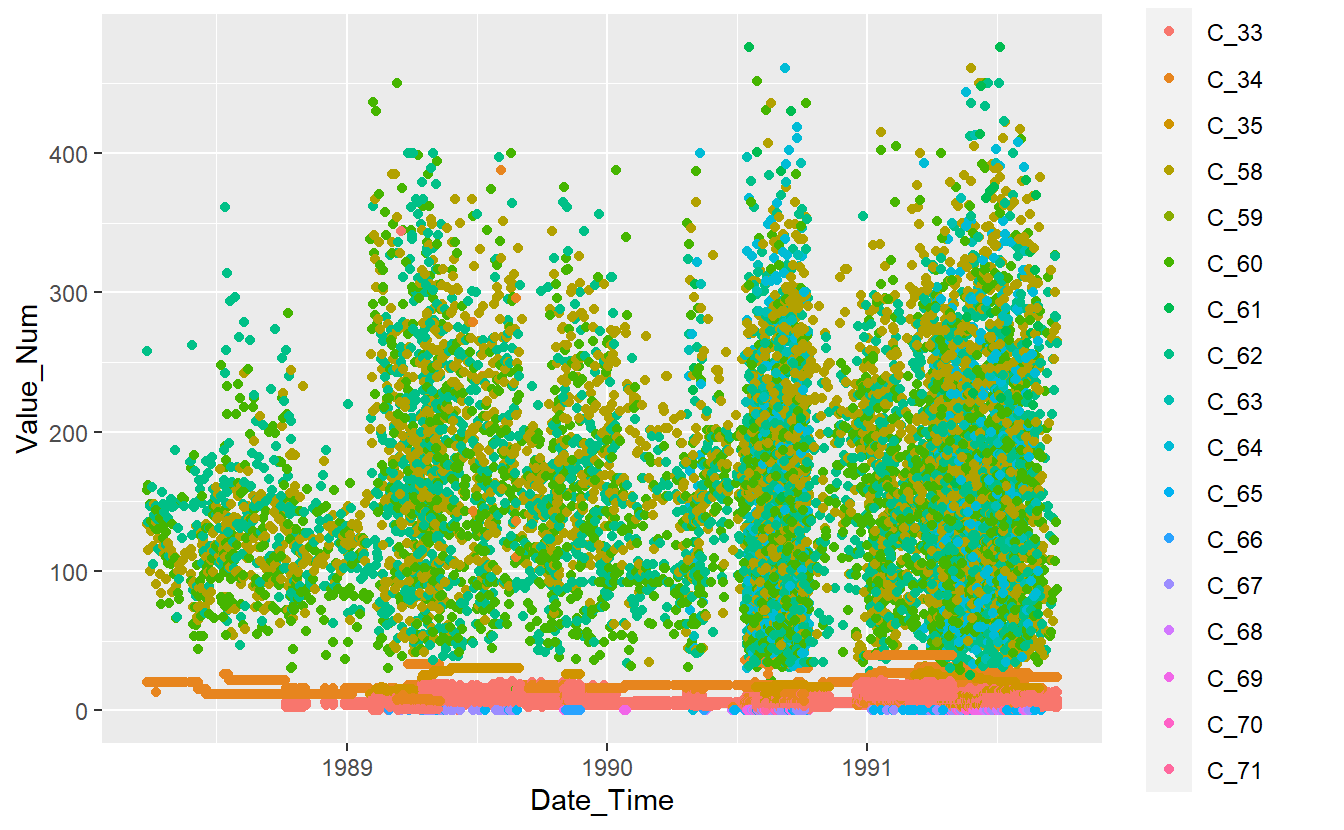
Features_Percent_Complete <- function(data, Percent_Complete = 0){
SumNa <- function(col){sum(is.na(col))}
na_sums <- data %>%
summarise_all(SumNa) %>%
tidyr::pivot_longer(everything() ,names_to = 'feature', values_to = 'SumNa') %>%
arrange(-SumNa) %>%
mutate(PctNa = SumNa/nrow(data)) %>%
mutate(PctComp = (1 - PctNa)*100)
data_out <- na_sums %>%
filter(PctComp >= Percent_Complete)
return(data_out)
}
Feature_Percent_Complete_Graph <- function(data, Percent_Complete = 0 ){
table <- Features_Percent_Complete(data, Percent_Complete)
plot1 <- table %>%
ggplot(aes(x=reorder(feature, PctComp), y =PctComp, fill=feature)) +
geom_bar(stat = "identity") +
coord_flip() +
theme(legend.position = "none")
return(plot1)
}
Feature_Percent_Complete_Graph(data = Known_Measurements %>% select(Date_Time, Value_Num, Code_Factor) )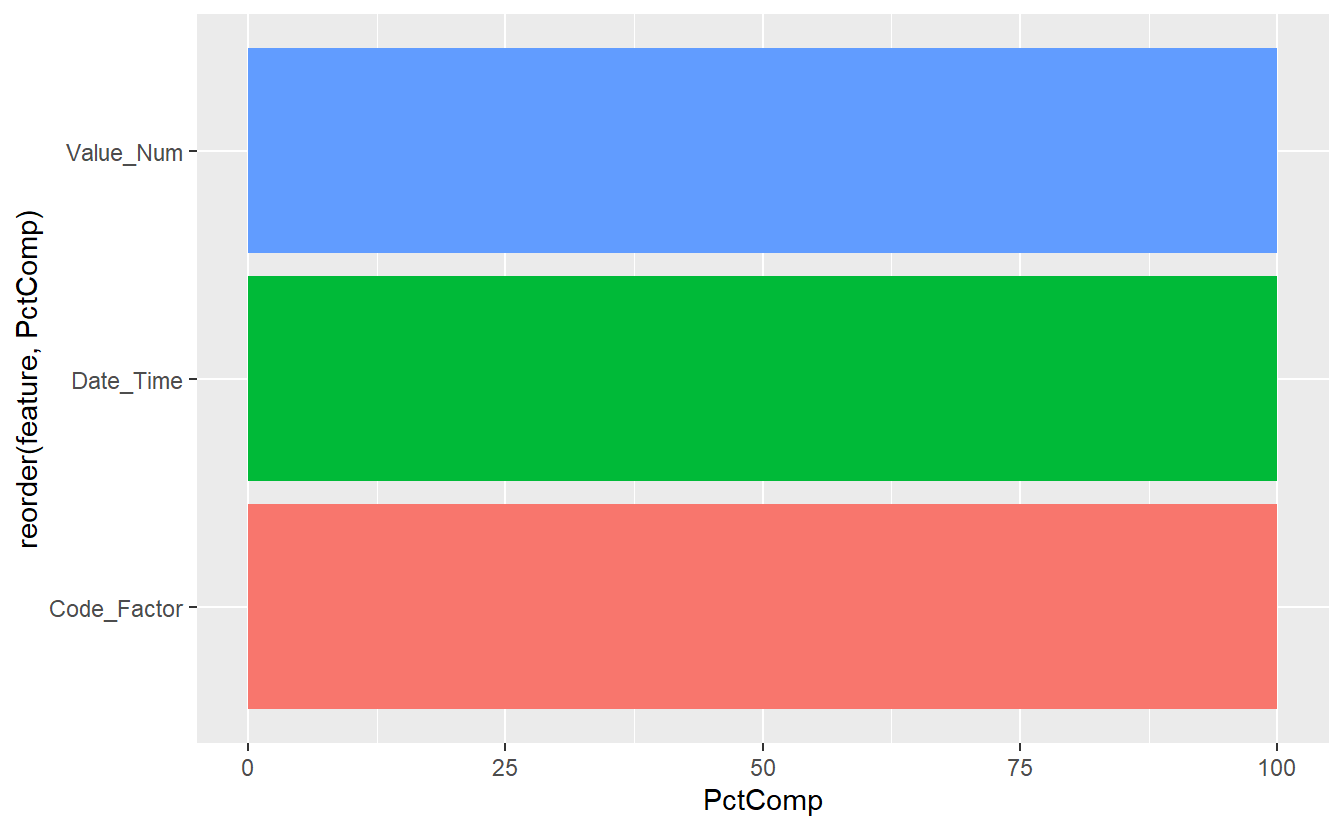
(#fig:5-percent-complete-graph-1 )Known Measurements Feature % Complete Graph
\(~\)
\(~\)
13.5 UnKnown Measurements
UnKnown_Measurements %>%
ggplot(aes(x=Date_Time, y=Value_Num)) +
geom_point()
#> Warning: Removed 86 rows containing missing values (`geom_point()`).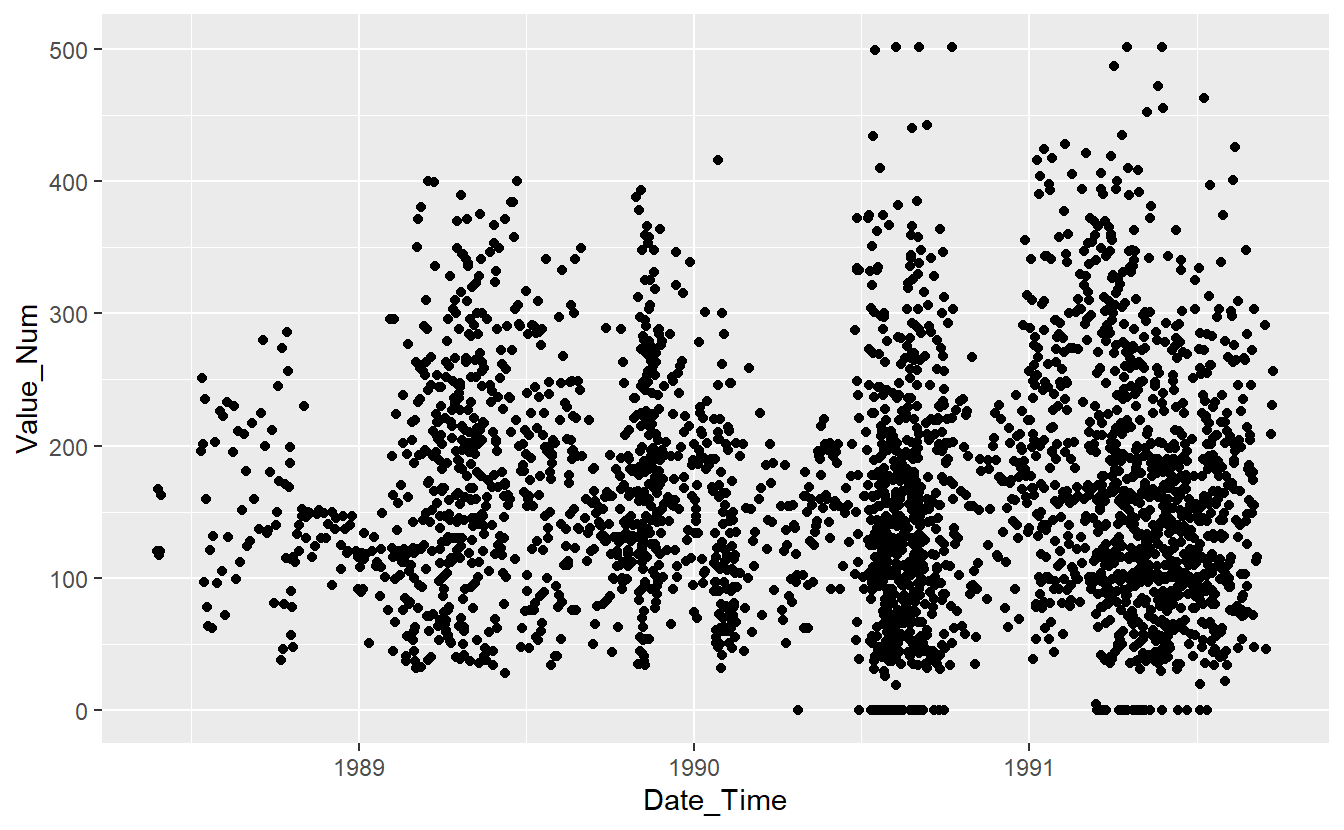
(#fig:5-unknown-fig-1 )UnKnown Measurements
\(~\)
\(~\)
13.6 Split Training Data
set.seed(123)
train_records <- sample(Known_Measurements$Record_ID,
size = nrow(Known_Measurements)*.6,
replace = FALSE)
DATA.train <- Known_Measurements %>%
filter(Record_ID %in% train_records)
DATA.test <- Known_Measurements %>%
filter(!(Record_ID %in% train_records))
DATA.train %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=Code_Factor)) +
geom_point() 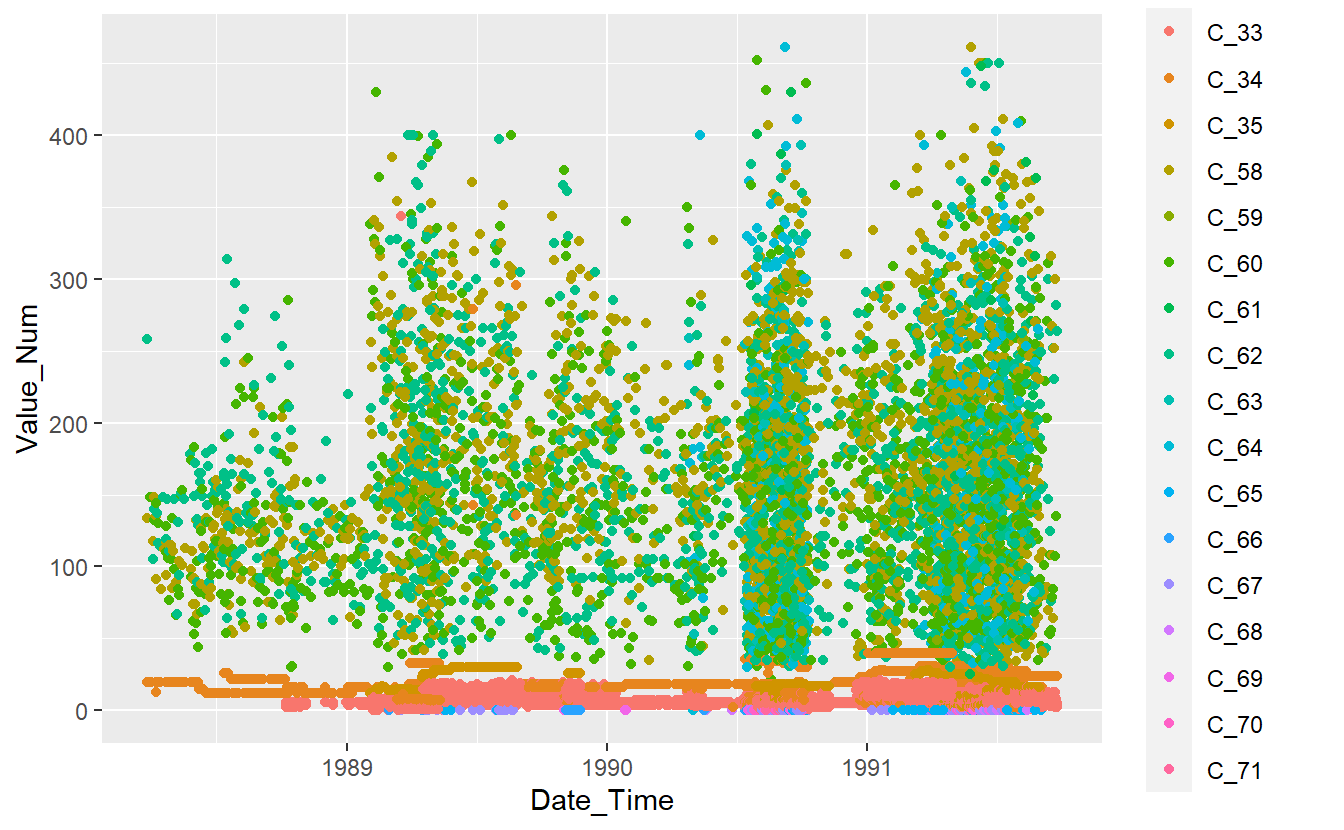
Figure 13.1: Plot of Training Data
DATA.test %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=Code_Factor)) +
geom_point() 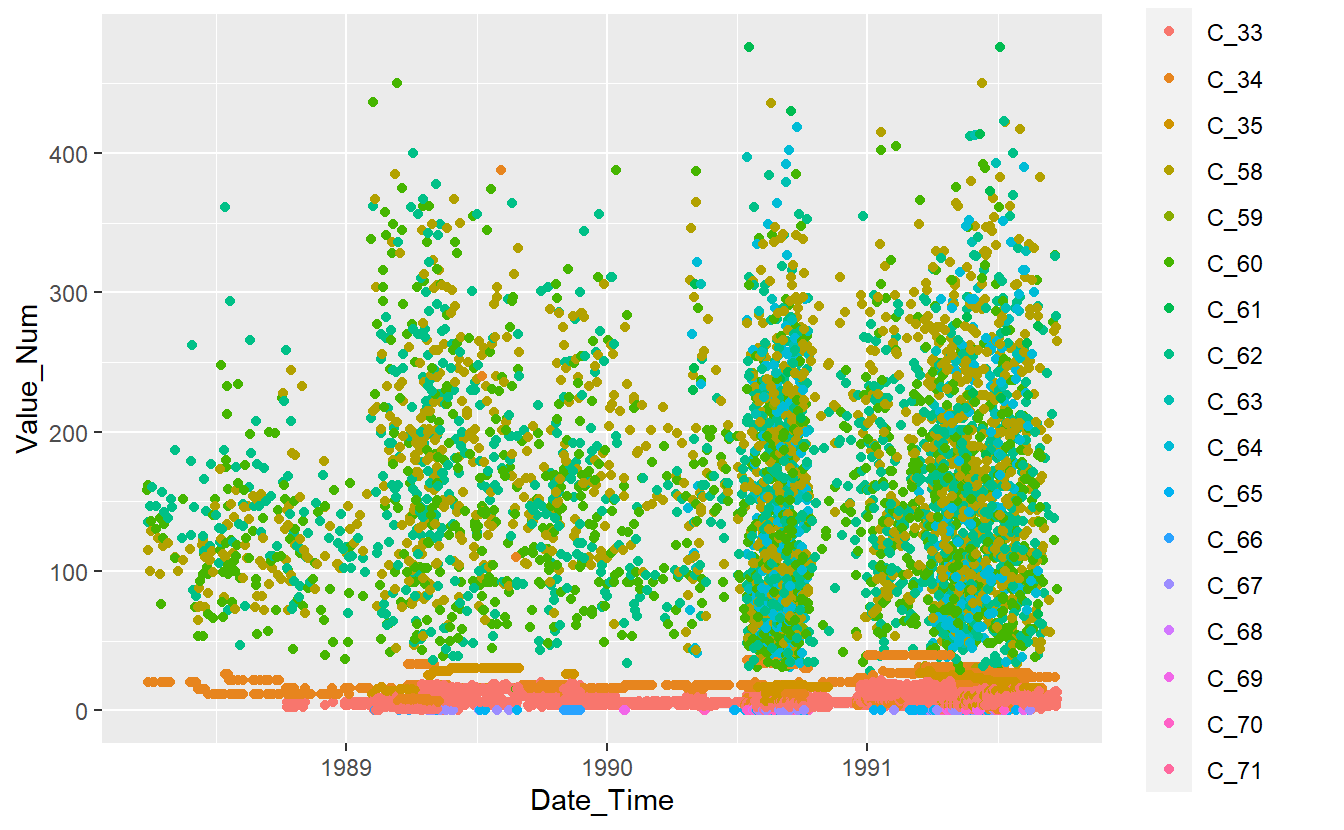
Figure 13.2: Plot of Test Data
\(~\)
\(~\)
13.7 Fit a Model
library('randomForest')
#> randomForest 4.7-1.1
#> Type rfNews() to see new features/changes/bug fixes.
#>
#> Attaching package: 'randomForest'
#> The following object is masked from 'package:dplyr':
#>
#> combine
#> The following object is masked from 'package:ggplot2':
#>
#> margin
rf_model <- randomForest(Code_Factor ~ Date_Time + Value_Num,
data = DATA.train,
ntree = 659,
importance = TRUE)13.7.1 Random Forest Ouput
Here’s the output of Random Forest
rf_model
#>
#> Call:
#> randomForest(formula = Code_Factor ~ Date_Time + Value_Num, data = DATA.train, ntree = 659, importance = TRUE)
#> Type of random forest: classification
#> Number of trees: 659
#> No. of variables tried at each split: 1
#>
#> OOB estimate of error rate: 36.11%
#> Confusion matrix:
#> C_33 C_34 C_35 C_58 C_59 C_60 C_61 C_62 C_63 C_64 C_65 C_66 C_67 C_68 C_69
#> C_33 5377 231 59 0 0 0 0 2 0 0 1 1 3 0 0
#> C_34 669 1586 59 4 0 2 0 1 0 0 0 0 0 0 0
#> C_35 167 88 398 0 0 0 0 0 0 0 0 0 0 0 0
#> C_58 0 5 3 969 1 473 6 551 21 79 0 0 0 0 0
#> C_59 0 0 0 3 0 3 0 5 0 0 0 0 0 0 0
#> C_60 0 10 1 555 2 534 1 465 9 66 0 0 0 0 0
#> C_61 0 0 1 12 0 7 4 12 0 4 0 0 0 0 0
#> C_62 0 8 0 651 2 440 1 680 10 85 0 0 0 0 0
#> C_63 0 0 0 45 0 25 0 35 26 5 0 0 0 0 0
#> C_64 0 5 0 137 0 85 0 127 7 173 0 1 0 0 0
#> C_65 0 0 0 0 0 0 0 0 0 0 47 4 143 0 0
#> C_66 1 0 0 0 0 0 0 0 0 0 3 66 27 0 0
#> C_67 1 0 0 0 0 0 0 0 0 0 32 1 167 0 0
#> C_68 0 0 0 0 0 0 0 0 0 0 3 2 16 0 0
#> C_69 0 0 0 0 0 0 0 0 0 0 3 11 25 0 0
#> C_70 0 0 0 0 0 0 0 0 0 0 4 1 85 0 0
#> C_71 0 0 0 0 0 0 0 0 0 0 8 0 47 0 0
#> C_70 C_71 class.error
#> C_33 0 0 0.05234403
#> C_34 0 0 0.31667385
#> C_35 0 0 0.39050536
#> C_58 0 0 0.54032258
#> C_59 0 0 1.00000000
#> C_60 0 0 0.67498478
#> C_61 0 0 0.90000000
#> C_62 0 0 0.63771977
#> C_63 0 0 0.80882353
#> C_64 0 0 0.67663551
#> C_65 0 0 0.75773196
#> C_66 0 0 0.31958763
#> C_67 0 0 0.16915423
#> C_68 0 0 1.00000000
#> C_69 0 0 1.00000000
#> C_70 0 0 1.00000000
#> C_71 0 0 1.00000000The Confusion Matrix that we see above is the confusion matrix from fitting the model against the training data so it is overly optimistic in terms of predictions.
Random Forest is a collection of Decision Trees, we can look at the individual trees in the forest by using one of the two following functions.
library(ggraph)
library(igraph)
#>
#> Attaching package: 'igraph'
#> The following objects are masked from 'package:lubridate':
#>
#> %--%, union
#> The following objects are masked from 'package:future':
#>
#> %->%, %<-%
#> The following objects are masked from 'package:dplyr':
#>
#> as_data_frame, groups, union
#> The following objects are masked from 'package:purrr':
#>
#> compose, simplify
#> The following object is masked from 'package:tidyr':
#>
#> crossing
#> The following object is masked from 'package:tibble':
#>
#> as_data_frame
#> The following objects are masked from 'package:stats':
#>
#> decompose, spectrum
#> The following object is masked from 'package:base':
#>
#> union
plot_rf_tree <- function(final_model, tree_num, shorten_label = TRUE) {
# get tree by index
tree <- randomForest::getTree(final_model,
k = tree_num,
labelVar = TRUE) %>%
tibble::rownames_to_column() %>%
# make leaf split points to NA, so the 0s won't get plotted
mutate(`split point` = ifelse(is.na(prediction), `split point`, NA))
# prepare data frame for graph
graph_frame <- data.frame(from = rep(tree$rowname, 2),
to = c(tree$`left daughter`, tree$`right daughter`))
# convert to graph and delete the last node that we don't want to plot
graph <- graph_from_data_frame(graph_frame) %>%
delete_vertices("0")
# set node labels
V(graph)$node_label <- gsub("_", " ", as.character(tree$`split var`))
if (shorten_label) {
V(graph)$leaf_label <- substr(as.character(tree$prediction), 1, 1)
}
V(graph)$split <- as.character(round(tree$`split point`, digits = 2))
# plot
plot <- ggraph(graph, 'tree') +
theme_graph() +
geom_edge_link() +
geom_node_point() +
geom_node_label(aes(label = leaf_label, fill = leaf_label), na.rm = TRUE,
repel = FALSE, colour = "white",
show.legend = FALSE)
print(plot)
}
plot_rf_tree(rf_model, 456)
#> Warning: Duplicated aesthetics after name standardisation: na.rm
#> Warning: Using the `size` aesthetic in this geom was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` in the `default_aes` field and elsewhere instead.
#> Warning: Removed 2855 rows containing missing values (`geom_label()`).
Random Forest models normally come equipped with feature importances:
varImpPlot(rf_model)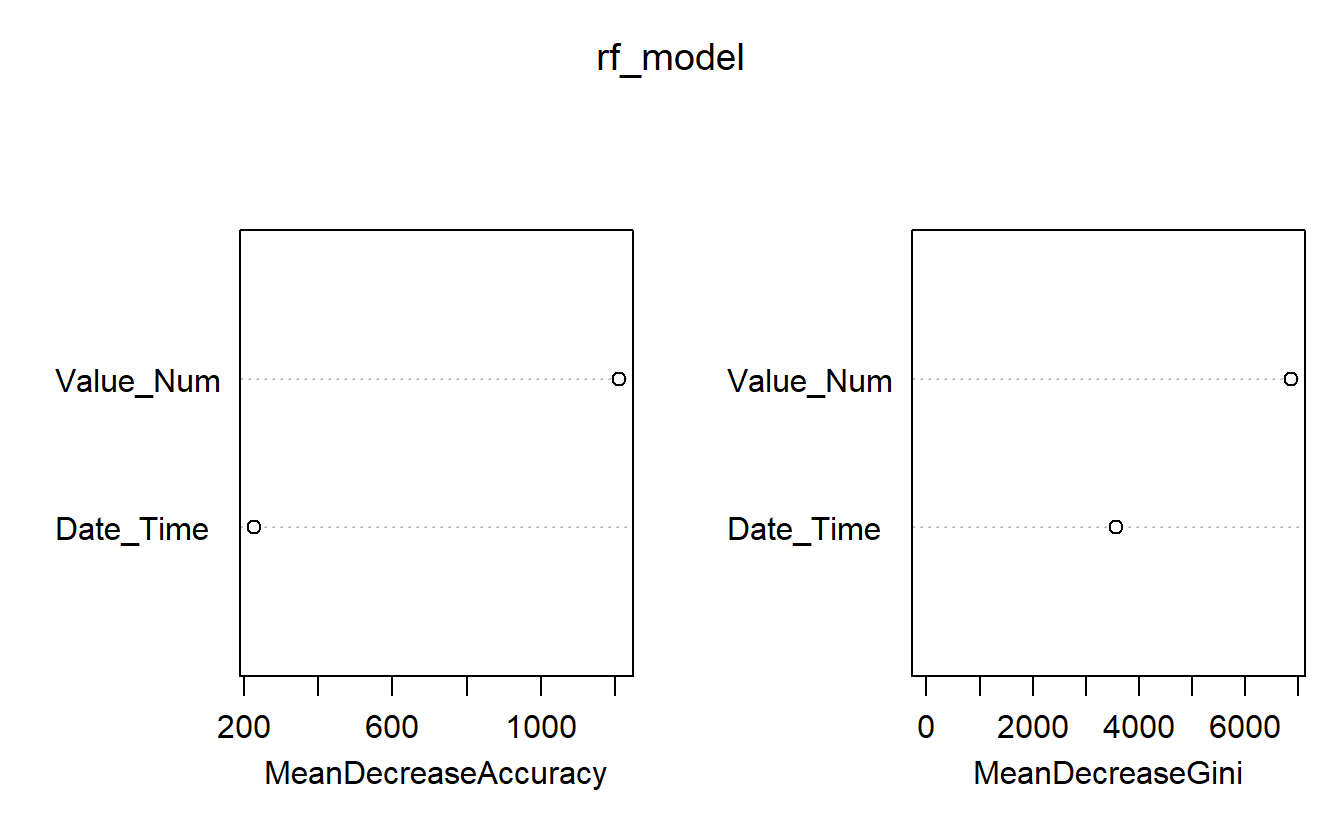
Figure 13.3: Feature Importances of Base Model
For instance, above we see that Date_Time is more in terms classifying the measurement as opposed to Value_Num
We can also extract the feature importances for each of the classes:
caret::varImp(rf_model, scale = FALSE) %>%
t() %>% # transpose the entire dataframe
as_tibble(rownames = 'Class' ) # turn it back to a tibble
#> # A tibble: 17 × 3
#> Class Date_Time Value_Num
#> <chr> <dbl> <dbl>
#> 1 C_33 0.0969 0.517
#> 2 C_34 0.264 0.509
#> 3 C_35 0.373 0.507
#> 4 C_58 0.0481 0.246
#> 5 C_59 -0.000723 -0.00106
#> 6 C_60 0.0413 0.194
#> # … with 11 more rowsThe Random Forest model also has a plot associated with it:
plot(rf_model)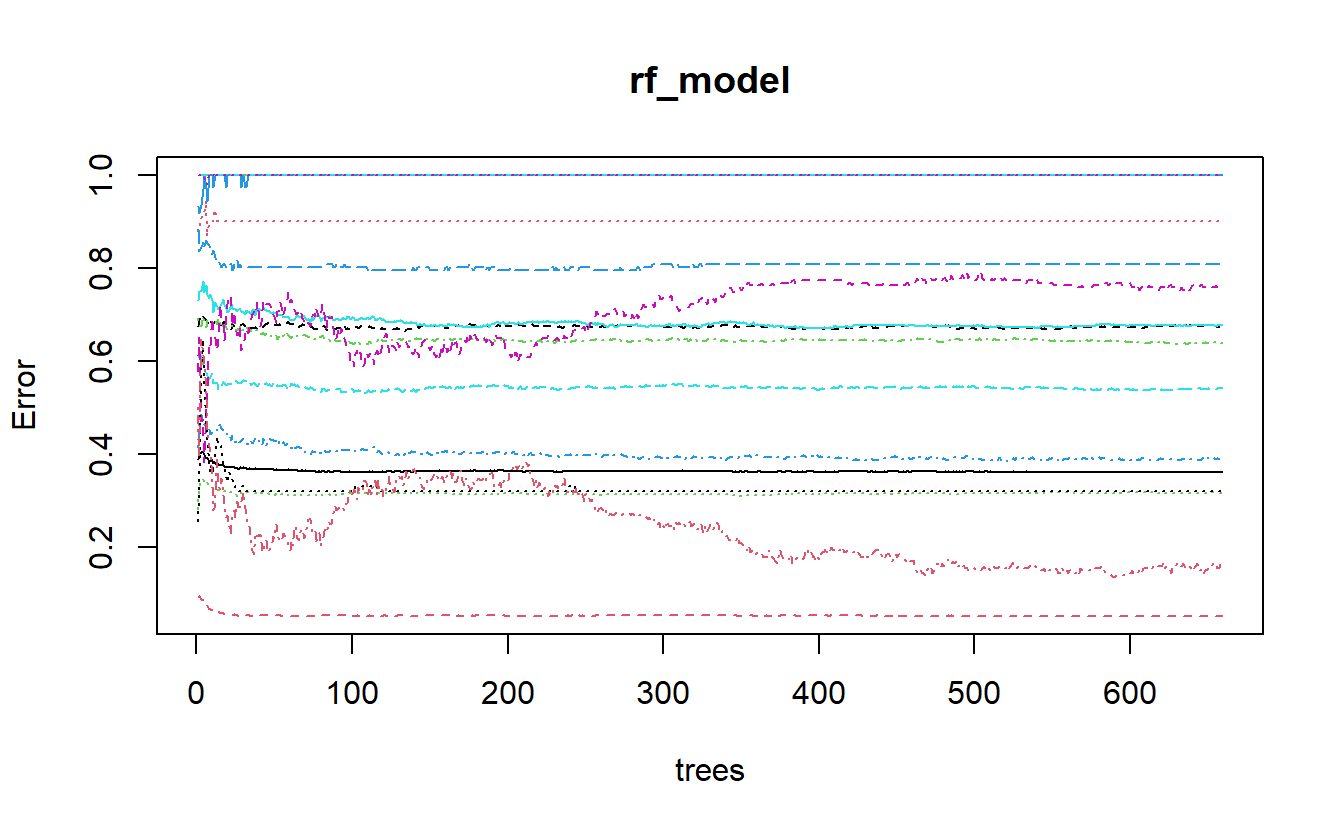
Figure 13.4: plot of randomForest model output
Let’s see if we can figure out what is going on with the above graph. We can extract the err.rate and ntree from the model object and place them in a matplot to match the above graph.
err <- rf_model$err.rate
N_trees <- 1:rf_model$ntree
matplot(N_trees , err, type = 'l', xlab="trees", ylab="Error")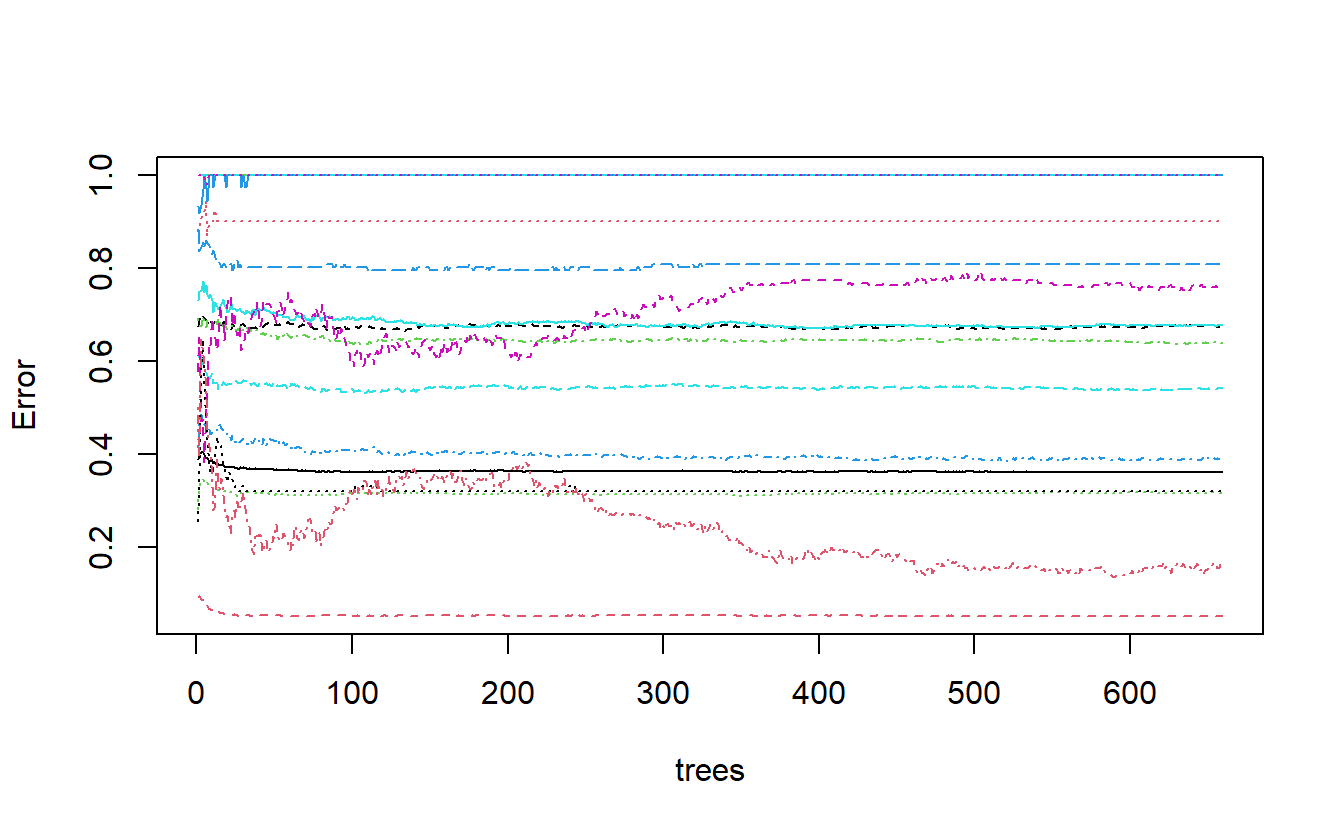
Figure 13.5: derivation of plot of randomForest model output
We can improve this a bit by performing the computations ourselves and keeping track of things:
err2 <- as_tibble(err, rownames='n_trees')
err2 %>%
glimpse()
#> Rows: 659
#> Columns: 19
#> $ n_trees <chr> "1", "2", "3", "4", "5", "6", "7", "8", "9", "10", "11", "12",…
#> $ OOB <dbl> 0.3888985, 0.4010195, 0.4031778, 0.4038520, 0.3991778, 0.39185…
#> $ C_33 <dbl> 0.09125475, 0.09435626, 0.08716876, 0.08636837, 0.08526068, 0.…
#> $ C_34 <dbl> 0.2818713, 0.3272194, 0.3405500, 0.3448990, 0.3397964, 0.33502…
#> $ C_35 <dbl> 0.4140625, 0.4623116, 0.4863732, 0.4646840, 0.4636678, 0.45928…
#> $ C_58 <dbl> 0.6084724, 0.6100235, 0.6091371, 0.5932394, 0.5891270, 0.58295…
#> $ C_59 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
#> $ C_60 <dbl> 0.6745363, 0.6883768, 0.7014563, 0.6958763, 0.6922034, 0.68802…
#> $ C_61 <dbl> 0.8823529, 0.8695652, 0.9062500, 0.9166667, 0.9166667, 0.94444…
#> $ C_62 <dbl> 0.6921986, 0.6770186, 0.6747851, 0.6957352, 0.6849882, 0.67257…
#> $ C_63 <dbl> 0.8837209, 0.8354430, 0.8446602, 0.8547009, 0.8412698, 0.85937…
#> $ C_64 <dbl> 0.7313433, 0.7507692, 0.7500000, 0.7707424, 0.7479508, 0.76247…
#> $ C_65 <dbl> 0.5774648, 0.6504065, 0.4697987, 0.4756098, 0.3815029, 0.47777…
#> $ C_66 <dbl> 0.2564103, 0.3000000, 0.5769231, 0.6470588, 0.5617978, 0.48351…
#> $ C_67 <dbl> 0.5000000, 0.3931624, 0.6466667, 0.6190476, 0.6032609, 0.53968…
#> $ C_68 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
#> $ C_69 <dbl> 0.9333333, 0.9166667, 0.9354839, 0.9696970, 1.0000000, 0.97142…
#> $ C_70 <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
#> $ C_71 <dbl> 1.0000000, 1.0000000, 1.0000000, 1.0000000, 1.0000000, 0.98076…
predicted_levels <- colnames(err2)[!colnames(err2) %in% c('n_trees','OOB')]
predicted_levels
#> [1] "C_33" "C_34" "C_35" "C_58" "C_59" "C_60" "C_61" "C_62" "C_63" "C_64"
#> [11] "C_65" "C_66" "C_67" "C_68" "C_69" "C_70" "C_71"
plot <- err2 %>%
select(-OOB) %>%
pivot_longer(all_of(predicted_levels), names_to = "Class", values_to = "Error") %>%
mutate(N_Trees = as.numeric(n_trees)) %>%
ggplot(aes(x=N_Trees, y=Error, color=Class)) +
geom_point() +
geom_smooth(method = lm, formula = y ~ splines::bs(x, 3), se = FALSE)
plot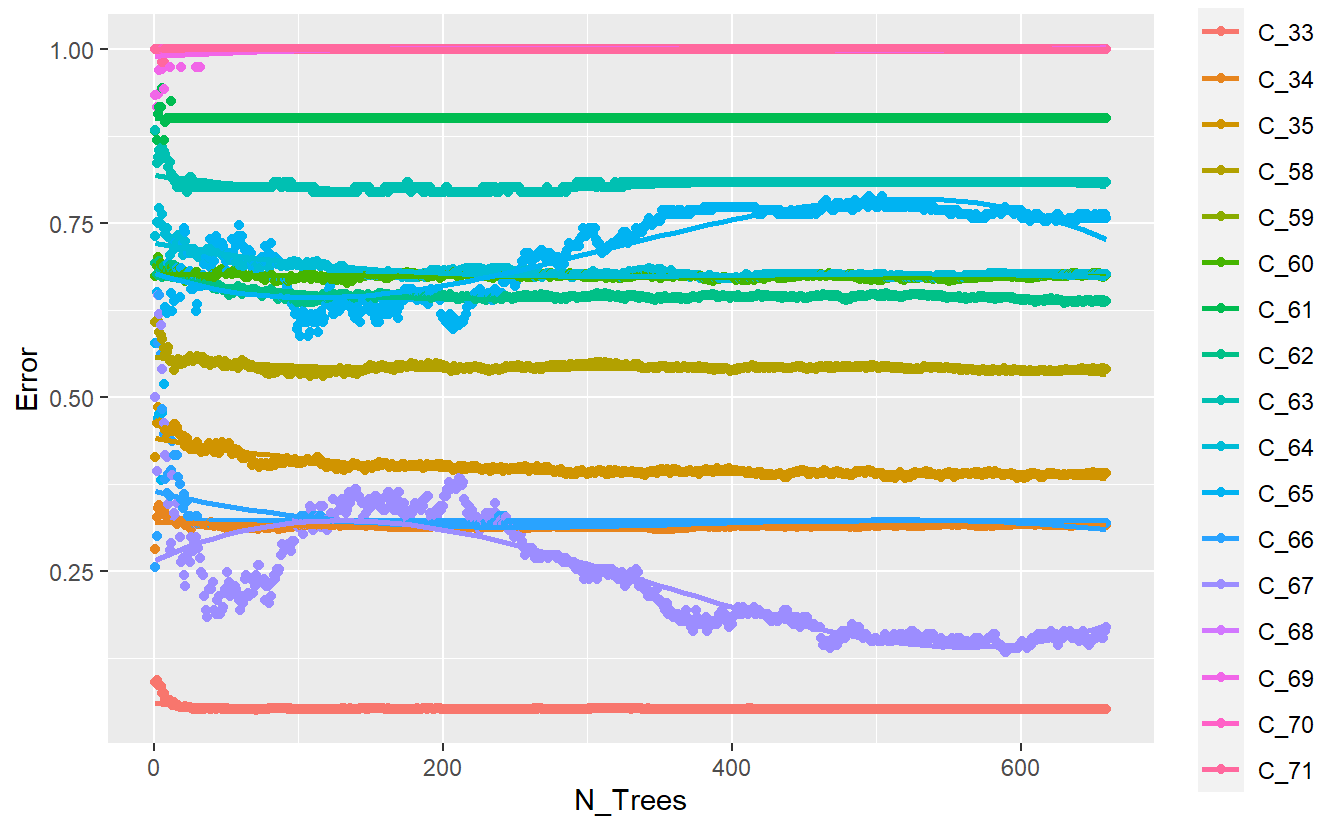
Figure 13.6: Improved plot of base randomForest model output
better_rand_forest_errors <- function(rf_model_obj){
err <- rf_model_obj$err.rate
err2 <- as_tibble(err, rownames='n_trees')
predicted_levels <- colnames(err2)[!colnames(err2) %in% c('n_trees','OOB')]
plot <- err2 %>%
select(-OOB) %>%
pivot_longer(all_of(predicted_levels), names_to = "Class", values_to = "Error") %>%
mutate(N_Trees = as.numeric(n_trees)) %>%
ggplot(aes(x=N_Trees, y=Error, color=Class)) +
geom_point() +
geom_smooth(method = lm, formula = y ~ splines::bs(x, 3), se = FALSE)
return(plot)
}\(~\)
\(~\)
13.8 Evaluate Model Performace
First, we will get the expected classes for the test data:
DATA.test.Scored$pred <- predict(rf_model, DATA.test , 'response')
DATA.test.Scored %>%
glimpse()
#> Rows: 10,464
#> Columns: 15
#> $ Date <chr> "04-21-1991", "04-21-1991", "04-22-1991", "04-22-1991…
#> $ Time <chr> "9:09", "9:09", "7:35", "7:35", "7:25", "7:29", "7:29…
#> $ Code <dbl> 58, 34, 58, 34, 58, 58, 33, 34, 33, 62, 33, 62, 33, 3…
#> $ Value <chr> "100", "013", "216", "013", "257", "067", "009", "014…
#> $ file_no <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
#> $ Date_Time_Char <chr> "04-21-1991 9:09", "04-21-1991 9:09", "04-22-1991 7:3…
#> $ Date_Time <dttm> 1991-04-21 09:09:00, 1991-04-21 09:09:00, 1991-04-22…
#> $ Value_Num <dbl> 100, 13, 216, 13, 257, 67, 9, 14, 2, 228, 7, 256, 8, …
#> $ Record_ID <int> 1, 3, 7, 9, 13, 25, 26, 27, 32, 37, 38, 42, 43, 45, 5…
#> $ Code_Description <chr> "Pre-breakfast blood glucose measurement", "NPH insul…
#> $ Code_Invalid <lgl> FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALSE, FALS…
#> $ Code_New <dbl> 58, 34, 58, 34, 58, 58, 33, 34, 33, 62, 33, 62, 33, 3…
#> $ Code_Factor <fct> C_58, C_34, C_58, C_34, C_58, C_58, C_33, C_34, C_33,…
#> $ model <chr> "base", "base", "base", "base", "base", "base", "base…
#> $ pred <fct> C_60, C_35, C_58, C_35, C_62, C_63, C_33, C_33, C_33,…
library('yardstick')
#> For binary classification, the first factor level is assumed to be the event.
#> Use the argument `event_level = "second"` to alter this as needed.
#>
#> Attaching package: 'yardstick'
#> The following object is masked from 'package:readr':
#>
#> spec
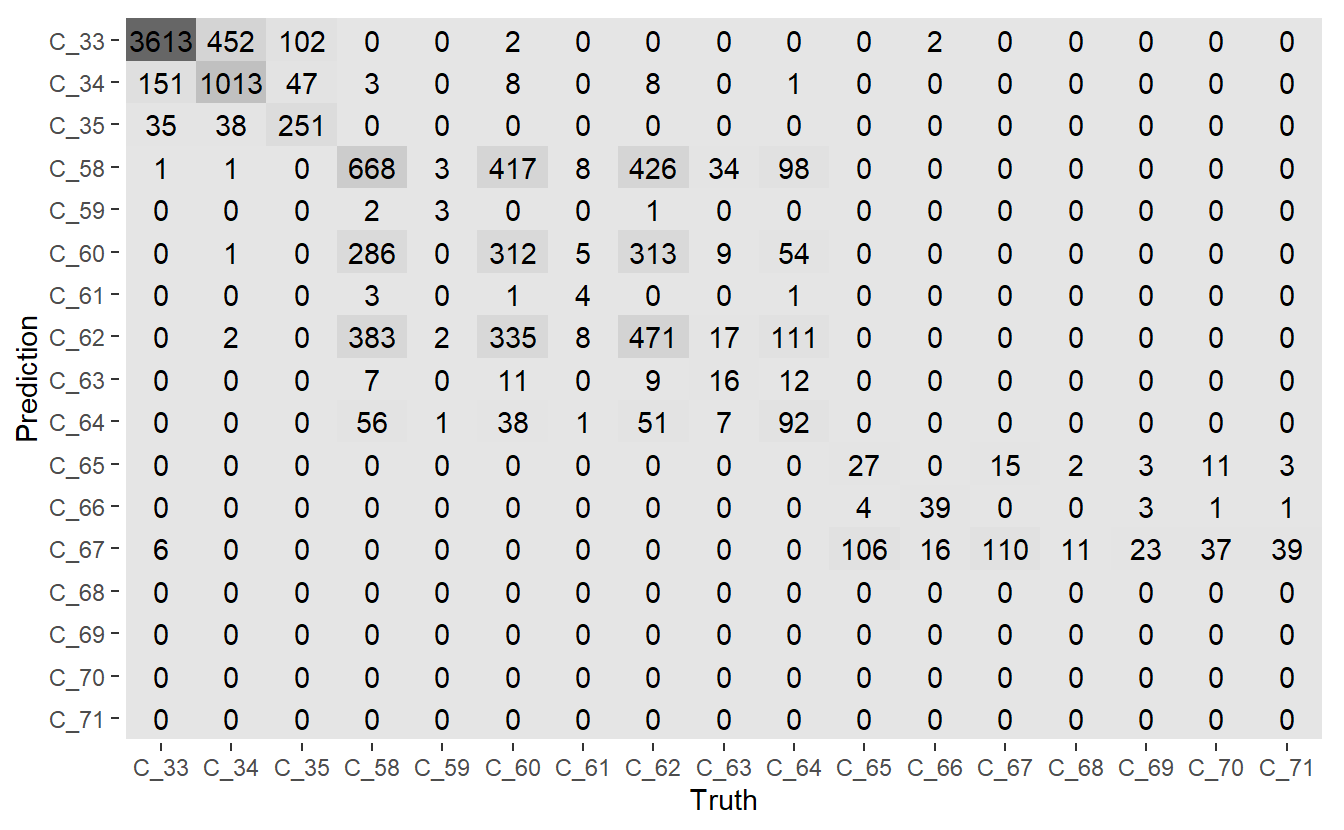
conf_mat.DATA.test.Scored <- conf_mat(DATA.test.Scored,
truth = Code_Factor, pred)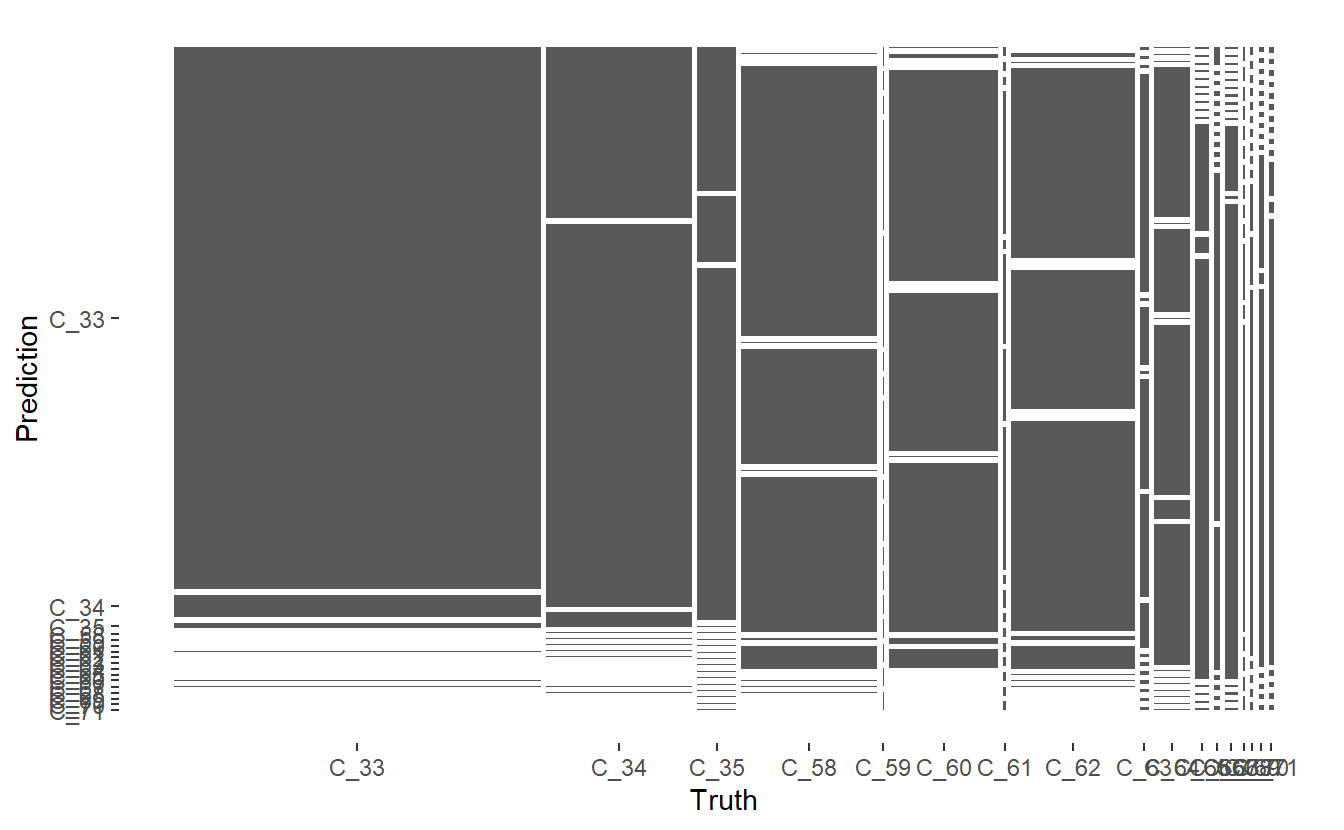
Figure 13.7: Mosaic Confusion Matrix of Base Model
DATA.test.Scored %>%
conf_mat(truth= Code_Factor, pred) %>%
summary() %>%
ggplot(aes(x=.metric, y=.estimate, fill= .metric)) +
geom_bar(stat='identity', position = 'dodge') +
coord_flip()
#> Warning: While computing multiclass `precision()`, some levels had no predicted events (i.e. `true_positive + false_positive = 0`).
#> Precision is undefined in this case, and those levels will be removed from the averaged result.
#> Note that the following number of true events actually occured for each problematic event level:
#> 'C_68': 13
#> 'C_69': 29
#> 'C_70': 49
#> 'C_71': 43
#> While computing multiclass `precision()`, some levels had no predicted events (i.e. `true_positive + false_positive = 0`).
#> Precision is undefined in this case, and those levels will be removed from the averaged result.
#> Note that the following number of true events actually occured for each problematic event level:
#> 'C_68': 13
#> 'C_69': 29
#> 'C_70': 49
#> 'C_71': 43
#> Warning: Removed 1 rows containing missing values (`geom_bar()`).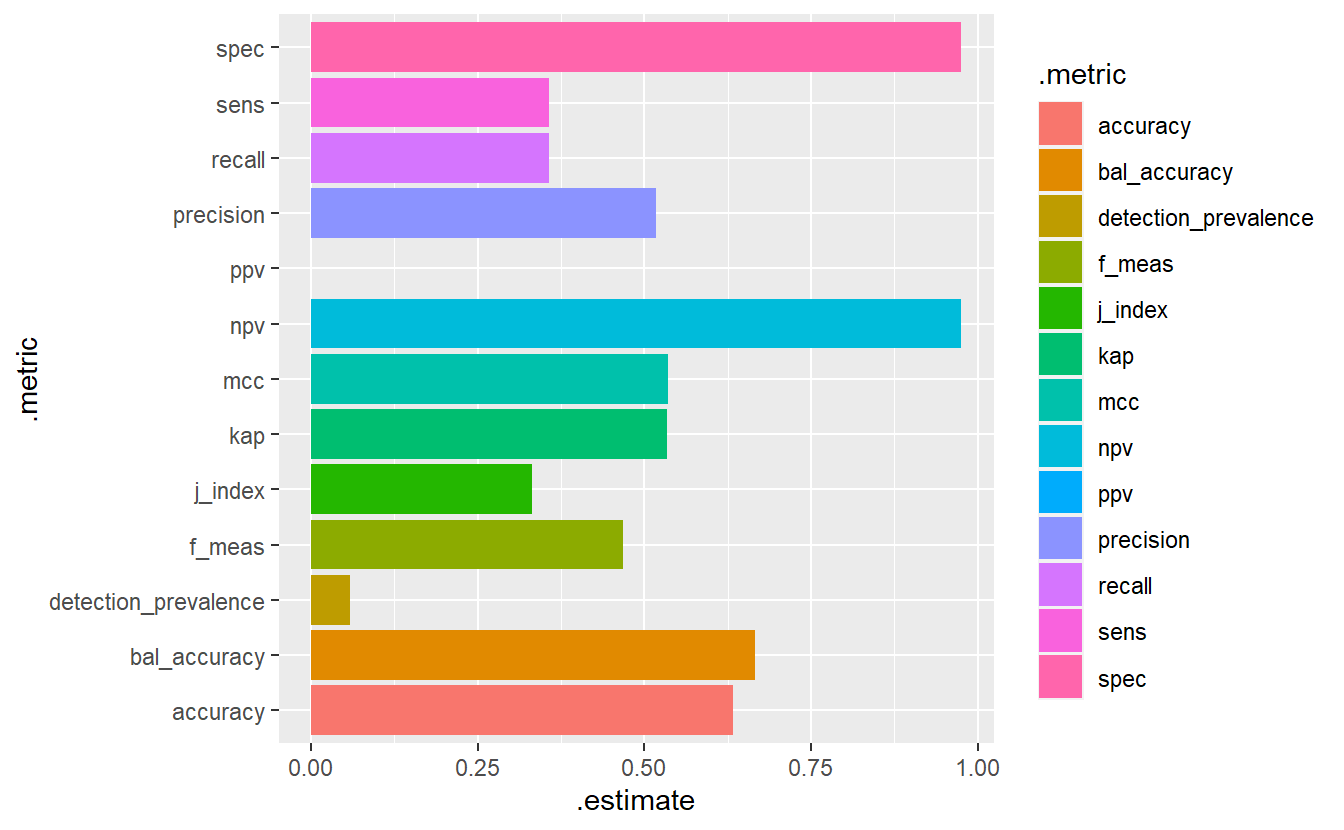
(#fig:5-mm-1 )Model Evaluation Metrics - Base Model
DATA.test.Scored %>%
mutate(Predict_Correct = if_else(Code_Factor == pred, TRUE, FALSE)) %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=Predict_Correct)) +
geom_point() 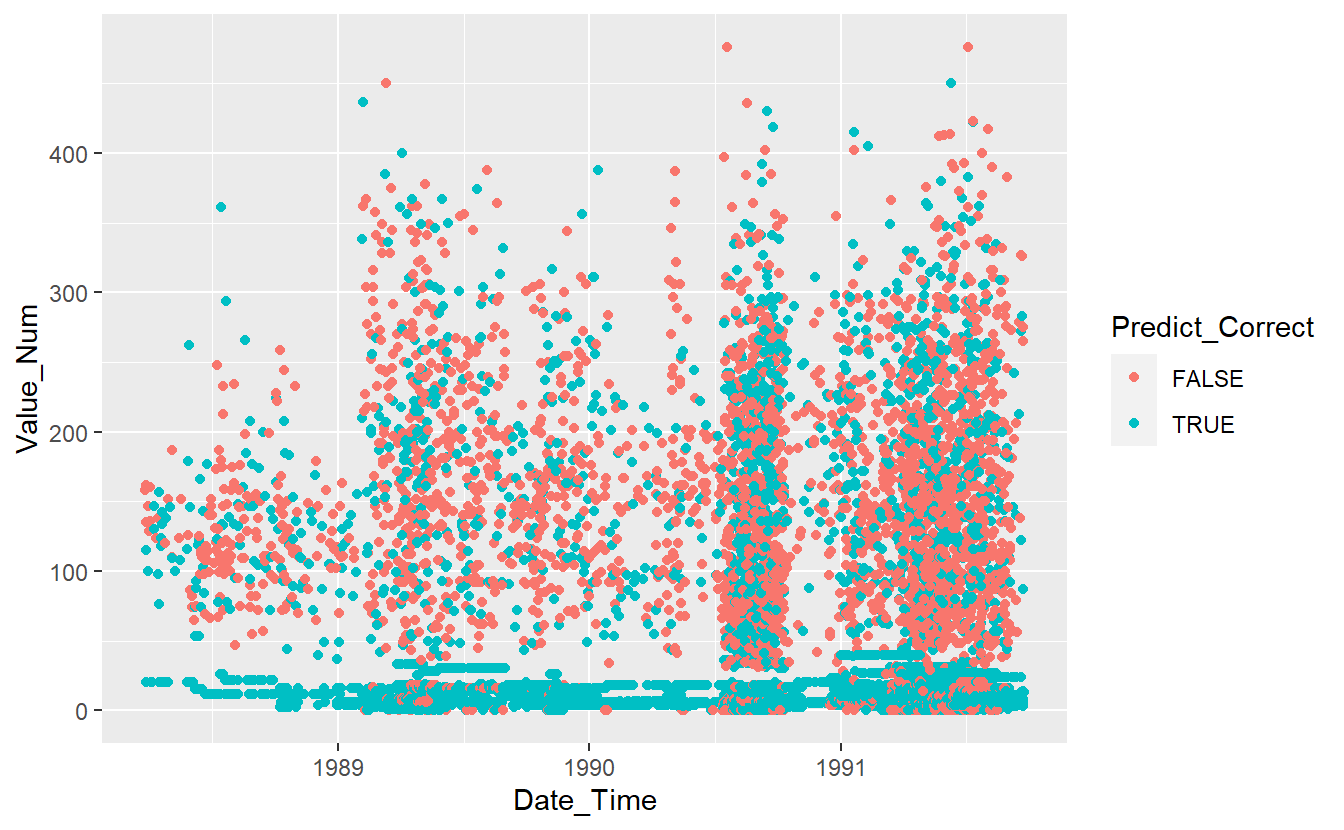
Figure 13.8: Graph of Correct Predictions on test data under base model
\(~\)
\(~\)
13.10 Make Adjustments
Our previous model has some issues, which we can attempt to correct.
Code_Table_Factor_2 <- Code_Table_Factor %>%
mutate(Code_Description_2 = case_when(
Code %in% c(33, 34, 35) ~ "insulin dose",
Code %in% c(58, 60, 62, 64) ~ "pre-meal",
Code %in% c(59, 61, 63) ~ "post-meal",
Code %in% c(66, 67, 68, 65) ~ "meal ingestion / Hypoglycemic",
Code %in% c(69, 70, 71) ~ "exercise activity")
) %>%
mutate(Code_Description_2 = factor(Code_Description_2)) %>%
select(Code, Code_Description_2)
Code_Table_Factor_2
#> # A tibble: 17 × 2
#> Code Code_Description_2
#> <dbl> <fct>
#> 1 33 insulin dose
#> 2 34 insulin dose
#> 3 35 insulin dose
#> 4 58 pre-meal
#> 5 59 post-meal
#> 6 60 pre-meal
#> # … with 11 more rows
DATA.train_2 <- DATA.train %>% left_join(Code_Table_Factor_2)
#> Joining, by = "Code"
rf_model_2 <- randomForest(Code_Description_2 ~ Date_Time + Value_Num,
data = DATA.train_2,
ntree = 659,
importance = TRUE)
better_rand_forest_errors(rf_model_2)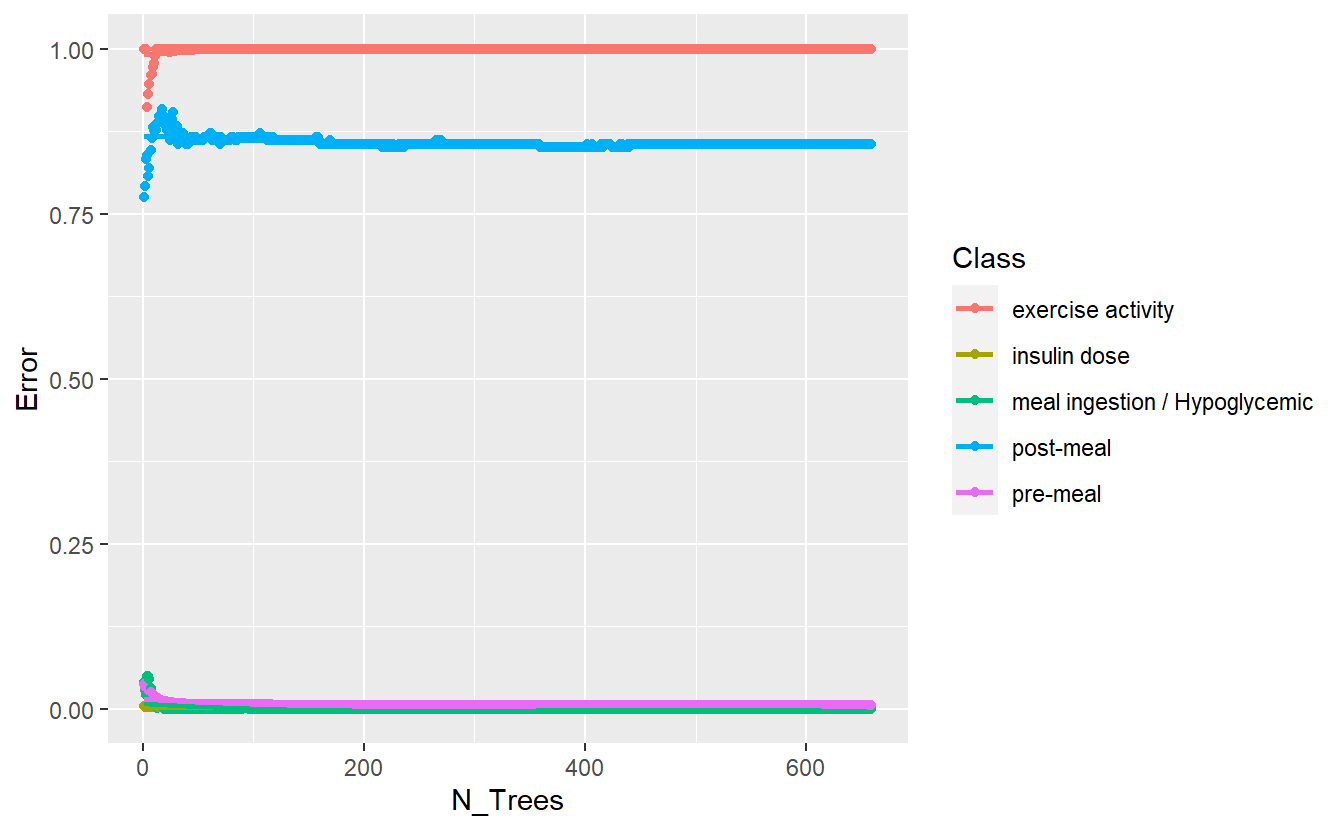
(#fig:5-better-rand-errors-rf-2 )Training Errors on New Model
DATA.test.Scored2 <- DATA.test %>%
left_join(Code_Table_Factor_2) %>%
mutate(model = 'new')
#> Joining, by = "Code"
DATA.test.Scored2$pred <- predict(rf_model_2, DATA.test.Scored2 , 'response')
conf_mat.DATA.test.Scored2 <- DATA.test.Scored2 %>%
conf_mat(truth = Code_Description_2, pred)
conf_mat.DATA.test.Scored2 %>%
autoplot('heatmap')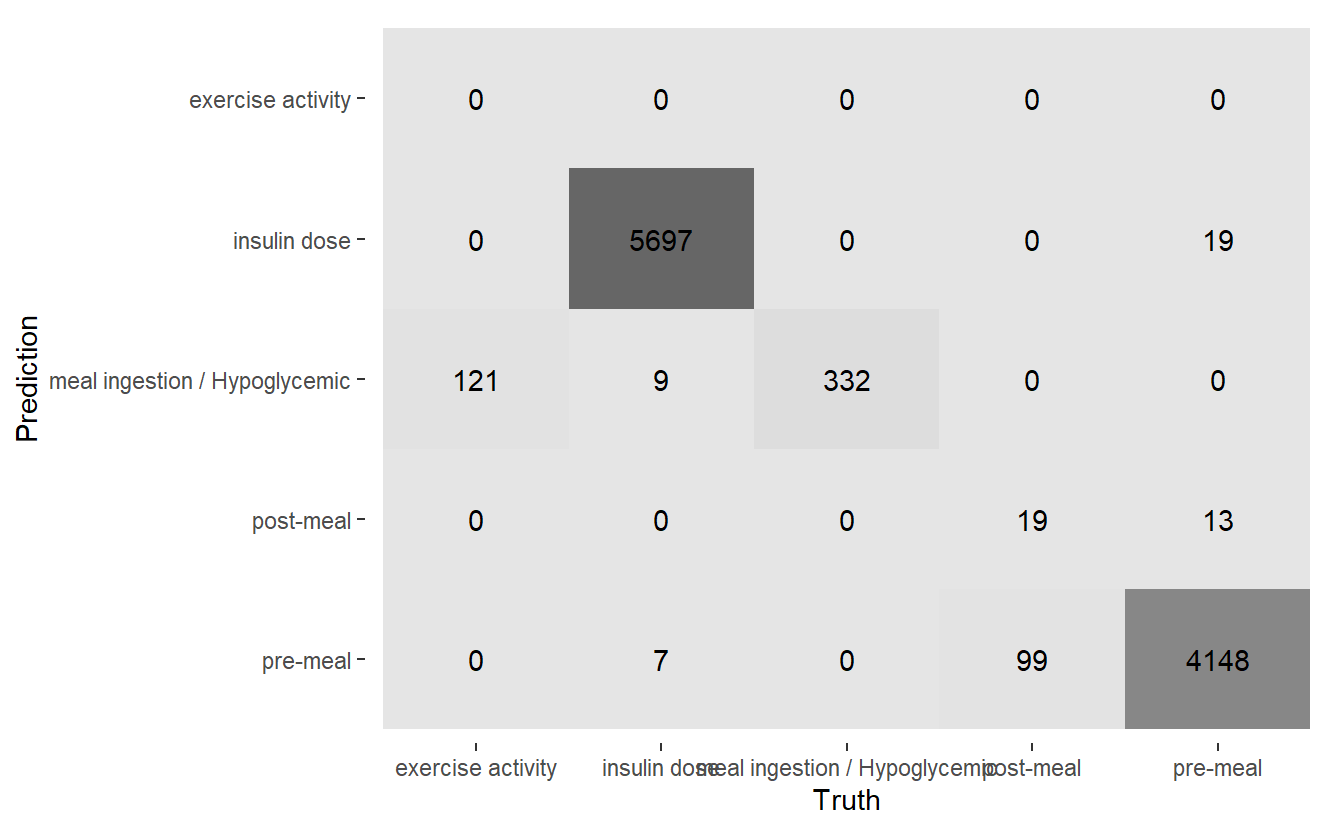
(#fig:5-heatmap-conf-mat-rf-2 )Heatmap Confusion Matrix of New Model
saveRDS(rf_model_2, 'DATA/Part_5/models/rf_model_2.RDS')
base <- DATA.test.Scored %>% conf_mat(truth= Code_Factor, pred) %>% summary() %>% mutate(model = 'base')
#> Warning: While computing multiclass `precision()`, some levels had no predicted events (i.e. `true_positive + false_positive = 0`).
#> Precision is undefined in this case, and those levels will be removed from the averaged result.
#> Note that the following number of true events actually occured for each problematic event level:
#> 'C_68': 13
#> 'C_69': 29
#> 'C_70': 49
#> 'C_71': 43
#> While computing multiclass `precision()`, some levels had no predicted events (i.e. `true_positive + false_positive = 0`).
#> Precision is undefined in this case, and those levels will be removed from the averaged result.
#> Note that the following number of true events actually occured for each problematic event level:
#> 'C_68': 13
#> 'C_69': 29
#> 'C_70': 49
#> 'C_71': 43
new <- DATA.test.Scored2 %>% conf_mat(truth= Code_Description_2, pred) %>% summary() %>% mutate(model = 'new')
#> Warning: While computing multiclass `precision()`, some levels had no predicted events (i.e. `true_positive + false_positive = 0`).
#> Precision is undefined in this case, and those levels will be removed from the averaged result.
#> Note that the following number of true events actually occured for each problematic event level:
#> 'exercise activity': 121
#> Warning: While computing multiclass `precision()`, some levels had no predicted events (i.e. `true_positive + false_positive = 0`).
#> Precision is undefined in this case, and those levels will be removed from the averaged result.
#> Note that the following number of true events actually occured for each problematic event level:
#> 'exercise activity': 121
comp <- bind_rows(base,
new)
comp %>%
ggplot(aes(x=model, y=.estimate, fill= model)) +
geom_bar(stat='identity', position = 'dodge') +
coord_flip() +
facet_wrap(.metric ~ .)
#> Warning: Removed 2 rows containing missing values (`geom_bar()`).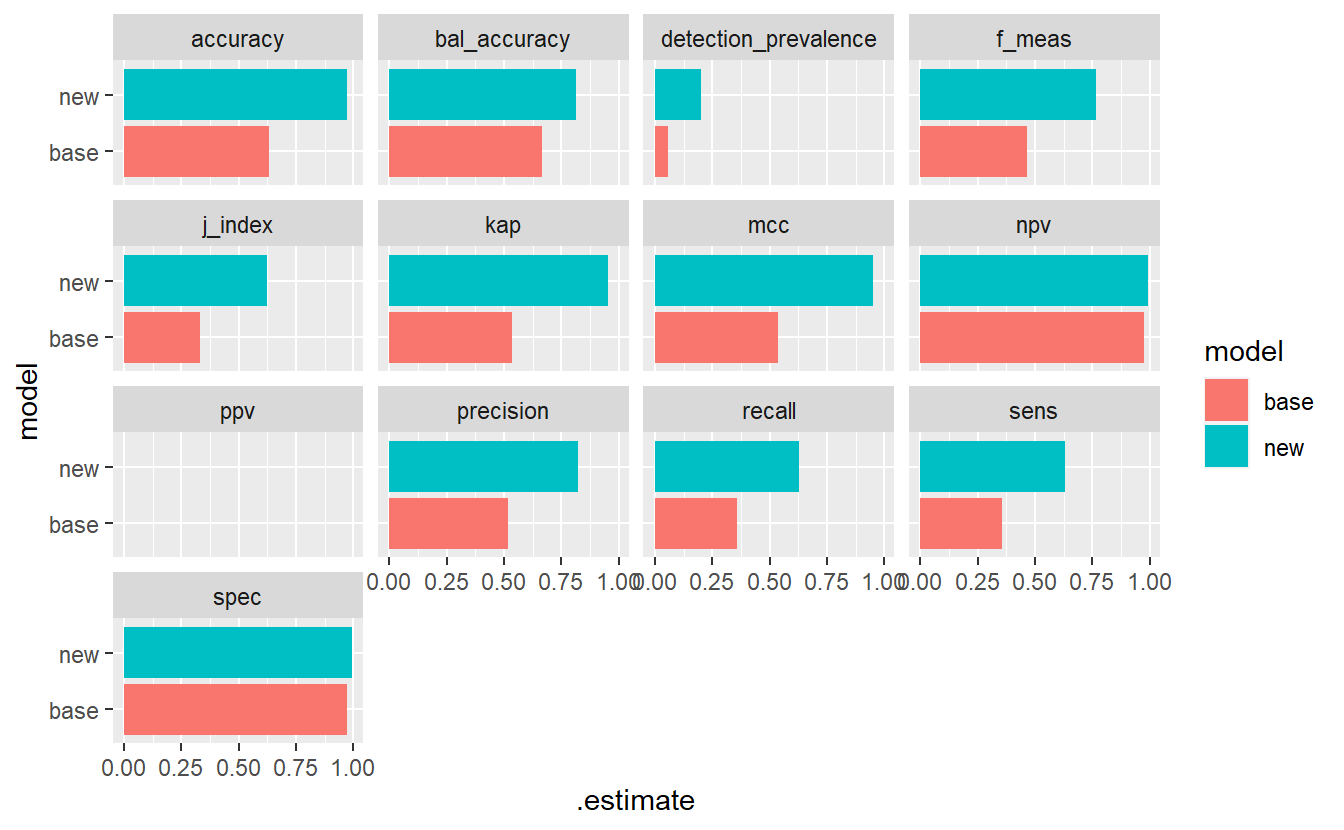
(#fig:5-compare-rf12 )Model Evaluation Metrics - base Vs new model
13.11 Make Adjustments, Again
Let’s make another pass at this, this time we will group together
Code_Table_Factor_3 <- Code_Table_Factor %>%
mutate(Code_Description_3 = case_when(
Code %in% c(33, 34, 35) ~ "insulin dose",
Code %in% c(58, 60, 62, 64, 59, 61, 63) ~ "pre / post meal",
Code %in% c(66, 67, 68, 65, 69, 70, 71) ~ "meal ingestion / Hypoglycemic / exercise activity")) %>%
mutate(Code_Description_3 = factor(Code_Description_3)) %>%
select(Code, Code_Description_3)
Code_Table_Factor_3
#> # A tibble: 17 × 2
#> Code Code_Description_3
#> <dbl> <fct>
#> 1 33 insulin dose
#> 2 34 insulin dose
#> 3 35 insulin dose
#> 4 58 pre / post meal
#> 5 59 pre / post meal
#> 6 60 pre / post meal
#> # … with 11 more rows
DATA.train_3 <- DATA.train %>% left_join(Code_Table_Factor_3)
#> Joining, by = "Code"
rf_model_3 <- randomForest(Code_Description_3 ~ Date_Time + Value_Num,
data = DATA.train_3,
ntree = 659,
importance = TRUE)
rm(DATA.train_3)
better_rand_forest_errors(rf_model_3)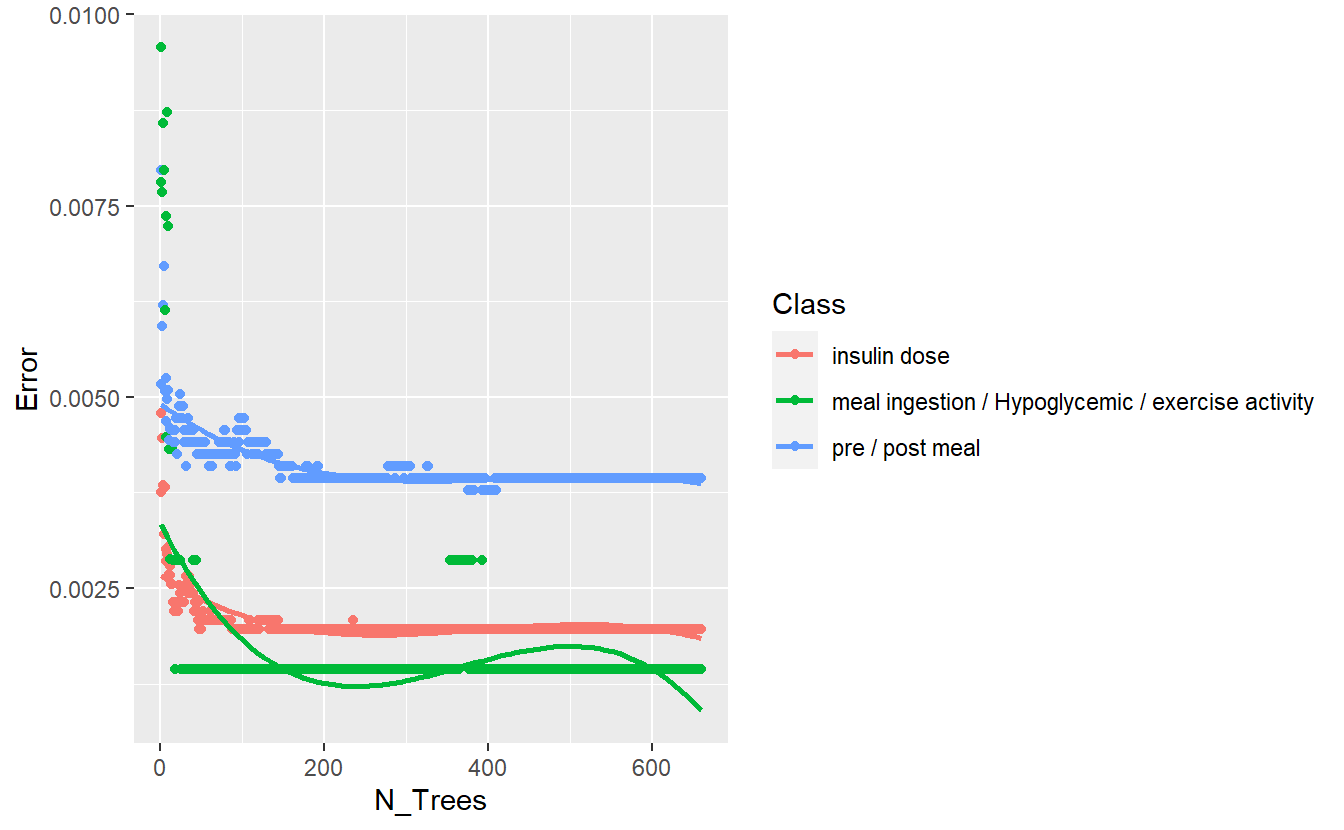
Figure 13.9: Training Errors of Final Model
DATA.test.Scored3 <- DATA.test %>%
left_join(Code_Table_Factor_3) %>%
mutate(model = 'final')
#> Joining, by = "Code"
DATA.test.Scored3$pred <- predict(rf_model_3, DATA.test.Scored3 , 'response')
conf_mat.DATA.test.Scored3 <- DATA.test.Scored3 %>%
conf_mat(truth = Code_Description_3, pred)
conf_mat.DATA.test.Scored3 %>%
autoplot('heatmap')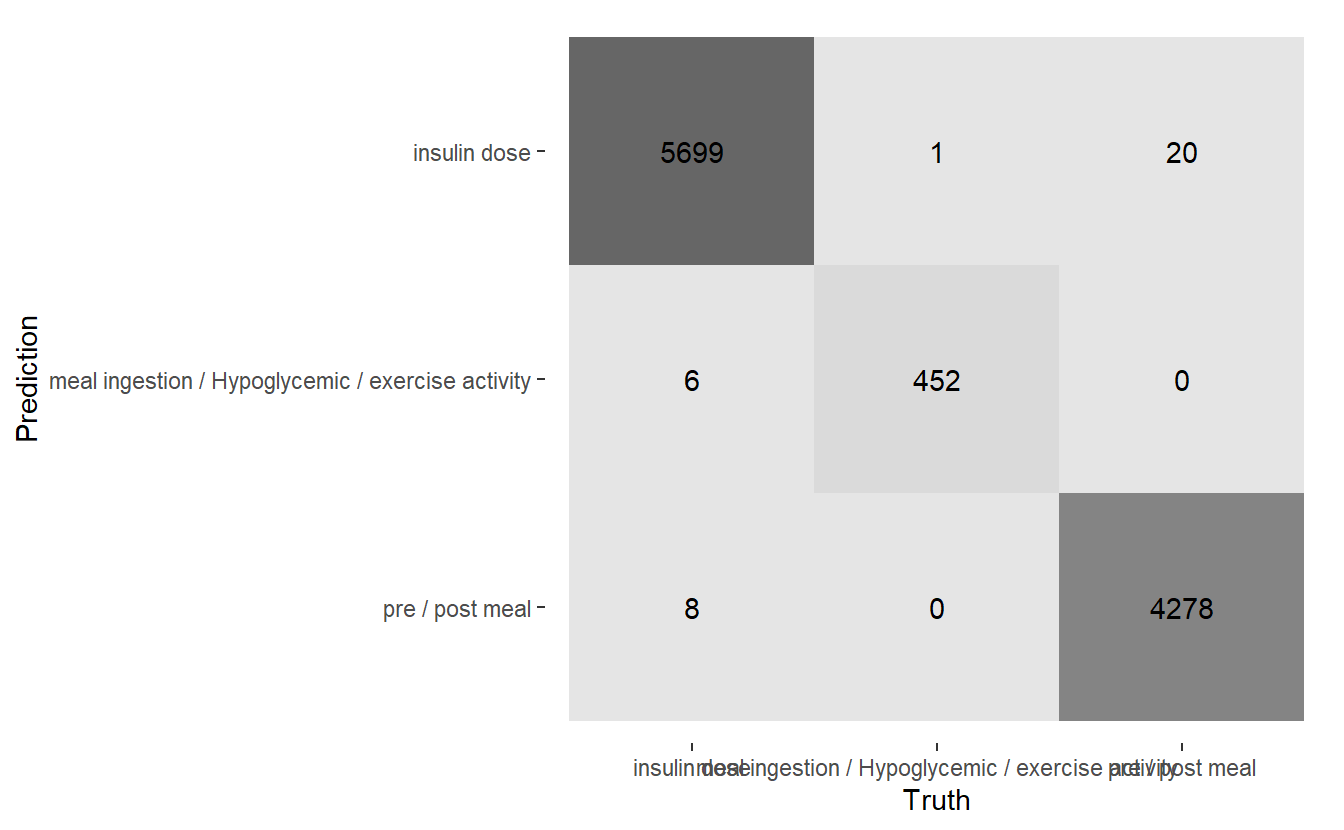
(#fig:5-heatmap-cf-model-rf3 )Heatmap Confusion Matrix of Final Model
13.12 Extract More Data
library('ggplot2')
library('ggdendro')
library('tree')
#>
#> Attaching package: 'tree'
#> The following object is masked from 'package:igraph':
#>
#> tree
tree_1 <- tree::tree(Code_Factor ~ Date_Time + Value_Num,
data = DATA.train)
#> Warning in tree::tree(Code_Factor ~ Date_Time + Value_Num, data = DATA.train):
#> NAs introduced by coercion
plot_dendro_tree <- function(model){
tree_data <- dendro_data(model)
p <- segment(tree_data) %>%
ggplot() +
geom_segment(aes(x = x,
y = y,
xend = xend,
yend = yend,
size = n),
colour = "blue", alpha = 0.5) +
scale_size("n") +
geom_text(data = label(tree_data),
aes(x = x, y = y, label = label), vjust = -0.5, size = 3) +
geom_text(data = leaf_label(tree_data),
aes(x = x, y = y, label = label), vjust = 0.5, size = 2) +
theme_dendro()
p
}
plot_dendro_tree(tree_1)
#> Warning: Using `size` aesthetic for lines was deprecated in ggplot2 3.4.0.
#> ℹ Please use `linewidth` instead.
tree_2 <- tree::tree(Code_Factor ~ Date_Time + Value_Num + h + day + month ,
data = DATA.train_2 %>%
mutate(h = factor(lubridate::hour(Date_Time))) %>%
select(Code_Factor, Date_Time, Value_Num, h) %>%
mutate(day = lubridate::day(Date_Time)) %>%
mutate(month = month(Date_Time, label = TRUE))
)
#> Warning in tree::tree(Code_Factor ~ Date_Time + Value_Num + h + day + month, :
#> NAs introduced by coercion
plot_dendro_tree(tree_2)
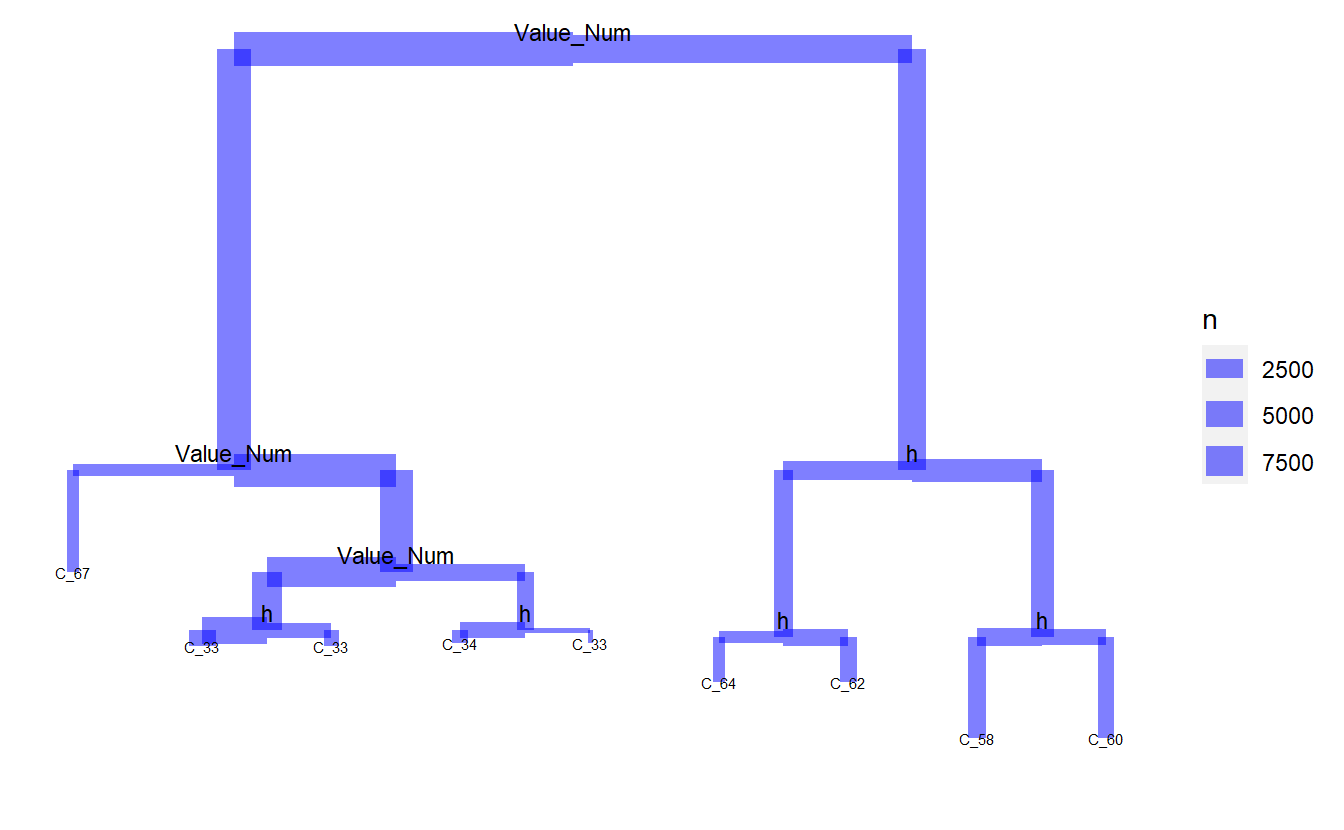
rf_model_4 <- randomForest(Code_Description_2 ~ Date_Time + Value_Num + h + day + month,
data = DATA.train_2 %>%
mutate(h = factor(lubridate::hour(Date_Time))) %>%
select(Code_Description_2, Code_Factor, Date_Time, Value_Num, h) %>%
mutate(day = lubridate::day(Date_Time)) %>%
mutate(month = month(Date_Time, label = TRUE)),
ntree = 659,
importance = TRUE)
better_rand_forest_errors(rf_model_4)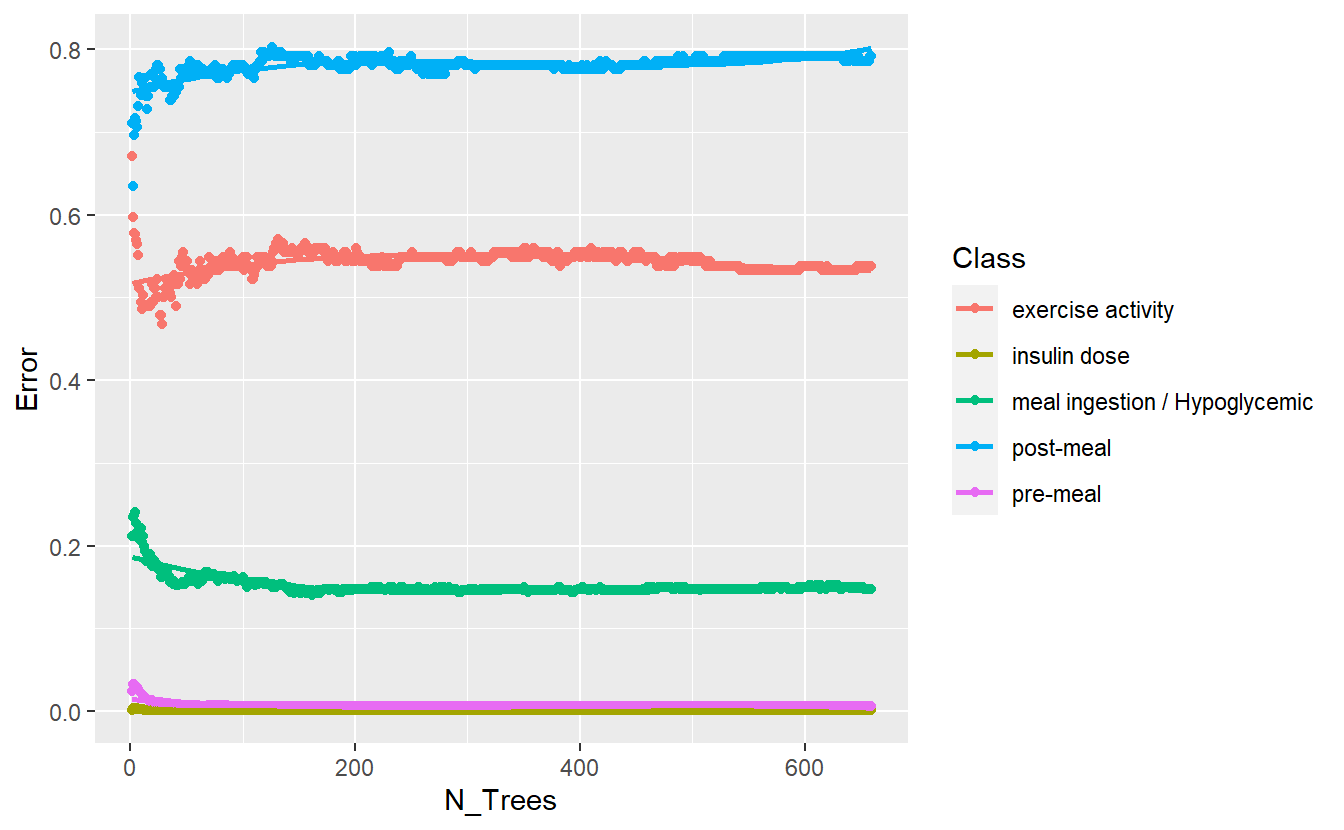
DATA.test.Scored4 <- DATA.test %>%
left_join(Code_Table_Factor_2) %>%
mutate(model = 'final') %>%
mutate(h = factor(lubridate::hour(Date_Time))) %>%
mutate(day = lubridate::day(Date_Time)) %>%
mutate(month = month(Date_Time, label = TRUE)) %>%
select(Code_Description_2, Code_Factor, Date_Time, Value_Num, day, month, h)
#> Joining, by = "Code"
DATA.test.Scored4$pred <- predict(rf_model_4, DATA.test.Scored4 , 'response')
conf_mat.DATA.test.Scored4 <- DATA.test.Scored4 %>%
conf_mat(truth = Code_Description_2, pred)
slim <- conf_mat.DATA.test.Scored3 %>% summary() %>% mutate(model = 'slim')
final <- conf_mat.DATA.test.Scored4 %>% summary() %>% mutate(model = 'final')
comp_f <- bind_rows(comp,
slim,
final)
comp_f %>%
ggplot(aes(x=model, y=.estimate, fill = model)) +
geom_bar(stat='identity', position = 'dodge') +
coord_flip() +
facet_wrap(.metric ~ .)
#> Warning: Removed 2 rows containing missing values (`geom_bar()`).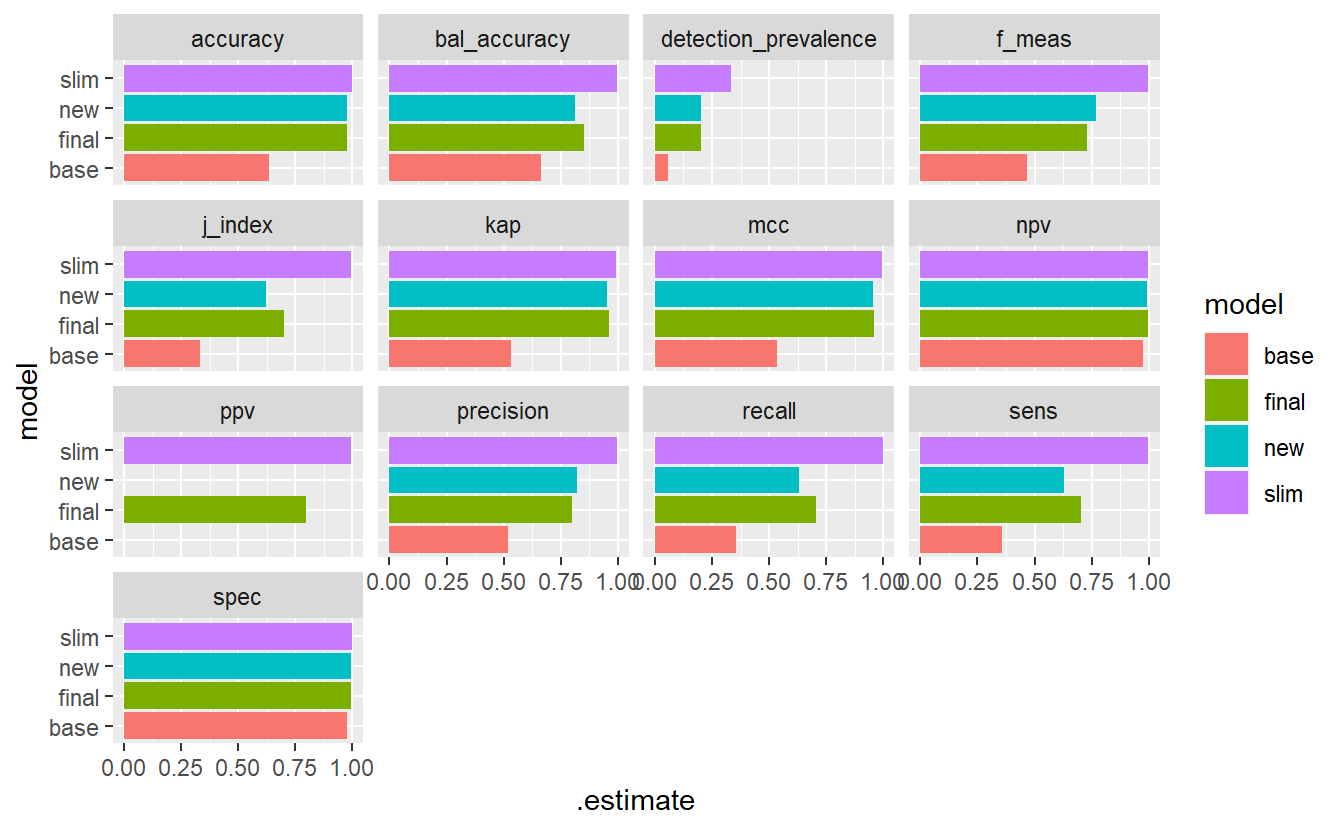
Figure 13.10: Model Evaluation Metrics - base Vs new model Vs Final
13.13 Predict UnKnown Data
keep = c('UnKnown_Measurements','rf_model','rf_model_2','rf_model_3','rf_model_4','keep')
rm(list = setdiff(ls(), keep))
rf_model <- readRDS('DATA/Part_5/models/rf_model.RDS')
rf_model_2 <- readRDS('DATA/Part_5/models/rf_model_2.RDS')
rf_model_3 <- readRDS('DATA/Part_5/models/rf_model_3.RDS')
UnKnown_Measurements$pred_base <- predict(rf_model, UnKnown_Measurements , 'response')
UnKnown_Measurements$pred_new <- predict(rf_model_2, UnKnown_Measurements , 'response')
UnKnown_Measurements$pred_slim <- predict(rf_model_3, UnKnown_Measurements , 'response')
UnKnown_Measurements <- UnKnown_Measurements %>%
mutate(h = factor(lubridate::hour(Date_Time))) %>%
mutate(day = lubridate::day(Date_Time)) %>%
mutate(month = month(Date_Time, label = TRUE))
UnKnown_Measurements$pred_final <- predict(rf_model_4, UnKnown_Measurements, 'response')
UnKnown_Measurements %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=pred_base)) +
geom_point()
#> Warning: Removed 86 rows containing missing values (`geom_point()`).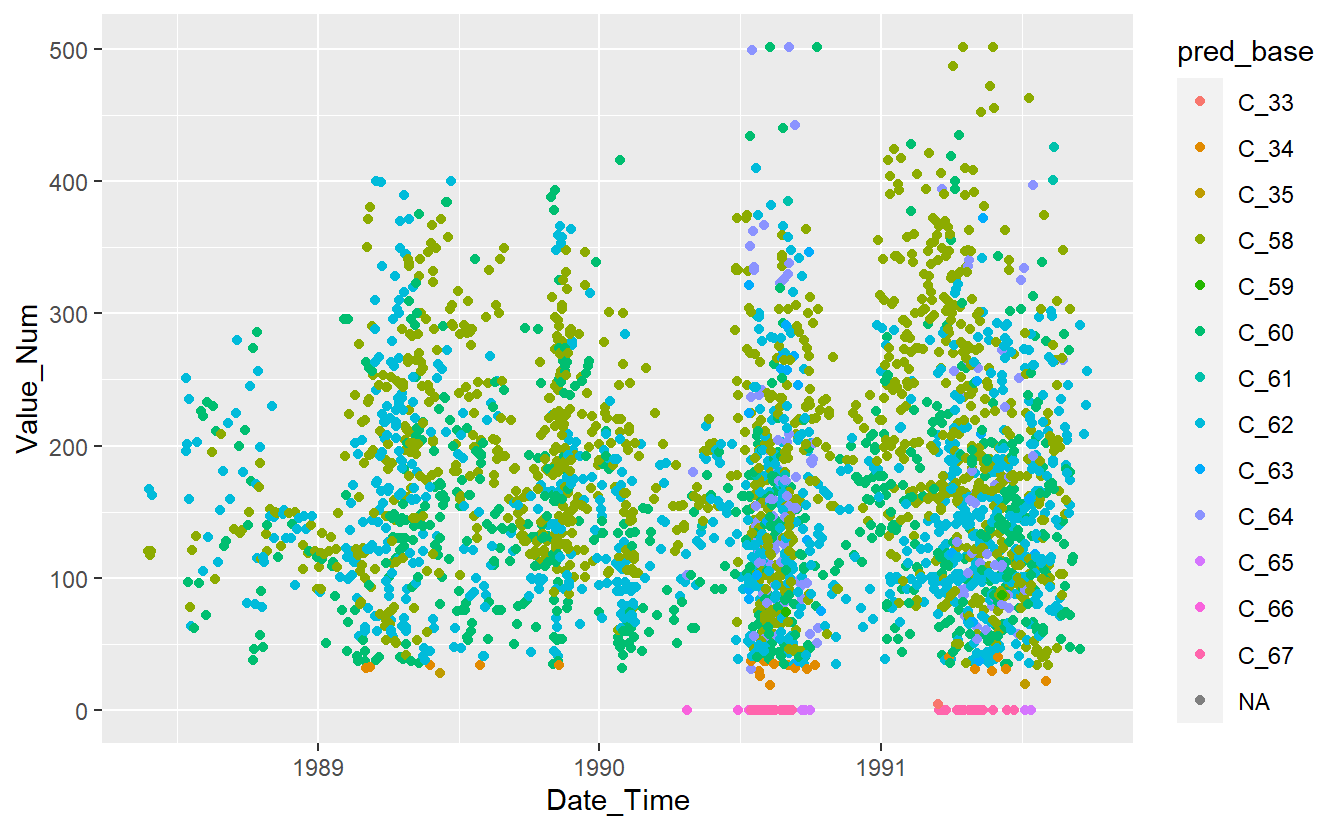
Figure 13.11: Preject UnKnown Data with Base Model
UnKnown_Measurements %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=pred_new)) +
geom_point()
#> Warning: Removed 86 rows containing missing values (`geom_point()`).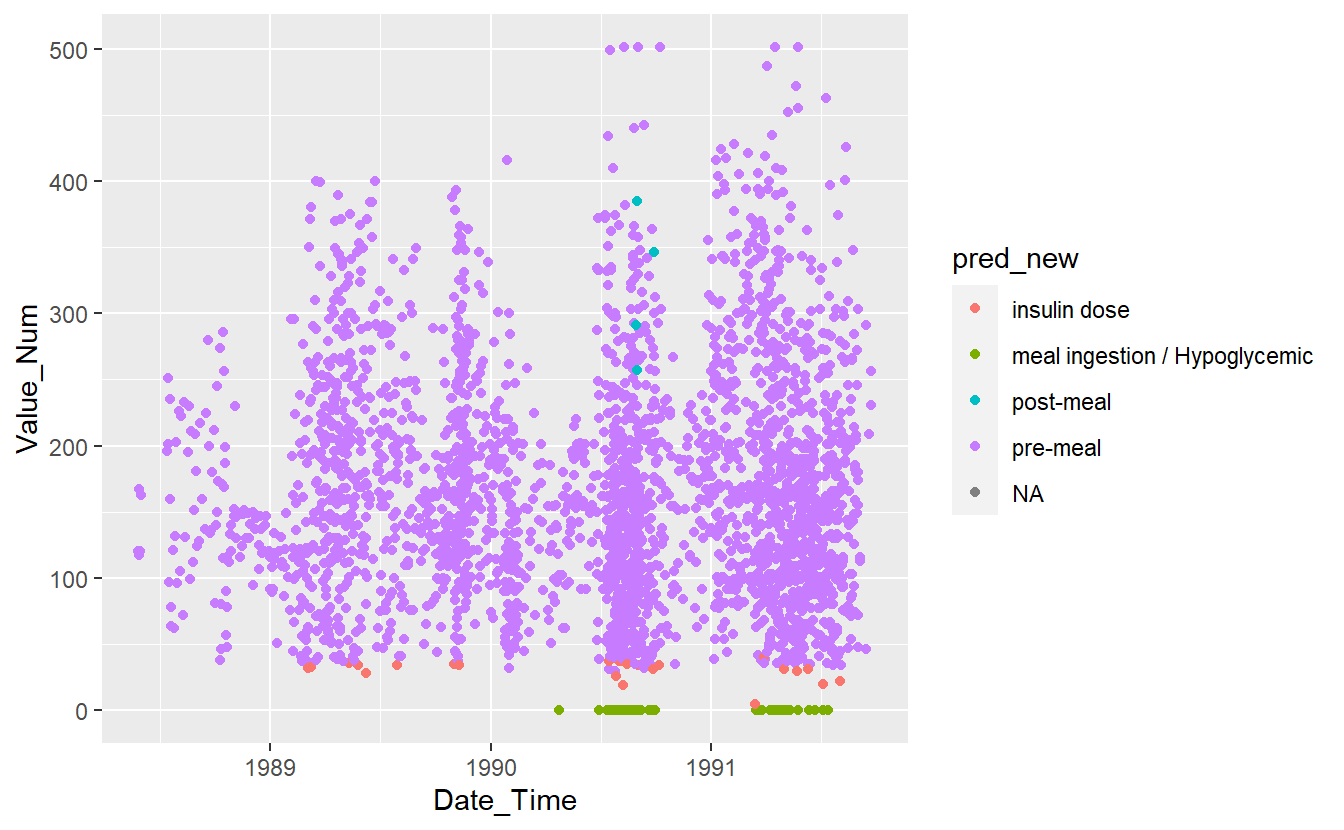
Figure 13.12: Preject UnKnown Data with New Model
UnKnown_Measurements %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=pred_slim)) +
geom_point()
#> Warning: Removed 86 rows containing missing values (`geom_point()`).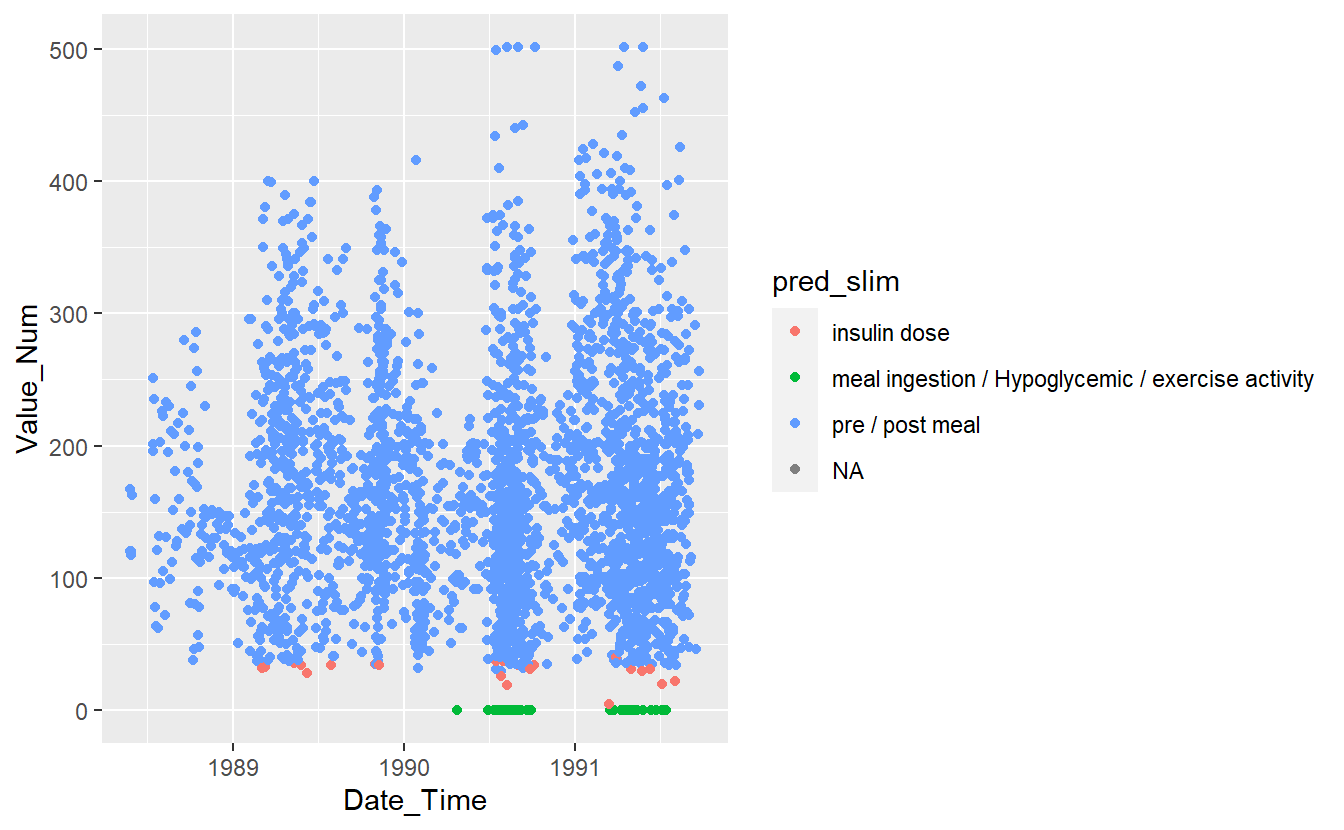
Figure 13.13: Preject UnKnown Data with slim Model
UnKnown_Measurements %>%
ggplot(aes(x=Date_Time, y=Value_Num, color=pred_final)) +
geom_point()
#> Warning: Removed 86 rows containing missing values (`geom_point()`).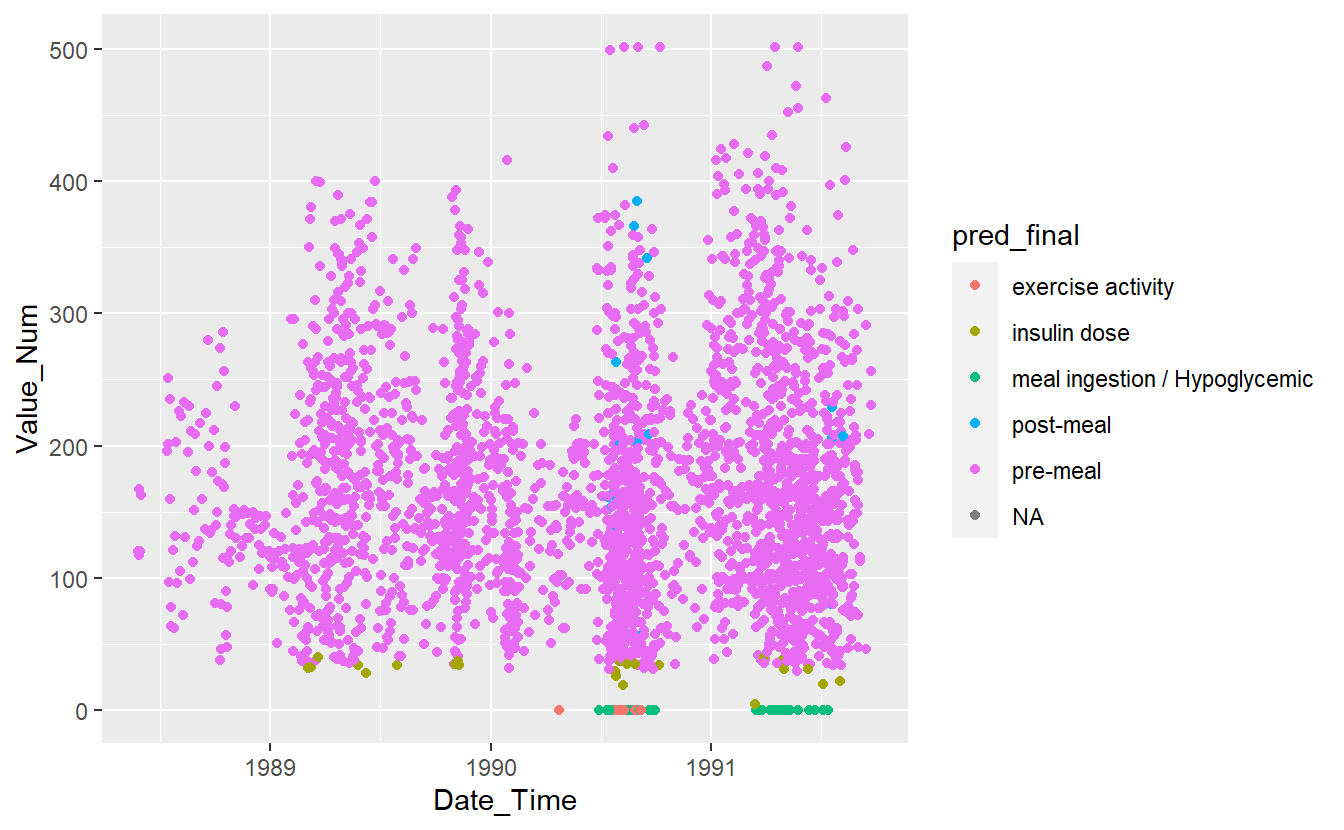
Figure 13.14: Preject UnKnown Data with Final Model