9 Curve fitting, with examples from Red Clover Valley carbon flux and gullies
This section will explore various curve fitting methods, starting with some carbon flux tower data from Red Clover Valley. There’s not a lot of interpretation, as this is basically a sandbox for now…
9.1 Red Clover Valley flux data
- Initial data read and time series charting
Some ideas for processing are from Mason Kealy. Data are from Andrew Oliphant.
9.1.1 Reading in the Excel File
First create a function to convert the first two rows into individual variables. We won’t include the units.
read_excel2 <- function(xl){
vnames <- read_excel(xl, n_max=0) |> names()
vunits <- read_excel(xl, n_max=0, skip=1) |> names()
read_excel(xl, skip=2, col_names=vnames)
}In reading the data, we’ll remove the zero GPP_sum values.
## New names:
## • `gCO2 m-2 d-1` -> `gCO2 m-2 d-1...2`
## • `gCO2 m-2 d-1` -> `gCO2 m-2 d-1...3`
## • `gCO2 m-2 d-1` -> `gCO2 m-2 d-1...4`
## • `degC` -> `degC...5`
## • `degC` -> `degC...6`
## • `degC` -> `degC...9`
## • `degC` -> `degC...10`
## • `degC` -> `degC...11`
## • `degC` -> `degC...12`
## • `0-1` -> `0-1...13`
## • `0-1` -> `0-1...14`
## • `0-1` -> `0-1...15`
## • `0-1` -> `0-1...16`Mountflux <- Carbon_Flux %>%
mutate_all(as.numeric) %>%
mutate(DOY = as.Date(Date,format = "%m-%d")) %>%
select(Date:PAR_ave) %>%
mutate(NEE = round(NEE_sum, digits = 3),
GPP = round(GPP_sum, digits = 3),
RE = round(RE_sum, digits = 3)) %>%
#select(-NEE_sum, -GPP_sum, -RE_sum) %>%
pivot_longer(cols = c("NEE", "RE", "GPP"), names_to = "CO2_FLUX_COMPONENTS", values_to ="value") %>%
filter(GPP_sum != 0, !is.na(PAR_ave))
print(Mountflux)## # A tibble: 906 × 9
## Date GPP_sum NEE_sum RE_sum Tair_min Tair_max PAR_ave CO2_FLUX_COMPONENTS
## <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr>
## 1 61 0.215 3.74 3.96 -8.32 13.0 420. NEE
## 2 61 0.215 3.74 3.96 -8.32 13.0 420. RE
## 3 61 0.215 3.74 3.96 -8.32 13.0 420. GPP
## 4 62 0.405 3.71 4.11 -6.75 14.8 424. NEE
## 5 62 0.405 3.71 4.11 -6.75 14.8 424. RE
## 6 62 0.405 3.71 4.11 -6.75 14.8 424. GPP
## 7 63 0.620 3.35 3.97 -7.84 9.47 480. NEE
## 8 63 0.620 3.35 3.97 -7.84 9.47 480. RE
## 9 63 0.620 3.35 3.97 -7.84 9.47 480. GPP
## 10 64 0.799 3.57 4.37 -4.99 11.8 474. NEE
## # ℹ 896 more rows
## # ℹ 1 more variable: value <dbl>9.1.2 Time series plot
Then I used ggplot to create a visualization of how NEE, GPP, an RE changed over a yearly time scale.
Mountflux %>%
ggplot(aes(x = Date, y= value, color = CO2_FLUX_COMPONENTS))+
geom_line()+
geom_hline(yintercept = 0, linetype = "longdash", color = "black")+
ggtitle(" Red Clover Valley: Interannual Components of Carbon Flux Measurements") +
xlab("Day of Year") +
ylab("g C/m^2")+
labs(caption =
"Figure 1: Components of Carbon Flux measurements collected in 30 minute intervals in 2021
and added into ‘sum daily value’ , from flux tower in Red Clover Valley,California. Components included NEE (Net Ecosystem Exchange), RE (Respiration Efficiency), GPP (Gross Primary Productivity) all measured in g C m^2 and are plotted against an annual time scale with days as the unit of measure.")
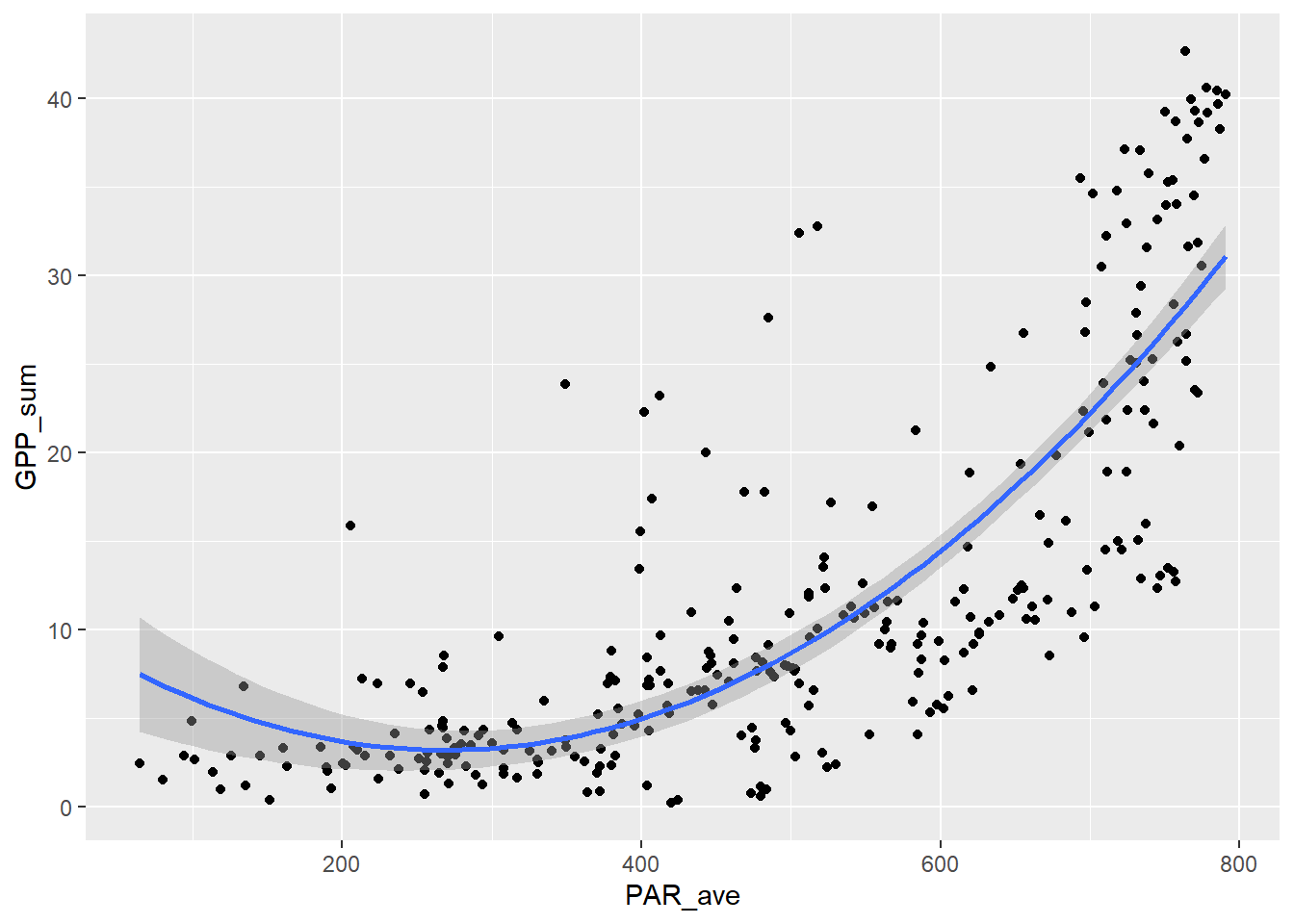
9.1.4 Non-linear model of PAR_ave ~ GPP_sum, using Michaelis-Menten model
Not the right goal for a model between these variables, since GPP should derive from PAR, not the other way around. But it does illustrate asymptotic data.
First the scatter plot:
9.1.5 Try fitting a non-linear model (SSmicmen from base R)
Based on https://www.datacamp.com/tutorial/introduction-to-non-linear-model-and-insights-using-r
Using start values for Vm and K based on interpreting the graph. We’ll use nonlinear least squares (nls) and starting values of the two coefficients.
micmenMod <- nls(PAR_ave~(Vm)*GPP_sum/(K+GPP_sum), data=Mountflux, start=list(Vm=750,K=1))
summary(micmenMod)##
## Formula: PAR_ave ~ (Vm) * GPP_sum/(K + GPP_sum)
##
## Parameters:
## Estimate Std. Error t value Pr(>|t|)
## Vm 819.0045 13.4911 60.71 <2e-16 ***
## K 5.0071 0.2573 19.46 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 121.1 on 904 degrees of freedom
##
## Number of iterations to convergence: 7
## Achieved convergence tolerance: 8.843e-06Calculate residual error
sse <- micmenMod$m$deviance()
null <- lm(PAR_ave~1, data=Mountflux)
sst <- data.frame(summary.aov(null)[[1]])$Sum.Sq
percent_variation_explained <- 100*(sst-sse)/sst
percent_variation_explained## [1] 59.97658plot(Mountflux$GPP_sum, Mountflux$PAR_ave,xlab="GPP_sum", ylab="PAR_ave")
lines(Mountflux$GPP_sum,
predict(micmenMod,Mountflux),col=2)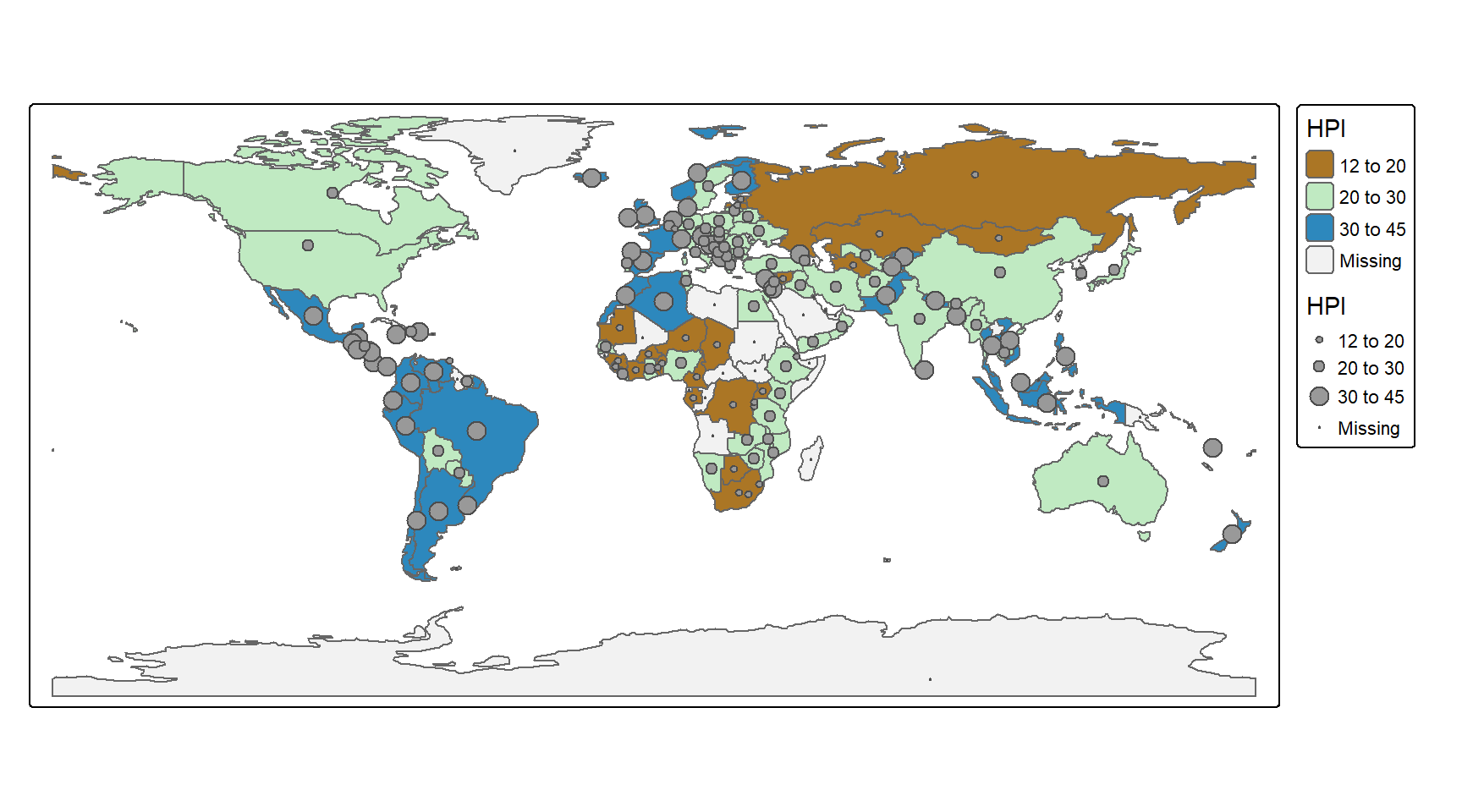
9.1.6 polynomial models
Ok, enough of that deceptive model of PAR predicted by GPP – not the way it works – and instead we’ll look at models that predict GPP from PAR, not asymptotic
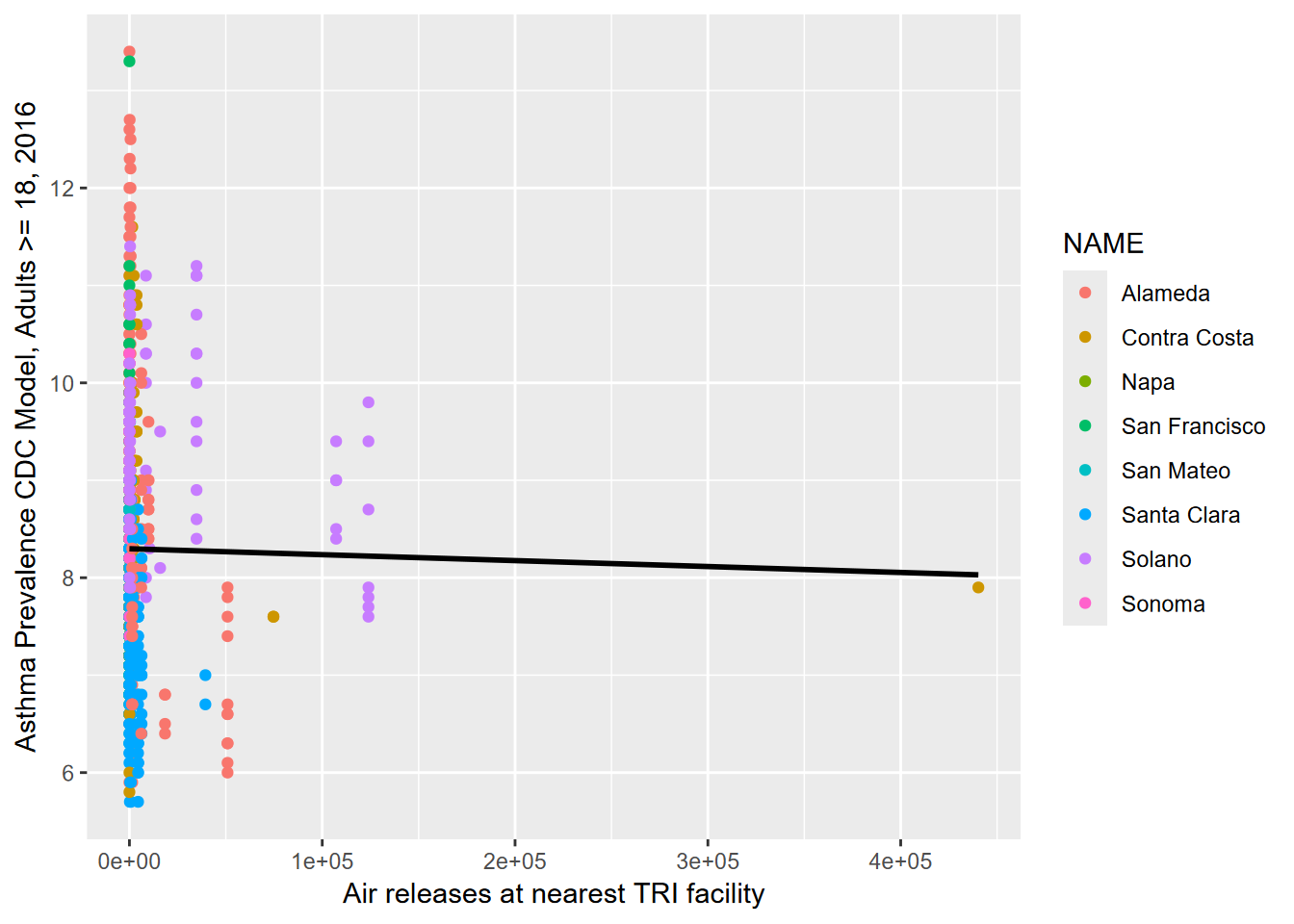
9.1.6.1 poly 1
##
## Call:
## lm(formula = GPP_sum ~ poly(PAR_ave, ord, raw = T), data = Mountflux)
##
## Residuals:
## Min 1Q Median 3Q Max
## -11.944 -4.931 -1.131 4.287 19.711
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -8.979814 0.669906 -13.40 <2e-16 ***
## poly(PAR_ave, ord, raw = T) 0.042818 0.001258 34.03 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 7.242 on 904 degrees of freedom
## Multiple R-squared: 0.5616, Adjusted R-squared: 0.5611
## F-statistic: 1158 on 1 and 904 DF, p-value: < 2.2e-16ggplot(Mountflux, aes(PAR_ave, GPP_sum)) + geom_point() +
stat_smooth(method=lm, formula=y~poly(x,ord,raw=T))
9.1.6.2 poly 2
##
## Call:
## lm(formula = GPP_sum ~ poly(PAR_ave, ord, raw = T), data = Mountflux)
##
## Residuals:
## Min 1Q Median 3Q Max
## -14.9170 -3.5363 -0.4922 2.4821 23.4111
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.062e+01 1.269e+00 8.368 <2e-16 ***
## poly(PAR_ave, ord, raw = T)1 -5.500e-02 5.737e-03 -9.587 <2e-16 ***
## poly(PAR_ave, ord, raw = T)2 1.023e-04 5.892e-06 17.367 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 6.273 on 903 degrees of freedom
## Multiple R-squared: 0.6714, Adjusted R-squared: 0.6706
## F-statistic: 922.3 on 2 and 903 DF, p-value: < 2.2e-16ggplot(Mountflux, aes(PAR_ave, GPP_sum)) + geom_point() +
stat_smooth(method=lm, formula=y~poly(x,ord,raw=T))
9.1.6.3 poly 3
##
## Call:
## lm(formula = GPP_sum ~ poly(PAR_ave, ord, raw = T), data = Mountflux)
##
## Residuals:
## Min 1Q Median 3Q Max
## -16.4635 -3.3477 -0.8198 2.3854 24.5018
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.290e+00 2.276e+00 -3.203 0.00141 **
## poly(PAR_ave, ord, raw = T)1 1.048e-01 1.804e-02 5.810 8.65e-09 ***
## poly(PAR_ave, ord, raw = T)2 -2.949e-04 4.309e-05 -6.845 1.41e-11 ***
## poly(PAR_ave, ord, raw = T)3 2.908e-07 3.127e-08 9.299 < 2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.996 on 902 degrees of freedom
## Multiple R-squared: 0.7001, Adjusted R-squared: 0.6991
## F-statistic: 701.9 on 3 and 902 DF, p-value: < 2.2e-16ggplot(Mountflux, aes(PAR_ave, GPP_sum)) + geom_point() +
stat_smooth(method=lm, formula=y~poly(x,ord,raw=T))
9.1.6.4 poly 4
##
## Call:
## lm(formula = GPP_sum ~ poly(PAR_ave, ord, raw = T), data = Mountflux)
##
## Residuals:
## Min 1Q Median 3Q Max
## -17.0946 -2.9525 -0.7201 2.0130 23.8099
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.346e+01 3.769e+00 3.570 0.000375 ***
## poly(PAR_ave, ord, raw = T)1 -1.768e-01 4.493e-02 -3.936 8.93e-05 ***
## poly(PAR_ave, ord, raw = T)2 9.001e-04 1.804e-04 4.990 7.25e-07 ***
## poly(PAR_ave, ord, raw = T)3 -1.692e-06 2.927e-07 -5.782 1.02e-08 ***
## poly(PAR_ave, ord, raw = T)4 1.122e-09 1.647e-10 6.812 1.75e-11 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.85 on 901 degrees of freedom
## Multiple R-squared: 0.7148, Adjusted R-squared: 0.7135
## F-statistic: 564.5 on 4 and 901 DF, p-value: < 2.2e-16ggplot(Mountflux, aes(PAR_ave, GPP_sum)) + geom_point() +
stat_smooth(method=lm, formula=y~poly(x,ord,raw=T))
9.1.6.5 poly 5
##
## Call:
## lm(formula = GPP_sum ~ poly(PAR_ave, ord, raw = T), data = Mountflux)
##
## Residuals:
## Min 1Q Median 3Q Max
## -17.0834 -2.7700 -0.7949 1.9559 23.4067
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 1.325e+00 6.746e+00 0.196 0.8443
## poly(PAR_ave, ord, raw = T)1 4.446e-02 1.116e-01 0.398 0.6904
## poly(PAR_ave, ord, raw = T)2 -4.500e-04 6.488e-04 -0.694 0.4881
## poly(PAR_ave, ord, raw = T)3 1.931e-06 1.698e-06 1.137 0.2557
## poly(PAR_ave, ord, raw = T)4 -3.295e-09 2.046e-09 -1.611 0.1076
## poly(PAR_ave, ord, raw = T)5 1.999e-12 9.226e-13 2.166 0.0306 *
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5.838 on 900 degrees of freedom
## Multiple R-squared: 0.7163, Adjusted R-squared: 0.7147
## F-statistic: 454.4 on 5 and 900 DF, p-value: < 2.2e-16ggplot(Mountflux, aes(PAR_ave, GPP_sum)) + geom_point() +
stat_smooth(method=lm, formula=y~poly(x,ord,raw=T))
9.1.7 power law relationship
##
## Call:
## lm(formula = log(GPP_sum) ~ log(PAR_ave), data = Mountflux)
##
## Residuals:
## Min 1Q Median 3Q Max
## -3.5030 -0.2995 0.0036 0.4460 1.8629
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -7.0150 0.3034 -23.12 <2e-16 ***
## log(PAR_ave) 1.4869 0.0495 30.04 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.7199 on 904 degrees of freedom
## Multiple R-squared: 0.4995, Adjusted R-squared: 0.499
## F-statistic: 902.2 on 1 and 904 DF, p-value: < 2.2e-16## `geom_smooth()` using formula = 'y ~ x' Now plot without the log transformed axes, we’ll need to create the curve with the model, using the coefficients.
Now plot without the log transformed axes, we’ll need to create the curve with the model, using the coefficients.
coeff = exp(powerModel$coefficients[1])
power = powerModel$coefficients[2]
fit_func <- function(x) coeff*x^power
fluxPlot <- ggplot(Mountflux) +
geom_point(aes(x=PAR_ave,y=GPP_sum))
fluxPlot <- fluxPlot +
stat_function(fun=fit_func, color="orange",lwd=2, alpha=0.5)
fluxPlot
fit_eq = paste("y = ", round(coeff,6), " x^", round(power,3))
fluxPlot <- fluxPlot +
annotate("text",x=200,y=40,label=fit_eq,size=4)
fluxPlot
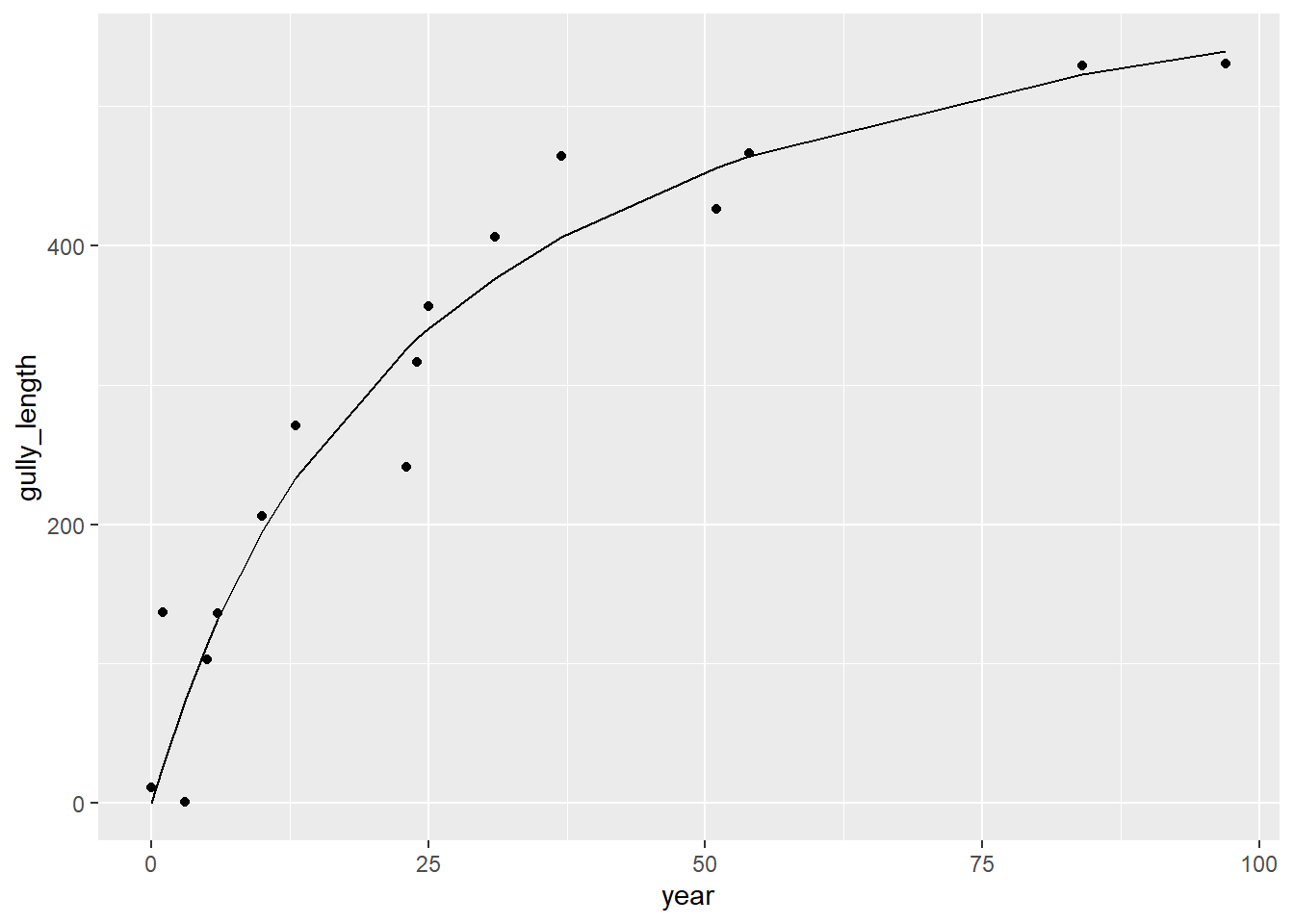
9.2 Another curve fit – of gullies that may follow a rate law of growth, or other asymptotic
from Graf WL (1977). The rate law in fluvial geomorphology. Am. J. Sci 277: 178-191.
The rate-law model that Graf used employs a half life, appropriate when depleting a quantity as will happen with an erosion phase following a disequilibrium (like a geomorphic drop in base level).
## Rows: 36 Columns: 3
## ── Column specification ────────────────────────────────────────────────────────
## Delimiter: ","
## chr (1): type
## dbl (2): distance, age_yrs
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
gully <- gullyGrowth |> filter(type=="gully")
gullyMod <- nls(age_yrs~(Vm)*distance/(K+distance), data=gully, start=list(Vm=1,K=1))
summary(gullyMod)##
## Formula: age_yrs ~ (Vm) * distance/(K + distance)
##
## Parameters:
## Estimate Std. Error t value Pr(>|t|)
## Vm 86.156 4.733 18.204 4.86e-13 ***
## K 152.503 23.546 6.477 4.31e-06 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.947 on 18 degrees of freedom
##
## Number of iterations to convergence: 9
## Achieved convergence tolerance: 8.225e-07Calculate residual error
sse <- gullyMod$m$deviance()
null <- lm(age_yrs~1, data=gully)
sst <- data.frame(summary.aov(null)[[1]])$Sum.Sq
percent_variation_explained <- 100*(sst-sse)/sst
percent_variation_explained## [1] 95.50215proto <- gullyGrowth |> filter(type=="proto-gully")
protoMod <- nls(age_yrs~(Vm)*distance/(K+distance), data=proto, start=list(Vm=1,K=1))
summary(protoMod)##
## Formula: age_yrs ~ (Vm) * distance/(K + distance)
##
## Parameters:
## Estimate Std. Error t value Pr(>|t|)
## Vm 141.36 12.66 11.162 2.35e-08 ***
## K 35.84 20.14 1.779 0.0969 .
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 21.17 on 14 degrees of freedom
##
## Number of iterations to convergence: 11
## Achieved convergence tolerance: 8.318e-06sse <- protoMod$m$deviance()
null <- lm(age_yrs~1, data=proto)
sst <- data.frame(summary.aov(null)[[1]])$Sum.Sq
percent_variation_explained <- 100*(sst-sse)/sst
percent_variation_explained## [1] 42.44041ggplot(data=gullyGrowth, aes(x=distance)) +
geom_point(aes(y=age_yrs, col=type)) +
geom_line(data=gully, aes(x=distance, y=predict(gullyMod,gully))) +
geom_line(data=proto, aes(x=distance, y=predict(protoMod,proto)))