4 Model diagnostics
4.1 Computational diagnostics
Current software still needs a decent amount of checks to make sure that the model we fitted produces reliable inference and predictions. Common points to check:
- Convergence (stationary distribution)
- Visually checking traceplots
- Rhat
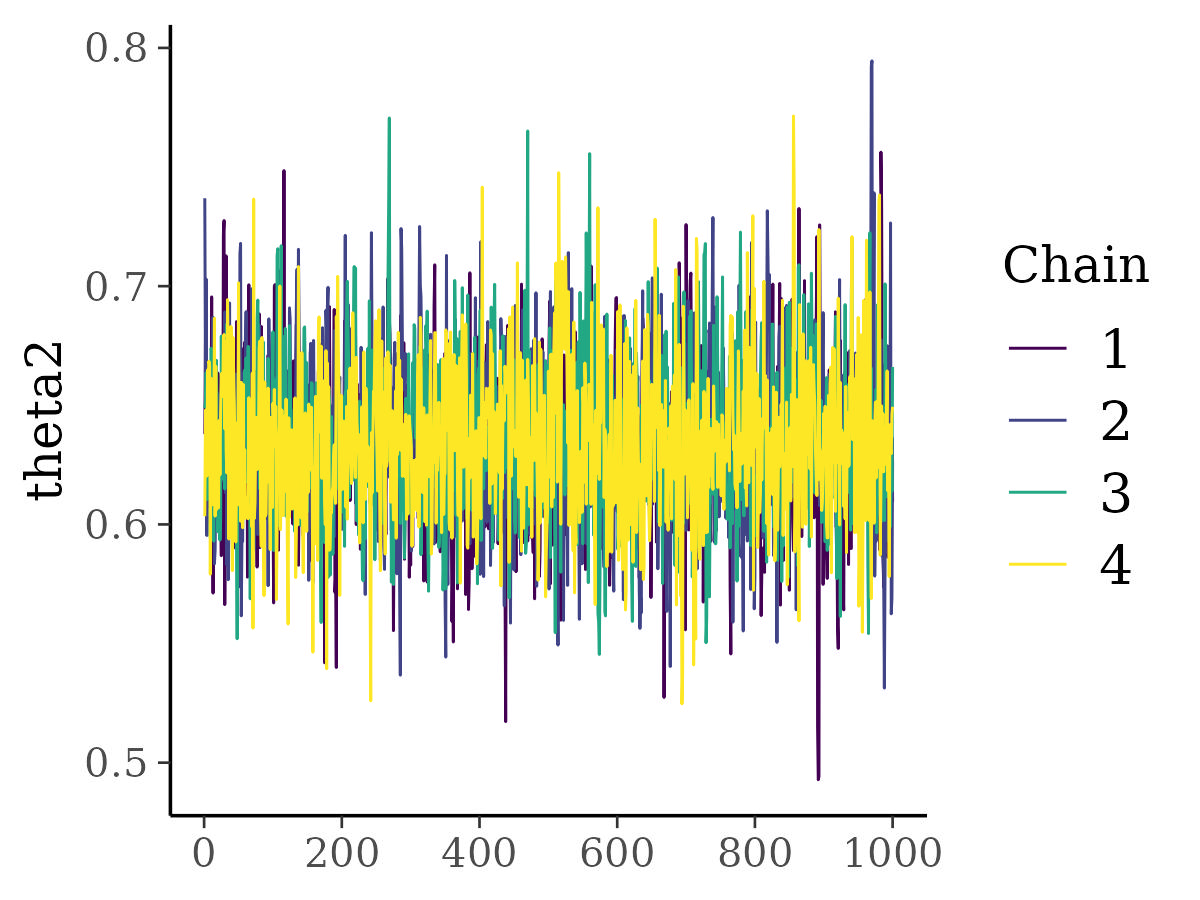

- Effective sample size.
- Others: read your software manual (e.g., divergent transitions in Stan).
- Good Bayesians do: simulate data with the same data generating process as the model to check that the program works.
- Be suspicious of undocumented R packages/software that don’t provide diagnostic tools.
4.2 Model checking and improvement
4.2.1 Evaluating the inference
- Do the parameters make sense according to the general consensus?
- How much information do the data contain?
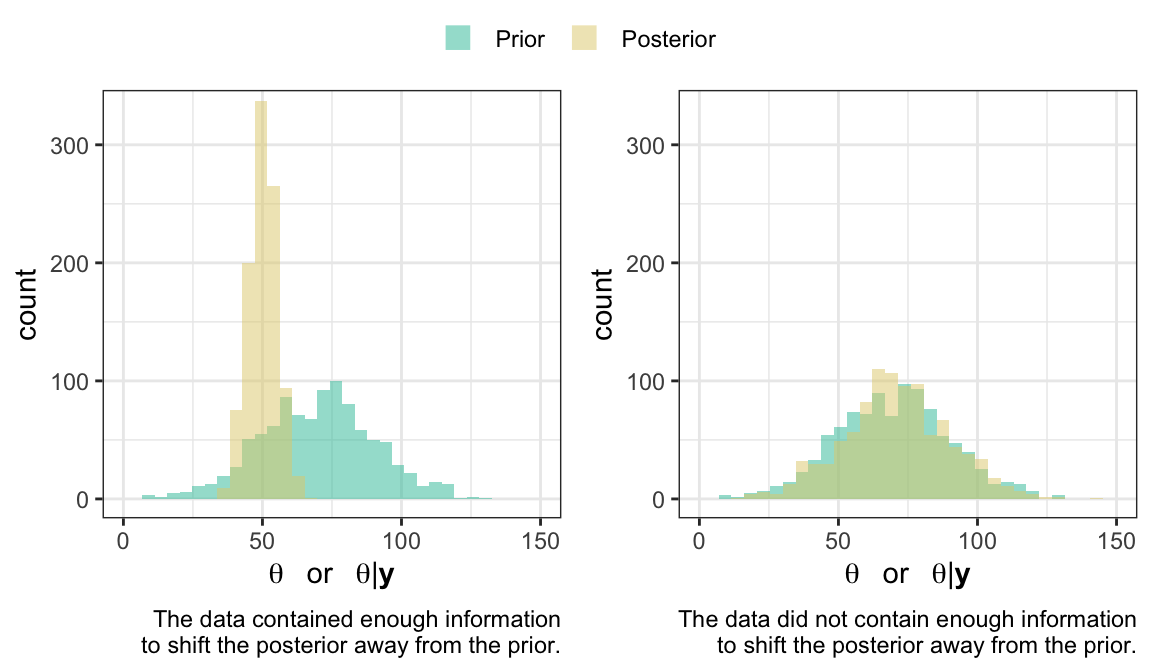
4.2.2 Posterior predictive checking
Simulate predictive distributions and compare them to the data.
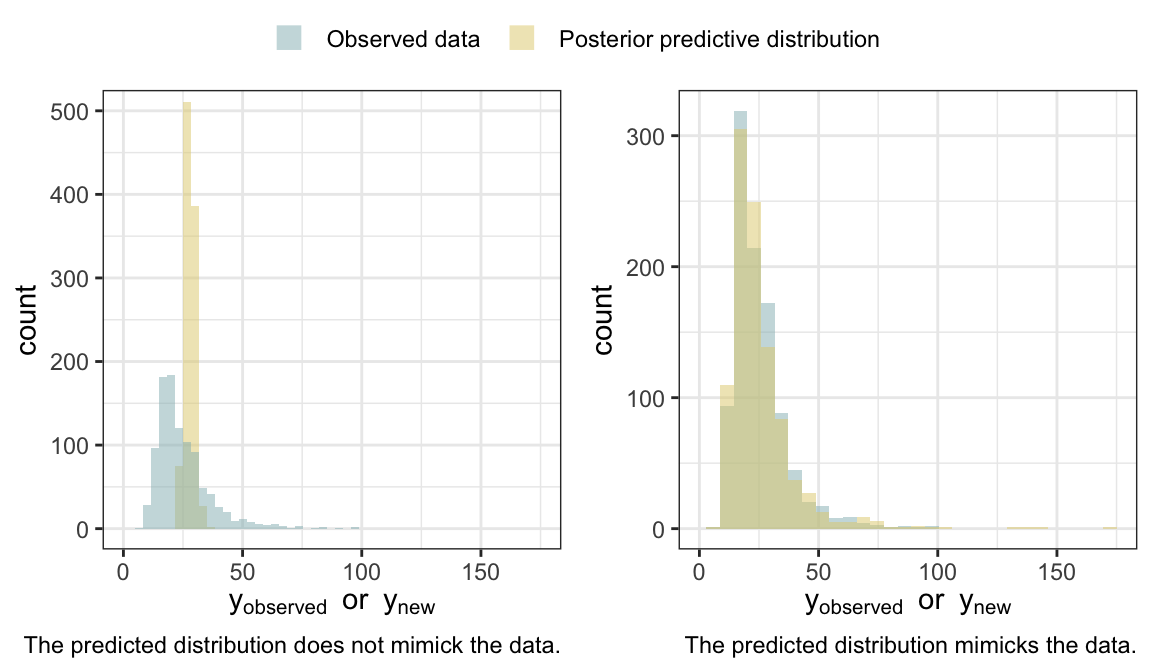
- Look for systematic discrepancies between model and data. See Gelman et al. (2020).
4.2.3 Model improvement:
- Model building as an iterative job.
- Select among the model that best fulfills your objectives.
- Conclusions based on many models.
4.2.3.1 Predictive performance metrics
- Information criteria
- Watanabe-Akaike or widely applicable information criterion (WAIC)
- Deviance information criterion (DIC)
- Akaike information criterion (AIC)
- More on predictive information criteria in Gelman et al. (2014).
- Watanabe-Akaike or widely applicable information criterion (WAIC)
- Cross validation
- Leave-one-out CV (see Vehtari et al., 2016).
- Leave-one-year/farm/field/herd-out. Diagnose your model.
Respect the structure of the data. Spatial, temporal, etc. See Roberts et al. (2017).
Keep in mind that all models are wrong. Rather than perfecting a model, learn how it performs under different scenarios to know its limitations.
4.3 Reporting a Bayesian model
- Communicate the strengths and weakpoints of our models.
- Example for standardized reporting: Bayesian Analysis Reporting Guidelines (BARG) in Kruschke (2021), summary in Table 1.
4.4 More useful references
- Gelman, A., Goegebeur, Y., Tuerlinckx, F. and Van Mechelen, I. (2000), Diagnostic checks for discrete data regression models using posterior predictive simulations. Journal of the Royal Statistical Society: Series C (Applied Statistics), 49: 247-268. [link]
- Gelman, A., Carlin, J. B., Stern, H. S., Dunson, D. B., Vehtari, A., & Rubin, D. B. (2013). Bayesian data analysis, Chapter 6. CRC press. [link]
- Gelman, A., Vehtari, A., Simpson, D., Margossian, C. C., Carpenter, B., Yao, Y., Kennedy, L., Gabry, J., Bürkner, P.-C., & Modrák, M. (2020). Bayesian Workflow. [link]
- Hobbs, N. T., & Hooten, M. B. (2015). Bayesian models: a statistical primer for ecologists. Princeton University Press. link
- Hooten, M.B. and Hobbs, N.T. (2015), A guide to Bayesian model selection for ecologists. Ecological Monographs, 85: 3-28. [link]
- Ver Hoef, J. M., & Boveng, P. L. (2015). Iterating on a single model is a viable alternative to multimodel inference. The Journal of Wildlife Management, 79(5), 719–729. [link]