Chapter 30 Statistical Tests
What You’ll Learn:
- t-tests (one-sample, two-sample, paired)
- Common statistical test errors
- Assumptions and violations
- Interpreting results
- Effect sizes
Key Errors Covered: 22+ statistical errors
Difficulty: ⭐⭐ Intermediate to ⭐⭐⭐ Advanced
30.1 Introduction
R excels at statistical testing, but errors are common:
# Simple t-test
t.test(mtcars$mpg[mtcars$am == 0],
mtcars$mpg[mtcars$am == 1])
#>
#> Welch Two Sample t-test
#>
#> data: mtcars$mpg[mtcars$am == 0] and mtcars$mpg[mtcars$am == 1]
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean of x mean of y
#> 17.14737 24.39231Let’s master statistical testing errors!
30.2 t-tests
💡 Key Insight: Types of t-tests
# One-sample t-test (compare to population mean)
t.test(mtcars$mpg, mu = 20)
#>
#> One Sample t-test
#>
#> data: mtcars$mpg
#> t = 0.08506, df = 31, p-value = 0.9328
#> alternative hypothesis: true mean is not equal to 20
#> 95 percent confidence interval:
#> 17.91768 22.26357
#> sample estimates:
#> mean of x
#> 20.09062
# Two-sample t-test (independent groups)
t.test(mpg ~ am, data = mtcars)
#>
#> Welch Two Sample t-test
#>
#> data: mpg by am
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean in group 0 mean in group 1
#> 17.14737 24.39231
# Paired t-test (same subjects, two conditions)
before <- c(120, 135, 140, 125, 130)
after <- c(115, 130, 135, 120, 128)
t.test(before, after, paired = TRUE)
#>
#> Paired t-test
#>
#> data: before and after
#> t = 7.3333, df = 4, p-value = 0.001841
#> alternative hypothesis: true mean difference is not equal to 0
#> 95 percent confidence interval:
#> 2.734133 6.065867
#> sample estimates:
#> mean difference
#> 4.4
# One-sided test
t.test(mpg ~ am, data = mtcars, alternative = "greater")
#>
#> Welch Two Sample t-test
#>
#> data: mpg by am
#> t = -3.7671, df = 18.332, p-value = 0.9993
#> alternative hypothesis: true difference in means between group 0 and group 1 is greater than 0
#> 95 percent confidence interval:
#> -10.57662 Inf
#> sample estimates:
#> mean in group 0 mean in group 1
#> 17.14737 24.39231
# Unequal variances (Welch's t-test, default)
t.test(mpg ~ am, data = mtcars, var.equal = FALSE)
#>
#> Welch Two Sample t-test
#>
#> data: mpg by am
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean in group 0 mean in group 1
#> 17.14737 24.39231
# Equal variances (Student's t-test)
t.test(mpg ~ am, data = mtcars, var.equal = TRUE)
#>
#> Two Sample t-test
#>
#> data: mpg by am
#> t = -4.1061, df = 30, p-value = 0.000285
#> alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
#> 95 percent confidence interval:
#> -10.84837 -3.64151
#> sample estimates:
#> mean in group 0 mean in group 1
#> 17.14737 24.3923130.3 Error #1: not enough 'x' observations
⭐ BEGINNER 📊 DATA
30.3.3 Common Causes
# Empty vector after filtering
data <- mtcars$mpg[mtcars$cyl == 99] # No cars with 99 cylinders
t.test(data)
#> Error in t.test.default(data): not enough 'x' observations
# Single group
t.test(mtcars$mpg[mtcars$am == 0], mtcars$mpg[mtcars$am == 99])
#> Error in t.test.default(mtcars$mpg[mtcars$am == 0], mtcars$mpg[mtcars$am == : not enough 'y' observations30.3.4 Solutions
✅ SOLUTION 1: Check Data First
# Check sample sizes
automatic <- mtcars$mpg[mtcars$am == 0]
manual <- mtcars$mpg[mtcars$am == 1]
cat("Automatic:", length(automatic), "observations\n")
#> Automatic: 19 observations
cat("Manual:", length(manual), "observations\n")
#> Manual: 13 observations
if (length(automatic) >= 2 && length(manual) >= 2) {
t.test(automatic, manual)
} else {
warning("Insufficient data for t-test")
}
#>
#> Welch Two Sample t-test
#>
#> data: automatic and manual
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean of x mean of y
#> 17.14737 24.39231✅ SOLUTION 2: Safe t-test Function
safe_t_test <- function(x, y = NULL, ...) {
if (is.null(y)) {
if (length(x) < 2) {
stop("x must have at least 2 observations")
}
} else {
if (length(x) < 2) {
stop("x must have at least 2 observations")
}
if (length(y) < 2) {
stop("y must have at least 2 observations")
}
}
t.test(x, y, ...)
}
# Test
safe_t_test(mtcars$mpg[mtcars$am == 0],
mtcars$mpg[mtcars$am == 1])
#>
#> Welch Two Sample t-test
#>
#> data: x and y
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean of x mean of y
#> 17.14737 24.3923130.4 Error #2: data are essentially constant
⭐⭐ INTERMEDIATE 📊 DATA
30.4.1 The Error
# All values the same
t.test(c(5, 5, 5, 5, 5))
#> Error in t.test.default(c(5, 5, 5, 5, 5)): data are essentially constant🔴 ERROR
Error in t.test.default(c(5, 5, 5, 5, 5)) :
data are essentially constant30.4.4 Solutions
✅ SOLUTION 1: Check for Variation
check_variation <- function(x) {
if (sd(x) == 0 || length(unique(x)) == 1) {
warning("No variation in data - t-test not appropriate")
return(FALSE)
}
TRUE
}
# Check before test
data <- c(10, 10, 10, 10)
if (check_variation(data)) {
t.test(data)
} else {
cat("Data has no variation\n")
}
#> Warning in check_variation(data): No variation in data - t-test not appropriate
#> Data has no variation✅ SOLUTION 2: Use Appropriate Test
# For proportions, use prop.test
successes <- c(1, 1, 1, 1) # All successes
prop.test(sum(successes), length(successes))
#> Warning in prop.test(sum(successes), length(successes)): Chi-squared
#> approximation may be incorrect
#>
#> 1-sample proportions test with continuity correction
#>
#> data: sum(successes) out of length(successes), null probability 0.5
#> X-squared = 2.25, df = 1, p-value = 0.1336
#> alternative hypothesis: true p is not equal to 0.5
#> 95 percent confidence interval:
#> 0.395773 1.000000
#> sample estimates:
#> p
#> 1
# Or check if variation exists
data <- mtcars$mpg
if (sd(data) > 0) {
t.test(data, mu = 20)
} else {
cat("Data is constant, mean =", mean(data), "\n")
}
#>
#> One Sample t-test
#>
#> data: data
#> t = 0.08506, df = 31, p-value = 0.9328
#> alternative hypothesis: true mean is not equal to 20
#> 95 percent confidence interval:
#> 17.91768 22.26357
#> sample estimates:
#> mean of x
#> 20.0906230.5 Formula Interface
💡 Key Insight: Formula vs Vector Interface
# Vector interface
group1 <- mtcars$mpg[mtcars$am == 0]
group2 <- mtcars$mpg[mtcars$am == 1]
t.test(group1, group2)
#>
#> Welch Two Sample t-test
#>
#> data: group1 and group2
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean of x mean of y
#> 17.14737 24.39231
# Formula interface (preferred)
t.test(mpg ~ am, data = mtcars)
#>
#> Welch Two Sample t-test
#>
#> data: mpg by am
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean in group 0 mean in group 1
#> 17.14737 24.39231
# Formula with subset
t.test(mpg ~ am, data = mtcars, subset = cyl == 4)
#>
#> Welch Two Sample t-test
#>
#> data: mpg by am
#> t = -2.8855, df = 8.9993, p-value = 0.01802
#> alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
#> 95 percent confidence interval:
#> -9.232108 -1.117892
#> sample estimates:
#> mean in group 0 mean in group 1
#> 22.900 28.075
# Advantages of formula:
# - Cleaner code
# - Automatic labeling
# - Works with model functions
# - Subset argument30.6 Error #3: grouping factor must have 2 levels
⭐ BEGINNER 📊 DATA
30.6.1 The Error
# Three levels in grouping variable
t.test(mpg ~ cyl, data = mtcars)
#> Error in t.test.formula(mpg ~ cyl, data = mtcars): grouping factor must have exactly 2 levels🔴 ERROR
Error in t.test.formula(mpg ~ cyl, data = mtcars) :
grouping factor must have exactly 2 levels30.6.3 Solutions
✅ SOLUTION 1: Filter to 2 Groups
# Compare only 4 vs 6 cylinders
mtcars_subset <- mtcars[mtcars$cyl %in% c(4, 6), ]
t.test(mpg ~ cyl, data = mtcars_subset)
#>
#> Welch Two Sample t-test
#>
#> data: mpg by cyl
#> t = 4.7191, df = 12.956, p-value = 0.0004048
#> alternative hypothesis: true difference in means between group 4 and group 6 is not equal to 0
#> 95 percent confidence interval:
#> 3.751376 10.090182
#> sample estimates:
#> mean in group 4 mean in group 6
#> 26.66364 19.74286
# Or use subset argument
t.test(mpg ~ cyl, data = mtcars, subset = cyl %in% c(4, 6))
#>
#> Welch Two Sample t-test
#>
#> data: mpg by cyl
#> t = 4.7191, df = 12.956, p-value = 0.0004048
#> alternative hypothesis: true difference in means between group 4 and group 6 is not equal to 0
#> 95 percent confidence interval:
#> 3.751376 10.090182
#> sample estimates:
#> mean in group 4 mean in group 6
#> 26.66364 19.74286✅ SOLUTION 2: Use ANOVA for >2 Groups
# For 3+ groups, use ANOVA
anova_result <- aov(mpg ~ factor(cyl), data = mtcars)
summary(anova_result)
#> Df Sum Sq Mean Sq F value Pr(>F)
#> factor(cyl) 2 824.8 412.4 39.7 4.98e-09 ***
#> Residuals 29 301.3 10.4
#> ---
#> Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
# Post-hoc tests
TukeyHSD(anova_result)
#> Tukey multiple comparisons of means
#> 95% family-wise confidence level
#>
#> Fit: aov(formula = mpg ~ factor(cyl), data = mtcars)
#>
#> $`factor(cyl)`
#> diff lwr upr p adj
#> 6-4 -6.920779 -10.769350 -3.0722086 0.0003424
#> 8-4 -11.563636 -14.770779 -8.3564942 0.0000000
#> 8-6 -4.642857 -8.327583 -0.9581313 0.0112287✅ SOLUTION 3: Multiple Comparisons
# Compare all pairs
library(dplyr)
# Get unique cylinder values
cyl_levels <- unique(mtcars$cyl)
comparisons <- combn(cyl_levels, 2)
# Perform all pairwise tests
results <- apply(comparisons, 2, function(pair) {
subset_data <- mtcars[mtcars$cyl %in% pair, ]
test <- t.test(mpg ~ cyl, data = subset_data)
data.frame(
comparison = paste(pair[1], "vs", pair[2]),
p_value = test$p.value,
statistic = test$statistic
)
})
do.call(rbind, results)
#> comparison p_value statistic
#> t 6 vs 4 4.048495e-04 4.719059
#> t1 6 vs 8 4.540355e-05 5.291135
#> t2 4 vs 8 1.641348e-06 7.59666430.7 Assumptions
💡 Key Insight: t-test Assumptions
# Assumptions:
# 1. Independence
# 2. Normality (esp. for small samples)
# 3. Equal variances (for Student's t-test)
# Check normality
shapiro.test(mtcars$mpg)
#>
#> Shapiro-Wilk normality test
#>
#> data: mtcars$mpg
#> W = 0.94756, p-value = 0.1229
# Visual check
qqnorm(mtcars$mpg)
qqline(mtcars$mpg)
# Check equal variances
var.test(mpg ~ am, data = mtcars)
#>
#> F test to compare two variances
#>
#> data: mpg by am
#> F = 0.38656, num df = 18, denom df = 12, p-value = 0.06691
#> alternative hypothesis: true ratio of variances is not equal to 1
#> 95 percent confidence interval:
#> 0.1243721 1.0703429
#> sample estimates:
#> ratio of variances
#> 0.3865615
# If assumptions violated:
# - Use Welch's t-test (var.equal = FALSE, default)
# - Use Mann-Whitney U test (non-parametric)
wilcox.test(mpg ~ am, data = mtcars)
#> Warning in wilcox.test.default(x = DATA[[1L]], y = DATA[[2L]], ...): cannot
#> compute exact p-value with ties
#>
#> Wilcoxon rank sum test with continuity correction
#>
#> data: mpg by am
#> W = 42, p-value = 0.001871
#> alternative hypothesis: true location shift is not equal to 0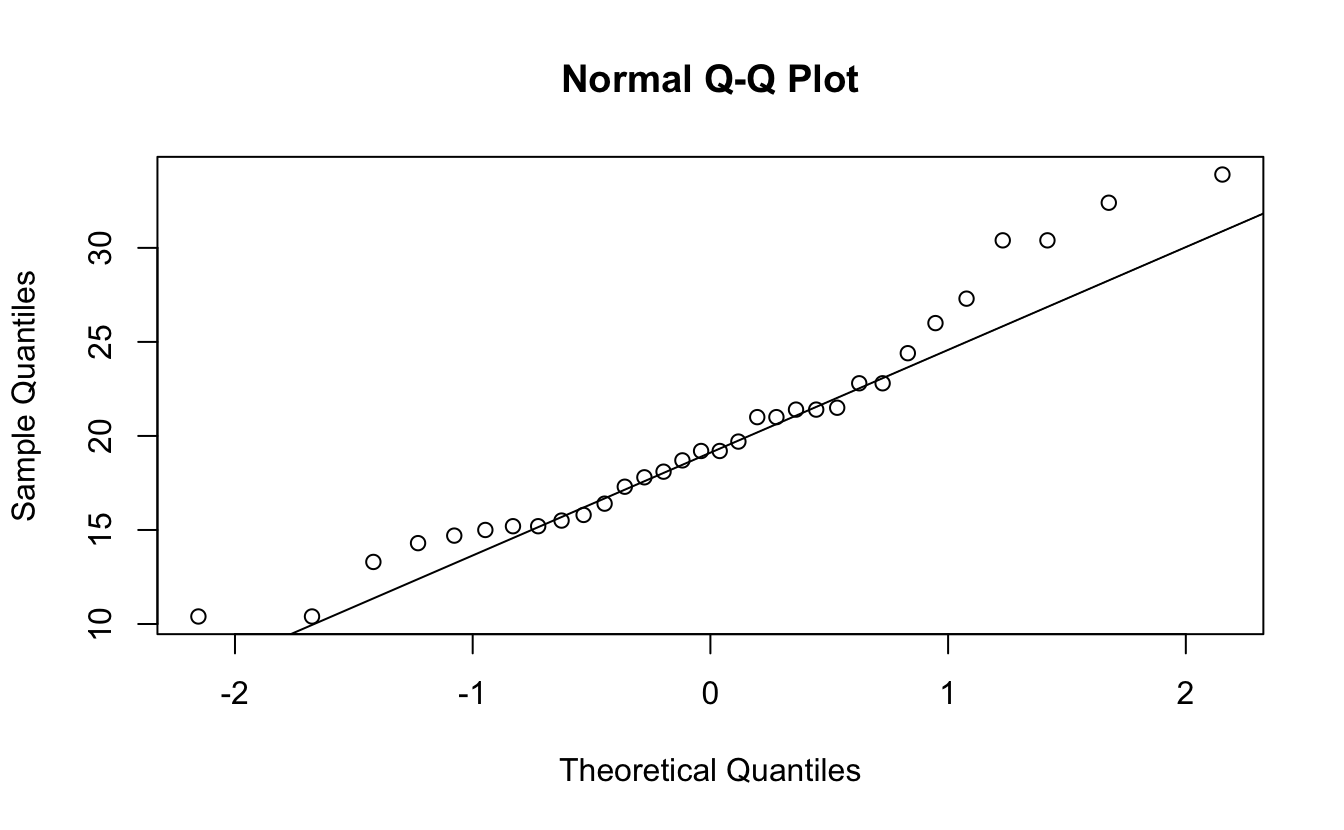
30.8 Interpreting Results
💡 Key Insight: Understanding Output
result <- t.test(mpg ~ am, data = mtcars)
result
#>
#> Welch Two Sample t-test
#>
#> data: mpg by am
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean in group 0 mean in group 1
#> 17.14737 24.39231
# Extract components
result$statistic # t-statistic
#> t
#> -3.767123
result$parameter # degrees of freedom
#> df
#> 18.33225
result$p.value # p-value
#> [1] 0.001373638
result$conf.int # confidence interval
#> [1] -11.280194 -3.209684
#> attr(,"conf.level")
#> [1] 0.95
result$estimate # group means
#> mean in group 0 mean in group 1
#> 17.14737 24.39231
# Interpretation
cat("t-statistic:", round(result$statistic, 3), "\n")
#> t-statistic: -3.767
cat("p-value:", round(result$p.value, 4), "\n")
#> p-value: 0.0014
cat("95% CI:", round(result$conf.int, 2), "\n")
#> 95% CI: -11.28 -3.21
cat("Mean difference:", round(diff(result$estimate), 2), "\n")
#> Mean difference: 7.24
if (result$p.value < 0.05) {
cat("\nSignificant difference between groups (p < 0.05)\n")
} else {
cat("\nNo significant difference between groups (p >= 0.05)\n")
}
#>
#> Significant difference between groups (p < 0.05)30.9 Effect Size
🎯 Best Practice: Report Effect Sizes
# Cohen's d for effect size
cohens_d <- function(x, y) {
n1 <- length(x)
n2 <- length(y)
# Pooled standard deviation
s_pooled <- sqrt(((n1 - 1) * var(x) + (n2 - 1) * var(y)) / (n1 + n2 - 2))
# Cohen's d
d <- (mean(x) - mean(y)) / s_pooled
# Interpretation
interpretation <- if (abs(d) < 0.2) "negligible"
else if (abs(d) < 0.5) "small"
else if (abs(d) < 0.8) "medium"
else "large"
list(
d = d,
interpretation = interpretation
)
}
# Calculate effect size
auto <- mtcars$mpg[mtcars$am == 0]
manual <- mtcars$mpg[mtcars$am == 1]
effect <- cohens_d(auto, manual)
cat("Cohen's d:", round(effect$d, 2), "\n")
#> Cohen's d: -1.48
cat("Effect size:", effect$interpretation, "\n")
#> Effect size: large30.10 Paired t-tests
💡 Key Insight: Paired vs Independent
# Paired: same subjects, two conditions
before <- c(120, 135, 140, 125, 130, 145, 150)
after <- c(115, 130, 135, 120, 128, 140, 145)
# Paired t-test
t.test(before, after, paired = TRUE)
#>
#> Paired t-test
#>
#> data: before and after
#> t = 10.667, df = 6, p-value = 4.004e-05
#> alternative hypothesis: true mean difference is not equal to 0
#> 95 percent confidence interval:
#> 3.522752 5.620105
#> sample estimates:
#> mean difference
#> 4.571429
# Calculate differences
differences <- before - after
cat("Mean difference:", mean(differences), "\n")
#> Mean difference: 4.571429
cat("SD of differences:", sd(differences), "\n")
#> SD of differences: 1.133893
# One-sample test on differences (equivalent)
t.test(differences, mu = 0)
#>
#> One Sample t-test
#>
#> data: differences
#> t = 10.667, df = 6, p-value = 4.004e-05
#> alternative hypothesis: true mean is not equal to 0
#> 95 percent confidence interval:
#> 3.522752 5.620105
#> sample estimates:
#> mean of x
#> 4.571429
# Independent would be WRONG here
t.test(before, after, paired = FALSE) # Ignores pairing
#>
#> Welch Two Sample t-test
#>
#> data: before and after
#> t = 0.79817, df = 11.997, p-value = 0.4403
#> alternative hypothesis: true difference in means is not equal to 0
#> 95 percent confidence interval:
#> -7.907792 17.050649
#> sample estimates:
#> mean of x mean of y
#> 135.0000 130.428630.11 Error #4: 'x' and 'y' must have the same length
⭐ BEGINNER 📊 DATA
30.11.1 The Error
before <- c(120, 135, 140, 125)
after <- c(115, 130, 135) # Only 3 values
t.test(before, after, paired = TRUE)
#> Error in complete.cases(x, y): not all arguments have the same length🔴 ERROR
Error in t.test.default(before, after, paired = TRUE) :
'x' and 'y' must have the same length30.11.3 Solutions
✅ SOLUTION 1: Check Lengths
before <- c(120, 135, 140, 125, 130)
after <- c(115, 130, 135, 120, 128)
if (length(before) != length(after)) {
stop("before and after must have same length for paired test")
}
t.test(before, after, paired = TRUE)
#>
#> Paired t-test
#>
#> data: before and after
#> t = 7.3333, df = 4, p-value = 0.001841
#> alternative hypothesis: true mean difference is not equal to 0
#> 95 percent confidence interval:
#> 2.734133 6.065867
#> sample estimates:
#> mean difference
#> 4.4✅ SOLUTION 2: Handle Missing Data
# Data with NAs
before <- c(120, 135, NA, 125, 130)
after <- c(115, 130, 135, 120, NA)
# Remove pairs with any NA
complete_cases <- complete.cases(before, after)
before_clean <- before[complete_cases]
after_clean <- after[complete_cases]
cat("Complete pairs:", sum(complete_cases), "\n")
#> Complete pairs: 3
t.test(before_clean, after_clean, paired = TRUE)
#> Error in t.test.default(before_clean, after_clean, paired = TRUE): data are essentially constant✅ SOLUTION 3: Use Data Frame Approach
library(dplyr)
library(tidyr)
# Store in data frame
data <- data.frame(
id = 1:5,
before = c(120, 135, NA, 125, 130),
after = c(115, 130, 135, 120, NA)
)
# Remove incomplete cases
data_complete <- data %>%
filter(!is.na(before) & !is.na(after))
cat("Complete cases:", nrow(data_complete), "\n")
#> Complete cases: 3
with(data_complete, t.test(before, after, paired = TRUE))
#> Error in t.test.default(before, after, paired = TRUE): data are essentially constant30.12 Power and Sample Size
🎯 Best Practice: Power Analysis
# Calculate required sample size
power.t.test(
delta = 5, # Expected difference
sd = 10, # Standard deviation
sig.level = 0.05, # Alpha
power = 0.80 # Desired power
)
#>
#> Two-sample t test power calculation
#>
#> n = 63.76576
#> delta = 5
#> sd = 10
#> sig.level = 0.05
#> power = 0.8
#> alternative = two.sided
#>
#> NOTE: n is number in *each* group
# Calculate power for given sample size
power.t.test(
n = 20,
delta = 5,
sd = 10,
sig.level = 0.05
)
#>
#> Two-sample t test power calculation
#>
#> n = 20
#> delta = 5
#> sd = 10
#> sig.level = 0.05
#> power = 0.3377084
#> alternative = two.sided
#>
#> NOTE: n is number in *each* group
# For existing study
auto <- mtcars$mpg[mtcars$am == 0]
manual <- mtcars$mpg[mtcars$am == 1]
power.t.test(
n = length(auto),
delta = abs(mean(auto) - mean(manual)),
sd = sd(c(auto, manual)),
sig.level = 0.05
)
#>
#> Two-sample t test power calculation
#>
#> n = 19
#> delta = 7.244939
#> sd = 6.026948
#> sig.level = 0.05
#> power = 0.9499733
#> alternative = two.sided
#>
#> NOTE: n is number in *each* group30.13 Summary
Key Takeaways:
- Check sample sizes - Need at least 2 observations per group
- Check variation - Data can’t be constant
- Exactly 2 groups - Use ANOVA for 3+
- Paired = same length - Remove incomplete pairs
- Check assumptions - Normality, equal variances
- Report effect sizes - Not just p-values
- Formula interface preferred - Cleaner code
Quick Reference:
| Error | Cause | Fix |
|---|---|---|
| not enough observations | n < 2 | Check sample sizes |
| data are constant | sd = 0 | Check for variation |
| must have 2 levels | 3+ groups | Filter or use ANOVA |
| must have same length | Paired mismatch | Remove incomplete pairs |
t-test Variations:
# One-sample
t.test(x, mu = 0)
# Two-sample (independent)
t.test(x, y)
t.test(y ~ x, data = df)
# Paired
t.test(x, y, paired = TRUE)
# One-sided
t.test(x, y, alternative = "greater")
t.test(x, y, alternative = "less")
# Equal variances
t.test(x, y, var.equal = TRUE)
# With subset
t.test(y ~ x, data = df, subset = condition)Best Practices:
# ✅ Good
Check sample sizes first
Check assumptions (normality, equal variance)
Use formula interface: t.test(y ~ x, data = df)
Report effect sizes (Cohen's d)
Use Welch's t-test (default) for unequal variances
Check for paired vs independent design
# ❌ Avoid
Assuming data meets assumptions
Using t-test for 3+ groups
Ignoring pairing in data
Only reporting p-values
Using with constant data
Not checking sample sizes30.14 Exercises
📝 Exercise 1: Basic t-test
Using mtcars: 1. Test if mean mpg differs by transmission (am) 2. Check assumptions 3. Calculate effect size 4. Interpret results
📝 Exercise 2: Paired t-test
Create before/after data and: 1. Perform paired t-test 2. Check if order matters 3. Calculate mean difference 4. Visualize differences
📝 Exercise 3: Safe t-test Function
Write safe_t_test() that:
1. Checks sample sizes
2. Checks for variation
3. Verifies assumptions
4. Performs test
5. Returns formatted results
📝 Exercise 4: Multiple Comparisons
Using iris dataset: 1. Compare petal length across species 2. Perform all pairwise t-tests 3. Adjust for multiple testing 4. Report results in table
30.15 Exercise Answers
Click to see answers
Exercise 1:
# 1. Test mean mpg by transmission
result <- t.test(mpg ~ am, data = mtcars)
result
#>
#> Welch Two Sample t-test
#>
#> data: mpg by am
#> t = -3.7671, df = 18.332, p-value = 0.001374
#> alternative hypothesis: true difference in means between group 0 and group 1 is not equal to 0
#> 95 percent confidence interval:
#> -11.280194 -3.209684
#> sample estimates:
#> mean in group 0 mean in group 1
#> 17.14737 24.39231
# 2. Check assumptions
# Normality
shapiro.test(mtcars$mpg[mtcars$am == 0])
#>
#> Shapiro-Wilk normality test
#>
#> data: mtcars$mpg[mtcars$am == 0]
#> W = 0.97677, p-value = 0.8987
shapiro.test(mtcars$mpg[mtcars$am == 1])
#>
#> Shapiro-Wilk normality test
#>
#> data: mtcars$mpg[mtcars$am == 1]
#> W = 0.9458, p-value = 0.5363
# Equal variances
var.test(mpg ~ am, data = mtcars)
#>
#> F test to compare two variances
#>
#> data: mpg by am
#> F = 0.38656, num df = 18, denom df = 12, p-value = 0.06691
#> alternative hypothesis: true ratio of variances is not equal to 1
#> 95 percent confidence interval:
#> 0.1243721 1.0703429
#> sample estimates:
#> ratio of variances
#> 0.3865615
# 3. Effect size
auto <- mtcars$mpg[mtcars$am == 0]
manual <- mtcars$mpg[mtcars$am == 1]
cohens_d <- function(x, y) {
n1 <- length(x)
n2 <- length(y)
s_pooled <- sqrt(((n1 - 1) * var(x) + (n2 - 1) * var(y)) / (n1 + n2 - 2))
(mean(x) - mean(y)) / s_pooled
}
d <- cohens_d(auto, manual)
# 4. Interpret
cat("\n=== Results ===\n")
#>
#> === Results ===
cat("t-statistic:", round(result$statistic, 3), "\n")
#> t-statistic: -3.767
cat("p-value:", format.pval(result$p.value, digits = 3), "\n")
#> p-value: 0.00137
cat("Mean difference:", round(diff(result$estimate), 2), "mpg\n")
#> Mean difference: 7.24 mpg
cat("95% CI:", round(result$conf.int, 2), "\n")
#> 95% CI: -11.28 -3.21
cat("Cohen's d:", round(d, 2), "\n")
#> Cohen's d: -1.48
if (result$p.value < 0.05) {
cat("\nConclusion: Significant difference in mpg between transmission types (p < 0.05)\n")
cat("Manual transmission has", round(abs(diff(result$estimate)), 1),
"higher mpg on average.\n")
}
#>
#> Conclusion: Significant difference in mpg between transmission types (p < 0.05)
#> Manual transmission has 7.2 higher mpg on average.Exercise 2:
# Create data
set.seed(123)
before <- c(120, 135, 140, 125, 130, 145, 150, 128, 138, 142)
after <- before - rnorm(10, mean = 8, sd = 3)
# 1. Paired t-test
result <- t.test(before, after, paired = TRUE)
result
#>
#> Paired t-test
#>
#> data: before and after
#> t = 9.0888, df = 9, p-value = 7.879e-06
#> alternative hypothesis: true mean difference is not equal to 0
#> 95 percent confidence interval:
#> 6.176989 10.270765
#> sample estimates:
#> mean difference
#> 8.223877
# 2. Order doesn't matter for pairing
t.test(after, before, paired = TRUE) # Same magnitude, opposite sign
#>
#> Paired t-test
#>
#> data: after and before
#> t = -9.0888, df = 9, p-value = 7.879e-06
#> alternative hypothesis: true mean difference is not equal to 0
#> 95 percent confidence interval:
#> -10.270765 -6.176989
#> sample estimates:
#> mean difference
#> -8.223877
# 3. Mean difference
differences <- before - after
cat("\nMean difference:", round(mean(differences), 2), "\n")
#>
#> Mean difference: 8.22
cat("SD of differences:", round(sd(differences), 2), "\n")
#> SD of differences: 2.86
# 4. Visualize
library(ggplot2)
data_df <- data.frame(
id = rep(1:10, 2),
time = rep(c("Before", "After"), each = 10),
value = c(before, after)
)
ggplot(data_df, aes(x = time, y = value, group = id)) +
geom_line(alpha = 0.5) +
geom_point(size = 3) +
labs(title = "Before vs After Comparison",
subtitle = paste("Mean difference:", round(mean(differences), 1)),
y = "Value") +
theme_minimal()
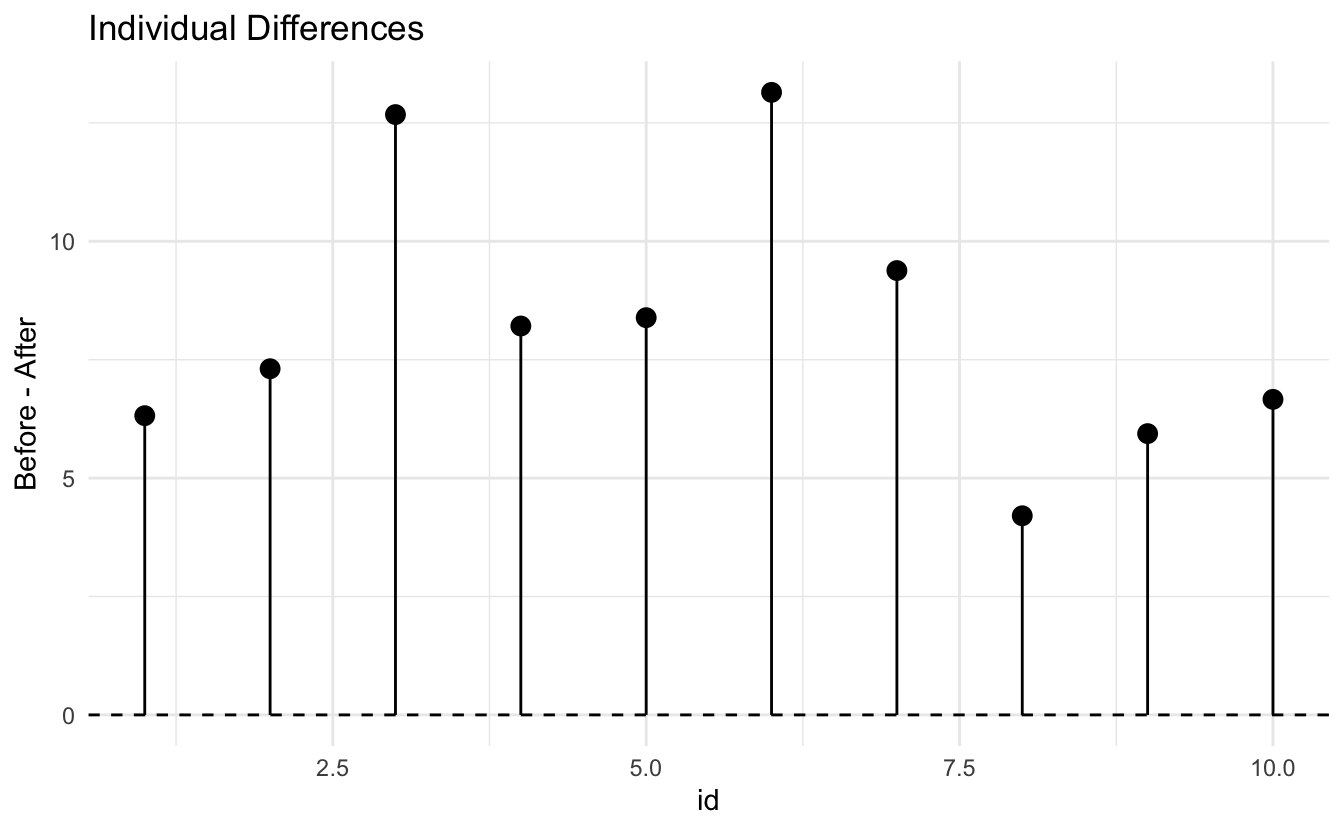
# Difference plot
diff_df <- data.frame(
id = 1:10,
difference = differences
)
ggplot(diff_df, aes(x = id, y = difference)) +
geom_hline(yintercept = 0, linetype = "dashed") +
geom_point(size = 3) +
geom_segment(aes(xend = id, yend = 0)) +
labs(title = "Individual Differences",
y = "Before - After") +
theme_minimal()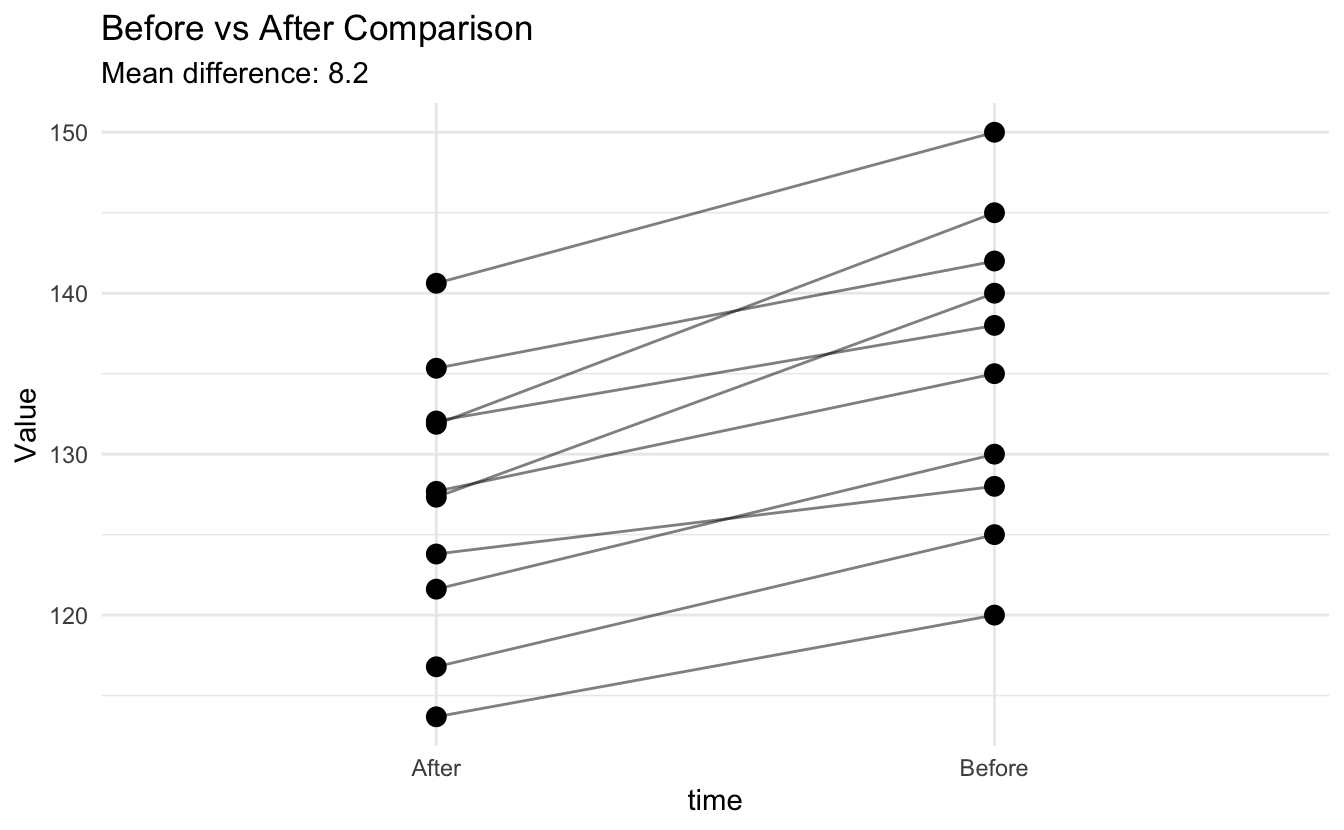
Exercise 3:
safe_t_test <- function(x, y = NULL, paired = FALSE, ...) {
# Check inputs
if (is.null(y)) {
# One-sample test
if (length(x) < 2) {
stop("Need at least 2 observations")
}
if (sd(x) == 0) {
stop("Data has no variation")
}
} else {
# Two-sample test
if (length(x) < 2) stop("x needs at least 2 observations")
if (length(y) < 2) stop("y needs at least 2 observations")
if (sd(x) == 0) stop("x has no variation")
if (sd(y) == 0) stop("y has no variation")
# Check paired
if (paired && length(x) != length(y)) {
stop("For paired test, x and y must have same length")
}
}
# Perform test
result <- t.test(x, y, paired = paired, ...)
# Format output
output <- list(
test = result,
summary = data.frame(
statistic = result$statistic,
df = result$parameter,
p_value = result$p.value,
mean_diff = ifelse(is.null(y),
result$estimate - result$null.value,
diff(result$estimate)),
ci_lower = result$conf.int[1],
ci_upper = result$conf.int[2]
)
)
# Print summary
cat("=== t-test Results ===\n")
cat("t =", round(output$summary$statistic, 3), "\n")
cat("df =", round(output$summary$df, 1), "\n")
cat("p-value =", format.pval(output$summary$p_value, digits = 3), "\n")
cat("Mean difference =", round(output$summary$mean_diff, 2), "\n")
cat("95% CI: [", round(output$summary$ci_lower, 2), ",",
round(output$summary$ci_upper, 2), "]\n")
if (output$summary$p_value < 0.05) {
cat("\nSignificant at α = 0.05\n")
} else {
cat("\nNot significant at α = 0.05\n")
}
invisible(output)
}
# Test it
safe_t_test(mtcars$mpg[mtcars$am == 0], mtcars$mpg[mtcars$am == 1])
#> === t-test Results ===
#> t = -3.767
#> df = 18.3
#> p-value = 0.00137
#> Mean difference = 7.24
#> 95% CI: [ -11.28 , -3.21 ]
#>
#> Significant at α = 0.05Exercise 4:
library(dplyr)
# 1 & 2. All pairwise comparisons
species <- levels(iris$Species)
results_list <- list()
for (i in 1:(length(species) - 1)) {
for (j in (i + 1):length(species)) {
sp1 <- species[i]
sp2 <- species[j]
data_subset <- iris %>%
filter(Species %in% c(sp1, sp2))
test <- t.test(Petal.Length ~ Species, data = data_subset)
results_list[[paste(sp1, "vs", sp2)]] <- data.frame(
comparison = paste(sp1, "vs", sp2),
mean_1 = test$estimate[1],
mean_2 = test$estimate[2],
mean_diff = diff(test$estimate),
t_stat = test$statistic,
p_value = test$p.value,
ci_lower = test$conf.int[1],
ci_upper = test$conf.int[2]
)
}
}
results_df <- do.call(rbind, results_list)
rownames(results_df) <- NULL
# 3. Adjust for multiple testing
results_df$p_adjusted <- p.adjust(results_df$p_value, method = "bonferroni")
# 4. Display results
cat("=== Pairwise Comparisons of Petal Length ===\n\n")
#> === Pairwise Comparisons of Petal Length ===
print(results_df, digits = 3)
#> comparison mean_1 mean_2 mean_diff t_stat p_value ci_lower
#> 1 setosa vs versicolor 1.46 4.26 2.80 -39.5 9.93e-46 -2.94
#> 2 setosa vs virginica 1.46 5.55 4.09 -50.0 9.27e-50 -4.25
#> 3 versicolor vs virginica 4.26 5.55 1.29 -12.6 4.90e-22 -1.50
#> ci_upper p_adjusted
#> 1 -2.66 2.98e-45
#> 2 -3.93 2.78e-49
#> 3 -1.09 1.47e-21
cat("\n=== Summary ===\n")
#>
#> === Summary ===
cat("Total comparisons:", nrow(results_df), "\n")
#> Total comparisons: 3
cat("Significant (unadjusted):", sum(results_df$p_value < 0.05), "\n")
#> Significant (unadjusted): 3
cat("Significant (Bonferroni):", sum(results_df$p_adjusted < 0.05), "\n")
#> Significant (Bonferroni): 3