Chapter 6 Predictive biomarkers
Estrogen receptor (ER) signaling is important in breast cancer cells. One approach of targeted therapy in breast cancer is the use of anti-ER agents. One question during the clinical development of anti-ER agents is the proposal of potential predictive biomarkers which might help with patient enrichment or stratification.
Let’s use the publicly available Depmap and CCLE data to identify the best gene expression predictors for ESR1 dependency (ESR1 is the gene that encodes the estrogen receptor), which can potentially serve as top predictive biomarkers. BTW, CCLE data contains the gene expression and sample annotations for a list of ~800 cancer cells lines, while the Depmap data contains , with both datasets available from the the Depmap website.
First, we will use glmnet (elastic net or lasso based methods to build model and also select features)
Then, we will use interpretable ML to understand the feature importance and obtain global/local model interpretation.
6.1 Download the Depmap and CCLE data from Depmap portal
The Depmap data could be downloaded from the following link: https://depmap.org/portal/download/all/
Three files are needed:
1. CCLE gene expression:
CCLE_expression.csv
2. Gene dependency scores for each gene in each cell line:
CRISPR_gene_effect.csv
3. metadata for the cell lines:
sample_info.csv
6.2 load required libraries
library(DescTools)## Warning: package 'DescTools' was built under R version 4.1.2##
## Attaching package: 'DescTools'## The following object is masked from 'package:data.table':
##
## %like%## The following objects are masked from 'package:psych':
##
## AUC, ICC, SD## The following objects are masked from 'package:caret':
##
## MAE, RMSErequire(RCurl); ## Loading required package: RCurl## Warning: package 'RCurl' was built under R version 4.1.2##
## Attaching package: 'RCurl'## The following object is masked from 'package:tidyr':
##
## completerequire(caret);
library(data.table) # provides enhanced data.frame
library(ggplot2) # plotting
library(glmnet) # ridge, elastic net, and lasso
library(gower)## Warning: package 'gower' was built under R version 4.1.2library(iml)## Warning: package 'iml' was built under R version 4.1.26.3 explore the data
6.3.2 CCLE
ccle_expr <- read.csv("CCLE_expression.csv")
ccle_expr[1:2,1:5]## X TSPAN6..7105. TNMD..64102. DPM1..8813. SCYL3..57147.
## 1 ACH-001113 4.331992 0.0000000 7.364397 2.792855
## 2 ACH-001289 4.566815 0.5849625 7.106537 2.5434966.3.3 Depmap crispr datt
achilles <- read.csv("CRISPR_gene_effect.csv")
achilles[1:3,1:5]## DepMap_ID A1BG..1. A1CF..29974. A2M..2. A2ML1..144568.
## 1 ACH-000001 -0.13480834 0.05976414 -0.008665348 -0.003572187
## 2 ACH-000004 0.08185267 -0.05640053 -0.106737674 -0.014498538
## 3 ACH-000005 -0.09419603 -0.01459842 0.100426025 0.169102790row.names(ccle_expr) <- ccle_expr$X
row.names(achilles) <- achilles$DepMap_ID#gene <- "ESR1"
target <- colnames(achilles)[colnames(achilles) %like any% paste0(gene,"\\..%")]
target## [1] "ESR1..2099."shared_samples = intersect(row.names(ccle_expr),row.names(achilles))6.4 calculate the correlation of CCLE gene expression with ESR1 dependency
corxy <- function(x){
Y<- as.vector(achilles[shared_samples,c(target)])
res <- cor.test(Y,x)
res$p.value ##return pvalue
}
X <- ccle_expr[shared_samples,-(1),drop=FALSE]
res_all <- apply(as.matrix(X), 2,corxy)## Warning in cor(x, y): the standard deviation is zeronames(res_all) <- colnames(X)6.4.1 Sorted the correlation results in descending order
We can see that the top 5 correlated gene expressions with ESR1 dependencies are PGR, ESR1, CST9, C5AR2, and OTOR.
ESR1 is the target Estrogen receptor itself, and PGR, which stands for progesterone receptor, is also a well-know marker in breast cancer classification and prognosis. This correlation analysis by itself is already quite interesting.
ordered_res_all <- sort(res_all, decreasing = FALSE)
ordered_res_all[1:5]## PGR..5241. ESR1..2099. CST9..128822. C5AR2..27202. OTOR..56914.
## 8.820861e-60 2.240082e-48 1.105729e-40 3.193223e-37 5.561849e-33names(ordered_res_all[1:5])## [1] "PGR..5241." "ESR1..2099." "CST9..128822." "C5AR2..27202."
## [5] "OTOR..56914."6.5 Predictive model building
Features: The CCLE gene expression for the top 100 genes whose expression correlated with ESR1 dependency in Depmap Response variable: ESR1 dependency score in Depmap
6.5.1 Let’s build a lasso model using the glmnet package
We can see that PGR and ESR1 expressions are the top predictors for ESR1 dependencies.
# glmnet requires x matrix (of predictors) and vector (values for y)
y = as.vector(achilles[shared_samples,c(target)])
#x = model.matrix(y~.,ccle_expr[shared_cell_lines,expr_genes_matched]) # matrix of predictors
expr_genes_matched <- names(ordered_res_all[1:100])
x=as.matrix(ccle_expr[shared_samples,expr_genes_matched])
scaled.x=scale(x)
set.seed(123) # replicate results
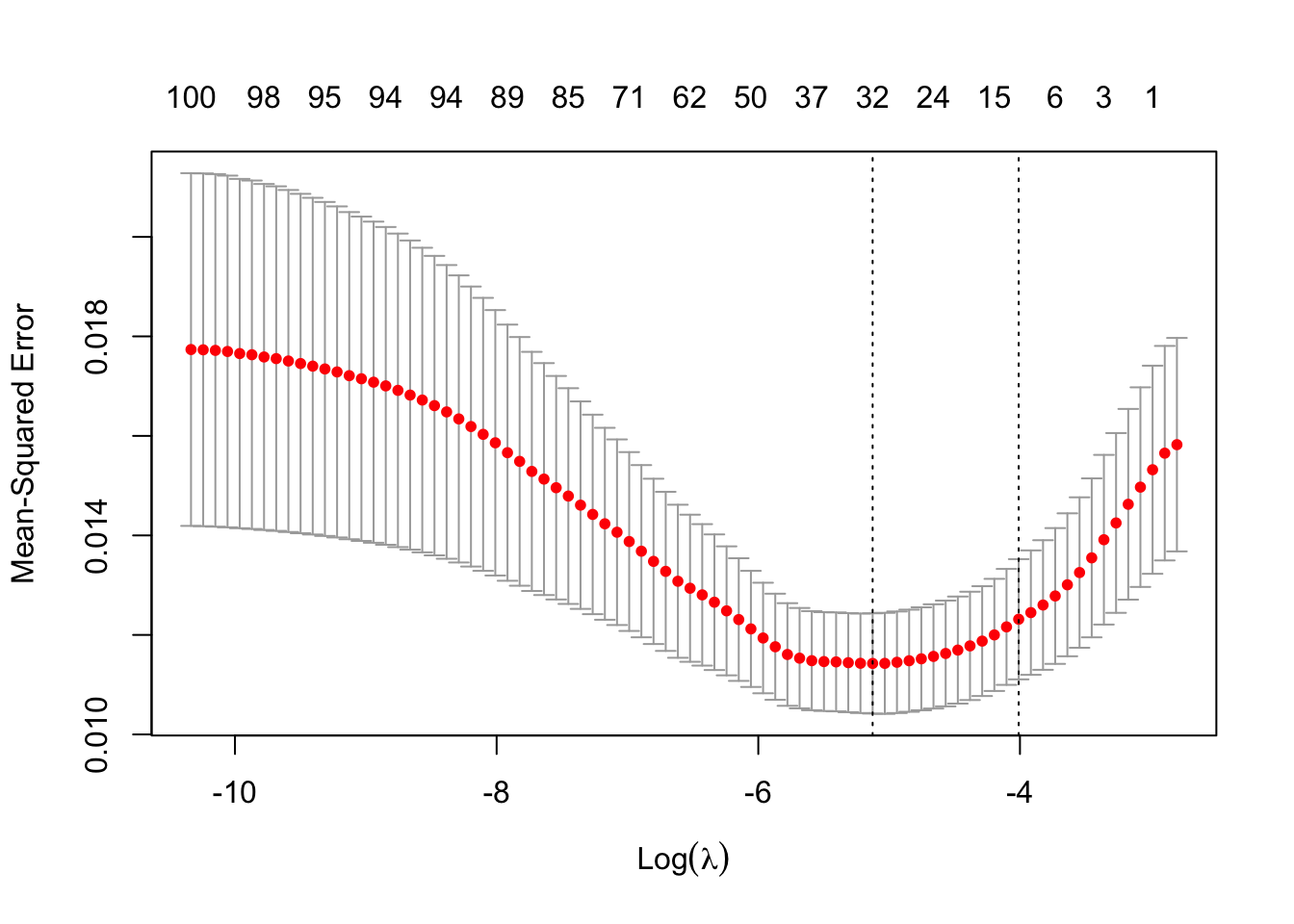
lasso_model <- cv.glmnet(scaled.x, y, alpha=1) # alpha = 1 lasso
best_lambda_la <- lasso_model$lambda.1se # largest lambda in 1 SE
lasso_coef <- lasso_model$glmnet.fit$beta[, # retrieve coefficients
lasso_model$glmnet.fit$lambda # at lambda.1se
== best_lambda_la]
coef_la = data.table(lassoReg = lasso_coef) # build table
coef_la[, feature := names(lasso_coef)] # add feature names
to_plot_r_la = melt(coef_la # label table
, id.vars='feature'
, variable.name = 'model'
, value.name = 'coefficient')
ggplot(data=to_plot_r_la, # plot coefficients
aes(x=feature, y=coefficient, fill=model)) +
coord_flip() +
geom_bar(stat='identity', fill='brown4', color='blue') +
facet_wrap(~ model) + guides(fill=FALSE) ## Warning: `guides(<scale> = FALSE)` is deprecated. Please use `guides(<scale>
## = "none")` instead.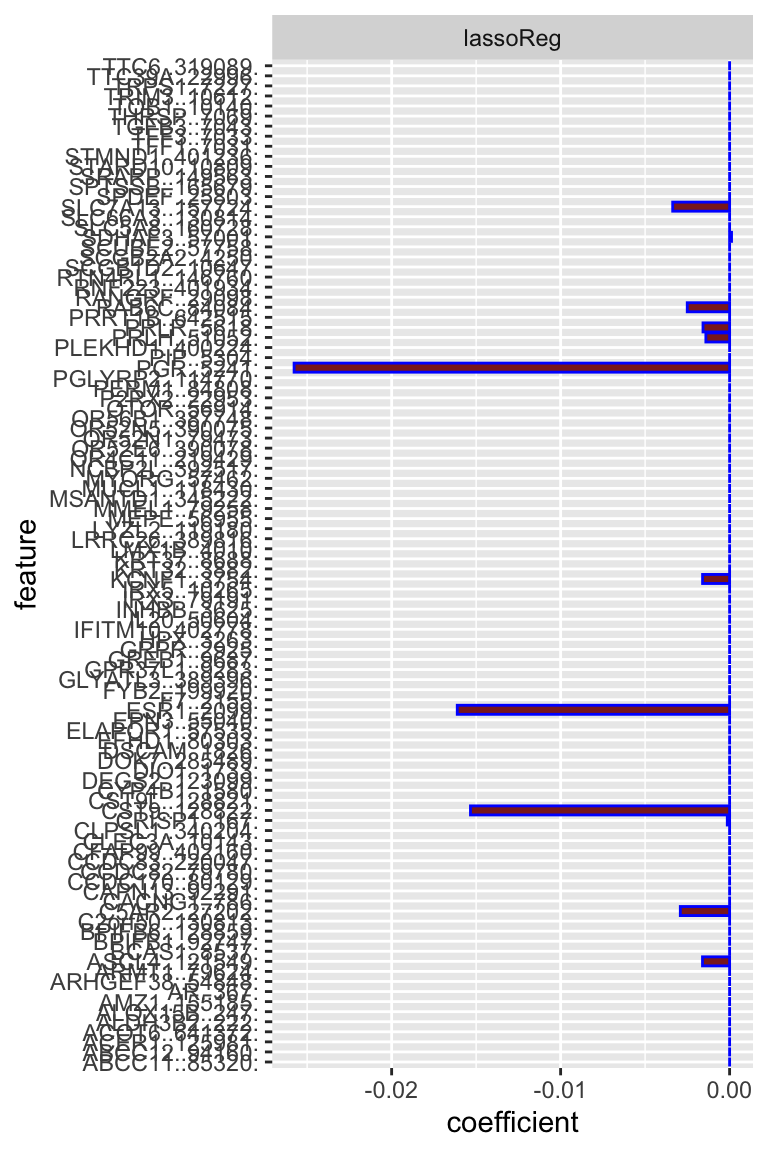 We can see the lasso model’s results are very similar to the elastic net model: PGR and ESR1 are the top predictors for ESR1 dependencies in CCLE cell lines.
We can see the lasso model’s results are very similar to the elastic net model: PGR and ESR1 are the top predictors for ESR1 dependencies in CCLE cell lines.
6.6 Let’s try to interpret the lasso model built using interpretable machine learning globally first
Use the interpretable machine learning library (iml) to illustrate the features.
Some technical details: iml needs data frame yet glmnet need matrix format input, and there needs to be a work-around. A work-around is available in the following link: https://github.com/christophM/iml/issues/29
6.6.1 Conduct global interpretation of the whole model
##adapted from the github repo above
predict.function=function(object, newdata){
newData_x = data.matrix(newdata)
results<-predict(lasso_model, newData_x)
return(results)
}
data1 <- as.data.frame(x)
colnames(data1) <- expr_genes_matched
iml_predictor <- Predictor$new(lasso_model, data = data1, y = y,
predict.fun = predict.function)
imp_features <- FeatureImp$new(iml_predictor, loss = "mse")## Warning: package 'tidyselect' was built under R version 4.1.2## Warning: package 'DT' was built under R version 4.1.2## Warning: package 'future' was built under R version 4.1.2## Warning: package 'withr' was built under R version 4.1.2## Warning: package 'Brobdingnag' was built under R version 4.1.2## Warning: package 'colorspace' was built under R version 4.1.2## Warning: package 'bayesplot' was built under R version 4.1.2## Warning: package 'rstan' was built under R version 4.1.3## Warning: package 'StanHeaders' was built under R version 4.1.3## Warning: package 'farver' was built under R version 4.1.2## Warning: package 'vctrs' was built under R version 4.1.2## Warning: package 'generics' was built under R version 4.1.2## Warning: package 'ipred' was built under R version 4.1.2## Warning: package 'xfun' was built under R version 4.1.2## Warning: package 'randomForest' was built under R version 4.1.2## Warning: package 'scales' was built under R version 4.1.2## Warning: package 'nnet' was built under R version 4.1.2## Warning: package 'globals' was built under R version 4.1.2## Warning: package 'processx' was built under R version 4.1.2## Warning: package 'lmom' was built under R version 4.1.2## Warning: package 'rlang' was built under R version 4.1.2## Warning: package 'broom' was built under R version 4.1.2## Warning: package 'checkmate' was built under R version 4.1.2## Warning: package 'inline' was built under R version 4.1.2## Warning: package 'yaml' was built under R version 4.1.2## Warning: package 'igraph' was built under R version 4.1.2## Warning: package 'httpuv' was built under R version 4.1.2## Warning: package 'posterior' was built under R version 4.1.2## Warning: package 'plyr' was built under R version 4.1.2## Warning: package 'ps' was built under R version 4.1.2## Warning: package 'rpart' was built under R version 4.1.2## Warning: package 'zoo' was built under R version 4.1.2## Warning: package 'haven' was built under R version 4.1.2## Warning: package 'magrittr' was built under R version 4.1.2## Warning: package 'matrixStats' was built under R version 4.1.2## Warning: package 'evaluate' was built under R version 4.1.2## Warning: package 'shinystan' was built under R version 4.1.2## Warning: package 'shiny' was built under R version 4.1.2## Warning: package 'readxl' was built under R version 4.1.2## Warning: package 'rstantools' was built under R version 4.1.2## Warning: package 'V8' was built under R version 4.1.2## Warning: package 'crayon' was built under R version 4.1.2## Warning: package 'htmltools' was built under R version 4.1.2## Warning: package 'tzdb' was built under R version 4.1.2## Warning: package 'RcppParallel' was built under R version 4.1.2## Warning: package 'dbplyr' was built under R version 4.1.2## Warning: package 'recipes' was built under R version 4.1.2## Warning: package 'foreach' was built under R version 4.1.2## Warning: package 'bslib' was built under R version 4.1.2## Warning: package 'hardhat' was built under R version 4.1.2## Warning: package 'distributional' was built under R version 4.1.2## Warning: package 'callr' was built under R version 4.1.2## Warning: package 'gtools' was built under R version 4.1.2## Warning: package 'nlme' was built under R version 4.1.2## Warning: package 'jsonlite' was built under R version 4.1.2## Warning: package 'fansi' was built under R version 4.1.2## Warning: package 'pillar' was built under R version 4.1.2## Warning: package 'loo' was built under R version 4.1.2## Warning: package 'httr' was built under R version 4.1.2## Warning: package 'survival' was built under R version 4.1.2## Warning: package 'glue' was built under R version 4.1.2## Warning: package 'iterators' was built under R version 4.1.2## Warning: package 'class' was built under R version 4.1.2## Warning: package 'stringi' was built under R version 4.1.2## Warning: package 'sass' was built under R version 4.1.2plot(imp_features)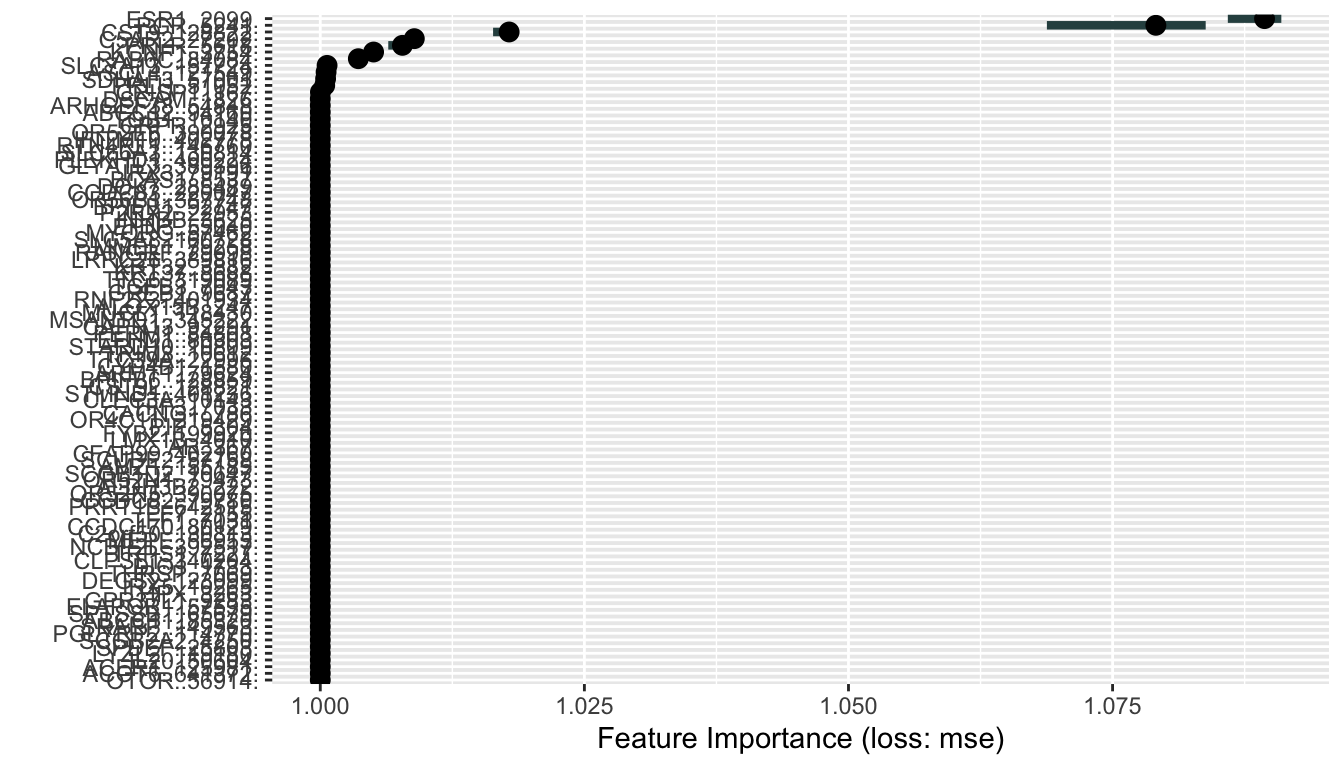
#shapley <- Shapley$new(predictor, x.interest = x[1,], sample.size = 10, run = TRUE)imp_features$results## feature importance.05 importance importance.95
## 1 ESR1..2099. 1.085909 1.089385 1.090974
## 2 PGR..5241. 1.068782 1.079097 1.083814
## 3 CST9..128822. 1.016358 1.017877 1.018489
## 4 C5AR2..27202. 1.008395 1.008901 1.009267
## 5 PRLR..5618. 1.006436 1.007775 1.008179
## 6 KCNF1..3754. 1.004201 1.005046 1.005851
## 7 RAB6C..84084. 1.003321 1.003602 1.003973
## 8 SLC7A13..157724. 1.000524 1.000642 1.000677
## 9 ASCL4..121549. 1.000434 1.000550 1.000572
## 10 SDHAF3..57001. 1.000445 1.000503 1.000683
## 11 PRLH..51052. 1.000355 1.000421 1.000472
## 12 CRISP1..167. 1.000011 1.000014 1.000016
## 13 OTOR..56914. 1.000000 1.000000 1.000000
## 14 ACOT6..641372. 1.000000 1.000000 1.000000
## 15 ACER1..125981. 1.000000 1.000000 1.000000
## 16 IL20..50604. 1.000000 1.000000 1.000000
## 17 LYZL2..119180. 1.000000 1.000000 1.000000
## 18 SPDEF..25803. 1.000000 1.000000 1.000000
## 19 SCGB2A2..4250. 1.000000 1.000000 1.000000
## 20 PGLYRP2..114770. 1.000000 1.000000 1.000000
## 21 SRARP..149563. 1.000000 1.000000 1.000000
## 22 ABCC11..85320. 1.000000 1.000000 1.000000
## 23 SPTSSB..165679. 1.000000 1.000000 1.000000
## 24 ELAPOR1..57535. 1.000000 1.000000 1.000000
## 25 GPR37L1..9283. 1.000000 1.000000 1.000000
## 26 HPX..3263. 1.000000 1.000000 1.000000
## 27 IRX5..10265. 1.000000 1.000000 1.000000
## 28 DEGS2..123099. 1.000000 1.000000 1.000000
## 29 THRSP..7069. 1.000000 1.000000 1.000000
## 30 DIO1..1733. 1.000000 1.000000 1.000000
## 31 CLPSL1..340204. 1.000000 1.000000 1.000000
## 32 TRPS1..7227. 1.000000 1.000000 1.000000
## 33 NCBP2L..392517. 1.000000 1.000000 1.000000
## 34 MEPE..56955. 1.000000 1.000000 1.000000
## 35 C2orf50..130813. 1.000000 1.000000 1.000000
## 36 CCDC170..80129. 1.000000 1.000000 1.000000
## 37 TFF1..7031. 1.000000 1.000000 1.000000
## 38 F7..2155. 1.000000 1.000000 1.000000
## 39 PRRT1B..642515. 1.000000 1.000000 1.000000
## 40 CCDC82..79780. 1.000000 1.000000 1.000000
## 41 OR52N5..390075. 1.000000 1.000000 1.000000
## 42 ALDH3B2..222. 1.000000 1.000000 1.000000
## 43 OR52N1..79473. 1.000000 1.000000 1.000000
## 44 SCGB1D2..10647. 1.000000 1.000000 1.000000
## 45 AMZ1..155185. 1.000000 1.000000 1.000000
## 46 SCUBE2..57758. 1.000000 1.000000 1.000000
## 47 CFAP99..402160. 1.000000 1.000000 1.000000
## 48 AR..367. 1.000000 1.000000 1.000000
## 49 LMX1B..4010. 1.000000 1.000000 1.000000
## 50 FYB2..199920. 1.000000 1.000000 1.000000
## 51 PIP..5304. 1.000000 1.000000 1.000000
## 52 OR4C11..219429. 1.000000 1.000000 1.000000
## 53 CACNG1..786. 1.000000 1.000000 1.000000
## 54 TFF3..7033. 1.000000 1.000000 1.000000
## 55 CLEC3A..10143. 1.000000 1.000000 1.000000
## 56 STMND1..401236. 1.000000 1.000000 1.000000
## 57 CST9L..128821. 1.000000 1.000000 1.000000
## 58 BPIFB6..128859. 1.000000 1.000000 1.000000
## 59 ARMT1..79624. 1.000000 1.000000 1.000000
## 60 CYP4B1..1580. 1.000000 1.000000 1.000000
## 61 TTC39A..22996. 1.000000 1.000000 1.000000
## 62 TRIM3..10612. 1.000000 1.000000 1.000000
## 63 STARD10..10809. 1.000000 1.000000 1.000000
## 64 EFHD1..80303. 1.000000 1.000000 1.000000
## 65 PERM1..84808. 1.000000 1.000000 1.000000
## 66 CAPN13..92291. 1.000000 1.000000 1.000000
## 67 MSANTD1..345222. 1.000000 1.000000 1.000000
## 68 MUCL1..118430. 1.000000 1.000000 1.000000
## 69 ALOX15B..247. 1.000000 1.000000 1.000000
## 70 RNF223..401934. 1.000000 1.000000 1.000000
## 71 GREB1..9687. 1.000000 1.000000 1.000000
## 72 TGFB3..7043. 1.000000 1.000000 1.000000
## 73 TTC6..319089. 1.000000 1.000000 1.000000
## 74 KRT37..8688. 1.000000 1.000000 1.000000
## 75 KRT32..3882. 1.000000 1.000000 1.000000
## 76 LRRC26..389816. 1.000000 1.000000 1.000000
## 77 RANGRF..29098. 1.000000 1.000000 1.000000
## 78 MMEL1..79258. 1.000000 1.000000 1.000000
## 79 SLC5A8..160728. 1.000000 1.000000 1.000000
## 80 MYORG..57462. 1.000000 1.000000 1.000000
## 81 EPN3..55040. 1.000000 1.000000 1.000000
## 82 INHBB..3625. 1.000000 1.000000 1.000000
## 83 P2RX2..22953. 1.000000 1.000000 1.000000
## 84 BPIFB1..92747. 1.000000 1.000000 1.000000
## 85 OR56B1..387748. 1.000000 1.000000 1.000000
## 86 CCDC83..220047. 1.000000 1.000000 1.000000
## 87 DOK7..285489. 1.000000 1.000000 1.000000
## 88 BCAS1..8537. 1.000000 1.000000 1.000000
## 89 IRX3..79191. 1.000000 1.000000 1.000000
## 90 GLYATL3..389396. 1.000000 1.000000 1.000000
## 91 PLEKHD1..400224. 1.000000 1.000000 1.000000
## 92 SLC66A3..130814. 1.000000 1.000000 1.000000
## 93 RTN4RL1..146760. 1.000000 1.000000 1.000000
## 94 IFITM10..402778. 1.000000 1.000000 1.000000
## 95 OR52E6..390078. 1.000000 1.000000 1.000000
## 96 GRPR..2925. 1.000000 1.000000 1.000000
## 97 TOB1..10140. 1.000000 1.000000 1.000000
## 98 ABCC12..94160. 1.000000 1.000000 1.000000
## 99 ARHGEF38..54848. 1.000000 1.000000 1.000000
## 100 DSCAM..1826. 1.000000 1.000000 1.000000
## permutation.error
## 1 0.01439574
## 2 0.01425979
## 3 0.01345079
## 4 0.01333217
## 5 0.01331730
## 6 0.01328124
## 7 0.01326215
## 8 0.01322304
## 9 0.01322182
## 10 0.01322121
## 11 0.01322012
## 12 0.01321474
## 13 0.01321455
## 14 0.01321455
## 15 0.01321455
## 16 0.01321455
## 17 0.01321455
## 18 0.01321455
## 19 0.01321455
## 20 0.01321455
## 21 0.01321455
## 22 0.01321455
## 23 0.01321455
## 24 0.01321455
## 25 0.01321455
## 26 0.01321455
## 27 0.01321455
## 28 0.01321455
## 29 0.01321455
## 30 0.01321455
## 31 0.01321455
## 32 0.01321455
## 33 0.01321455
## 34 0.01321455
## 35 0.01321455
## 36 0.01321455
## 37 0.01321455
## 38 0.01321455
## 39 0.01321455
## 40 0.01321455
## 41 0.01321455
## 42 0.01321455
## 43 0.01321455
## 44 0.01321455
## 45 0.01321455
## 46 0.01321455
## 47 0.01321455
## 48 0.01321455
## 49 0.01321455
## 50 0.01321455
## 51 0.01321455
## 52 0.01321455
## 53 0.01321455
## 54 0.01321455
## 55 0.01321455
## 56 0.01321455
## 57 0.01321455
## 58 0.01321455
## 59 0.01321455
## 60 0.01321455
## 61 0.01321455
## 62 0.01321455
## 63 0.01321455
## 64 0.01321455
## 65 0.01321455
## 66 0.01321455
## 67 0.01321455
## 68 0.01321455
## 69 0.01321455
## 70 0.01321455
## 71 0.01321455
## 72 0.01321455
## 73 0.01321455
## 74 0.01321455
## 75 0.01321455
## 76 0.01321455
## 77 0.01321455
## 78 0.01321455
## 79 0.01321455
## 80 0.01321455
## 81 0.01321455
## 82 0.01321455
## 83 0.01321455
## 84 0.01321455
## 85 0.01321455
## 86 0.01321455
## 87 0.01321455
## 88 0.01321455
## 89 0.01321455
## 90 0.01321455
## 91 0.01321455
## 92 0.01321455
## 93 0.01321455
## 94 0.01321455
## 95 0.01321455
## 96 0.01321455
## 97 0.01321455
## 98 0.01321455
## 99 0.01321455
## 100 0.013214556.6.2 Permutation-based feature importance measures (http://uc-r.github.io/iml-pkg)
The global interpreataion plot previously is not big enough, so it is plotted again here to clearly show that ESR1 and PGR are the two most important gene expression predictors for ESR1 dependencies in CCLE cell lines in the global model.
plot(imp_features)
6.6.3 sample info
cell_sampleinfo <- read.csv("sample_info.csv")
cell_sampleinfo[1:3,]## DepMap_ID cell_line_name stripped_cell_line_name
## 1 ACH-000016 SLR 21 SLR21
## 2 ACH-000032 MHH-CALL-3 MHHCALL3
## 3 ACH-000033 NCI-H1819 NCIH1819
## CCLE_Name alias COSMICID sex
## 1 SLR21_KIDNEY NA
## 2 MHHCALL3_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE NA Female
## 3 NCIH1819_LUNG NA Female
## source RRID WTSI_Master_Cell_ID sample_collection_site
## 1 Academic lab CVCL_V607 NA kidney
## 2 DSMZ CVCL_0089 NA bone_marrow
## 3 Academic lab CVCL_1497 NA lymph_node
## primary_or_metastasis primary_disease
## 1 Metastasis Kidney Cancer
## 2 Leukemia
## 3 Metastasis Lung Cancer
## Subtype age Sanger_Model_ID
## 1 Renal Cell Carcinoma
## 2 Acute Lymphoblastic Leukemia (ALL), B-cell 11
## 3 Non-Small Cell Lung Cancer (NSCLC), Adenocarcinoma 55
## depmap_public_comments lineage lineage_subtype lineage_sub_subtype
## 1 kidney renal_cell_carcinoma
## 2 blood ALL b_cell
## 3 lung NSCLC NSCLC_adenocarcinoma
## lineage_molecular_subtype default_growth_pattern model_manipulation
## 1
## 2
## 3
## model_manipulation_details patient_id parent_depmap_id
## 1 PT-JnARLB
## 2 PT-p2KOyI
## 3 PT-9p1WQv
## Cellosaurus_NCIt_disease Cellosaurus_NCIt_id
## 1 Clear cell renal cell carcinoma C4033
## 2 Childhood B acute lymphoblastic leukemia C9140
## 3 Lung adenocarcinoma C3512
## Cellosaurus_issues
## 1
## 2
## 3row.names(cell_sampleinfo) <- cell_sampleinfo$DepMap_IDnrow(ccle_expr[shared_samples,expr_genes_matched])## [1] 1005colnames(cell_sampleinfo)## [1] "DepMap_ID" "cell_line_name"
## [3] "stripped_cell_line_name" "CCLE_Name"
## [5] "alias" "COSMICID"
## [7] "sex" "source"
## [9] "RRID" "WTSI_Master_Cell_ID"
## [11] "sample_collection_site" "primary_or_metastasis"
## [13] "primary_disease" "Subtype"
## [15] "age" "Sanger_Model_ID"
## [17] "depmap_public_comments" "lineage"
## [19] "lineage_subtype" "lineage_sub_subtype"
## [21] "lineage_molecular_subtype" "default_growth_pattern"
## [23] "model_manipulation" "model_manipulation_details"
## [25] "patient_id" "parent_depmap_id"
## [27] "Cellosaurus_NCIt_disease" "Cellosaurus_NCIt_id"
## [29] "Cellosaurus_issues"target## [1] "ESR1..2099."k=ccle_expr[shared_samples,expr_genes_matched]
k$target_crispr <- achilles[shared_samples,]$`target`
k$ID <- row.names(k)
k<- merge(k, cell_sampleinfo,by.x="ID",by.y="DepMap_ID")k[1:5,]## ID PGR..5241. ESR1..2099. CST9..128822. C5AR2..27202.
## 1 ACH-000001 0.12432814 0.54596837 0 0.00000000
## 2 ACH-000004 0.00000000 0.12432814 0 0.02856915
## 3 ACH-000005 0.01435529 0.08406426 0 0.01435529
## 4 ACH-000007 0.01435529 0.01435529 0 0.01435529
## 5 ACH-000009 0.02856915 0.13750352 0 0.00000000
## OTOR..56914. ACOT6..641372. RAB6C..84084. ACER1..125981. PRLR..5618.
## 1 0.00000000 0.1375035 0.11103131 0 1.2868811
## 2 0.00000000 0.0000000 0.02856915 0 0.0000000
## 3 0.00000000 0.0000000 0.04264434 0 0.0000000
## 4 0.00000000 0.0000000 0.01435529 0 0.2141248
## 5 0.07038933 0.0000000 0.08406426 0 0.3673711
## IL20..50604. ASCL4..121549. LYZL2..119180. SPDEF..25803. SCGB2A2..4250.
## 1 0.11103131 0 0 0.83995959 0
## 2 0.00000000 0 0 0.18903382 0
## 3 0.00000000 0 0 0.08406426 0
## 4 0.00000000 0 0 2.81147103 0
## 5 0.02856915 0 0 0.12432814 0
## PGLYRP2..114770. SRARP..149563. ABCC11..85320. SPTSSB..165679.
## 1 0.00000000 0 0.1375035 2.28095631
## 2 0.07038933 0 0.1110313 0.01435529
## 3 0.02856915 0 0.1243281 0.00000000
## 4 0.00000000 0 0.1505597 1.69599381
## 5 0.00000000 0 0.2630344 5.52888383
## ELAPOR1..57535. GPR37L1..9283. HPX..3263. CRISP1..167. IRX5..10265.
## 1 0.3334237 0.20163386 1.00000000 0 0.0976108
## 2 0.2265085 0.07038933 0.08406426 0 0.7907720
## 3 0.1505597 0.07038933 0.02856915 0 0.7907720
## 4 2.5185351 0.09761080 0.13750352 0 1.1440464
## 5 0.1505597 0.52606881 1.35049725 0 1.7612853
## DEGS2..123099. THRSP..7069. DIO1..1733. CLPSL1..340204. TRPS1..7227.
## 1 0.2016339 1.64154603 0.1243281 0.00000000 3.64616266
## 2 1.5753123 0.16349873 0.3219281 0.00000000 0.04264434
## 3 0.4750849 0.00000000 0.0976108 0.00000000 0.09761080
## 4 4.3298412 0.02856915 0.2016339 0.00000000 0.01435529
## 5 0.3103401 0.17632277 1.7865964 0.08406426 0.13750352
## NCBP2L..392517. MEPE..56955. SLC7A13..157724. C2orf50..130813.
## 1 0.00000000 0.00000000 0 0.25096157
## 2 0.00000000 0.00000000 0 0.00000000
## 3 0.00000000 0.00000000 0 0.01435529
## 4 0.01435529 0.00000000 0 0.04264434
## 5 0.00000000 0.01435529 0 0.02856915
## CCDC170..80129. PRLH..51052. TFF1..7031. F7..2155. PRRT1B..642515.
## 1 0.1243281 0 0.4005379 0.11103131 0.0976108
## 2 0.0000000 0 0.0976108 0.04264434 0.0000000
## 3 0.0976108 0 0.2509616 0.00000000 0.0000000
## 4 2.7355222 0 4.6959938 0.07038933 0.5260688
## 5 0.5459684 0 4.5939513 3.22342255 0.7739963
## KCNF1..3754. SDHAF3..57001. CCDC82..79780. OR52N5..390075. ALDH3B2..222.
## 1 0.29865832 4.544733 3.377124 0 3.24184018
## 2 0.00000000 5.126395 5.411426 0 0.04264434
## 3 0.01435529 5.270155 5.698496 0 0.00000000
## 4 0.00000000 5.645875 3.495695 0 0.27500705
## 5 0.28688115 4.717539 5.075960 0 0.17632277
## OR52N1..79473. SCGB1D2..10647. AMZ1..155185. SCUBE2..57758.
## 1 0.00000000 1.339137 1.09085343 0.6322682
## 2 0.00000000 0.000000 0.02856915 0.1375035
## 3 0.02856915 0.000000 0.01435529 0.0000000
## 4 0.00000000 0.000000 0.16349873 0.1634987
## 5 0.00000000 0.000000 0.04264434 0.1505597
## CFAP99..402160. AR..367. LMX1B..4010. FYB2..199920. PIP..5304.
## 1 0.37851162 2.94673086 2.286881 0.7907720 0.0000000
## 2 0.25096157 0.00000000 0.000000 0.0000000 0.0000000
## 3 0.07038933 0.71369581 0.000000 0.0000000 0.0000000
## 4 0.47508488 0.02856915 0.000000 0.1505597 0.2509616
## 5 0.00000000 0.42223300 1.269033 0.6870607 0.0000000
## OR4C11..219429. CACNG1..786. TFF3..7033. CLEC3A..10143. STMND1..401236.
## 1 0 0.1890338 0.40053793 0.00000000 1.09761080
## 2 0 0.0000000 0.04264434 0.00000000 0.00000000
## 3 0 0.0000000 0.38956681 0.00000000 0.00000000
## 4 0 0.0000000 9.00292750 0.08406426 0.02856915
## 5 0 0.1110313 3.65191274 0.05658353 0.00000000
## CST9L..128821. BPIFB6..128859. ARMT1..79624. CYP4B1..1580. TTC39A..22996.
## 1 0.0000000 0.00000000 3.849999 1.79493566 2.4409522
## 2 0.8155754 0.02856915 3.989139 0.08406426 0.4541759
## 3 1.8479969 0.00000000 4.890933 0.00000000 0.3334237
## 4 0.0000000 0.00000000 4.598722 0.26303441 3.2585189
## 5 0.0000000 0.00000000 5.856239 0.00000000 3.1226727
## TRIM3..10612. STARD10..10809. EFHD1..80303. PERM1..84808. CAPN13..92291.
## 1 3.752749 5.321207 5.24260251 1.4005379 2.3561438
## 2 1.761285 2.792855 0.04264434 0.0000000 0.4005379
## 3 1.855990 2.319040 0.04264434 0.0000000 0.3673711
## 4 2.427606 7.732133 0.05658353 0.4436067 0.2387869
## 5 3.682573 9.090853 0.56559718 0.4956952 2.5921580
## MSANTD1..345222. MUCL1..118430. ALOX15B..247. RNF223..401934.
## 1 0.65076456 0.0000000 0.04264434 0.9634741
## 2 0.17632277 0.1110313 0.01435529 0.0000000
## 3 0.08406426 0.0000000 0.02856915 0.0000000
## 4 0.07038933 0.0000000 0.01435529 1.7180876
## 5 0.29865832 0.0000000 0.00000000 0.3103401
## GREB1..9687. TGFB3..7043. TTC6..319089. KRT37..8688. KRT32..3882.
## 1 1.83592407 2.3701643 0.1505597 0.0000000 0.01435529
## 2 0.18903382 0.2630344 0.0000000 0.0000000 0.00000000
## 3 0.52606881 0.4854268 0.0000000 0.0000000 0.00000000
## 4 0.02856915 0.8718436 0.0000000 0.4854268 1.07038933
## 5 0.35614381 0.9259994 1.6461627 0.0000000 0.00000000
## LRRC26..389816. RANGRF..29098. MMEL1..79258. SLC5A8..160728.
## 1 0.1763228 4.058316 1.4276062 0.1243281
## 2 0.0000000 5.603775 0.9335726 0.0000000
## 3 0.0000000 5.498570 0.2016339 0.0000000
## 4 1.0908534 4.769772 0.5655972 0.0000000
## 5 0.0000000 4.125155 0.3785116 0.0000000
## MYORG..57462. EPN3..55040. INHBB..3625. P2RX2..22953. BPIFB1..92747.
## 1 2.4672795 5.15218342 4.78083500 0.25096157 0.26303441
## 2 0.2265085 0.04264434 0.02856915 0.08406426 0.00000000
## 3 0.4646683 0.17632277 0.04264434 0.02856915 0.01435529
## 4 3.8083851 3.66220550 0.02856915 0.05658353 0.00000000
## 5 4.0338635 4.15542543 3.42088657 0.11103131 0.00000000
## OR56B1..387748. CCDC83..220047. DOK7..285489. BCAS1..8537. IRX3..79191.
## 1 0.01435529 0.00000000 3.75167795 0.36737107 0.27500705
## 2 0.00000000 0.00000000 0.04264434 0.05658353 0.00000000
## 3 0.00000000 0.01435529 0.00000000 0.00000000 0.01435529
## 4 0.00000000 0.27500705 3.34198575 2.89141919 0.02856915
## 5 0.00000000 0.04264434 0.23878686 0.26303441 1.51601515
## GLYATL3..389396. PLEKHD1..400224. SLC66A3..130814. RTN4RL1..146760.
## 1 0.00000000 1.68257330 3.082362 0.15055968
## 2 0.00000000 0.04264434 2.575312 0.00000000
## 3 0.02856915 0.00000000 3.097611 0.01435529
## 4 0.00000000 0.07038933 3.526069 0.00000000
## 5 0.02856915 0.08406426 3.176323 0.12432814
## IFITM10..402778. OR52E6..390078. GRPR..2925. TOB1..10140. ABCC12..94160.
## 1 0.40053793 0 0.07038933 5.338068 0.00000000
## 2 0.13750352 0 0.04264434 1.673556 0.00000000
## 3 0.01435529 0 0.01435529 1.941106 0.00000000
## 4 2.44095220 0 0.22650853 5.382321 0.07038933
## 5 0.05658353 0 0.04264434 5.677790 0.00000000
## ARHGEF38..54848. DSCAM..1826. cell_line_name stripped_cell_line_name
## 1 0.04264434 0.00000000 NIH:OVCAR-3 NIHOVCAR3
## 2 0.01435529 0.01435529 HEL HEL
## 3 0.00000000 0.00000000 HEL 92.1.7 HEL9217
## 4 2.41683974 0.00000000 LS513 LS513
## 5 0.83187724 0.00000000 C2BBe1 C2BBE1
## CCLE_Name alias COSMICID sex source
## 1 NIHOVCAR3_OVARY OVCAR3 905933 Female ATCC
## 2 HEL_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE 907053 Male DSMZ
## 3 HEL9217_HAEMATOPOIETIC_AND_LYMPHOID_TISSUE NA Male ATCC
## 4 LS513_LARGE_INTESTINE 907795 Male ATCC
## 5 C2BBE1_LARGE_INTESTINE 910700 Male ATCC
## RRID WTSI_Master_Cell_ID sample_collection_site
## 1 CVCL_0465 2201 ascites
## 2 CVCL_0001 783 haematopoietic_and_lymphoid_tissue
## 3 CVCL_2481 NA bone_marrow
## 4 CVCL_1386 569 large_intestine
## 5 CVCL_1096 2104 Colon
## primary_or_metastasis primary_disease
## 1 Metastasis Ovarian Cancer
## 2 Leukemia
## 3 Leukemia
## 4 Primary Colon/Colorectal Cancer
## 5 Primary Colon/Colorectal Cancer
## Subtype age
## 1 Adenocarcinoma, high grade serous 60
## 2 Acute Myelogenous Leukemia (AML), M6 (Erythroleukemia) 30
## 3 Acute Myelogenous Leukemia (AML), M6 (Erythroleukemia) 30
## 4 Adenocarcinoma 63
## 5 Adenocarcinoma 72
## Sanger_Model_ID depmap_public_comments lineage
## 1 SIDM00105 ovary
## 2 SIDM00594 blood
## 3 SIDM00593 blood
## 4 SIDM00677 colorectal
## 5 SIDM01233 colorectal
## lineage_subtype lineage_sub_subtype lineage_molecular_subtype
## 1 ovary_adenocarcinoma high_grade_serous
## 2 AML M6
## 3 AML M6
## 4 colorectal_adenocarcinoma
## 5 colorectal_adenocarcinoma
## default_growth_pattern model_manipulation model_manipulation_details
## 1 2D: adherent
## 2 2D: suspension
## 3 2D: suspension
## 4 2D: adherent
## 5 2D: adherent
## patient_id parent_depmap_id Cellosaurus_NCIt_disease
## 1 PT-gj46wT High grade ovarian serous adenocarcinoma
## 2 PT-q4K2cp Erythroleukemia
## 3 PT-q4K2cp ACH-000004 Erythroleukemia
## 4 PT-NOXwpH Cecum adenocarcinoma
## 5 PT-puKIyc ACH-000003 Colon adenocarcinoma
## Cellosaurus_NCIt_id Cellosaurus_issues
## 1 C105555
## 2 C7152
## 3 C7152
## 4 C5543
## 5 C4349colnames(cell_sampleinfo)## [1] "DepMap_ID" "cell_line_name"
## [3] "stripped_cell_line_name" "CCLE_Name"
## [5] "alias" "COSMICID"
## [7] "sex" "source"
## [9] "RRID" "WTSI_Master_Cell_ID"
## [11] "sample_collection_site" "primary_or_metastasis"
## [13] "primary_disease" "Subtype"
## [15] "age" "Sanger_Model_ID"
## [17] "depmap_public_comments" "lineage"
## [19] "lineage_subtype" "lineage_sub_subtype"
## [21] "lineage_molecular_subtype" "default_growth_pattern"
## [23] "model_manipulation" "model_manipulation_details"
## [25] "patient_id" "parent_depmap_id"
## [27] "Cellosaurus_NCIt_disease" "Cellosaurus_NCIt_id"
## [29] "Cellosaurus_issues"cell_sampleinfo_in_the_same_order<- cell_sampleinfo[shared_samples,]6.6.4 distribution of the Depmap crispr dependency score, the smaller the value, the bigger the dependency is
hist(y)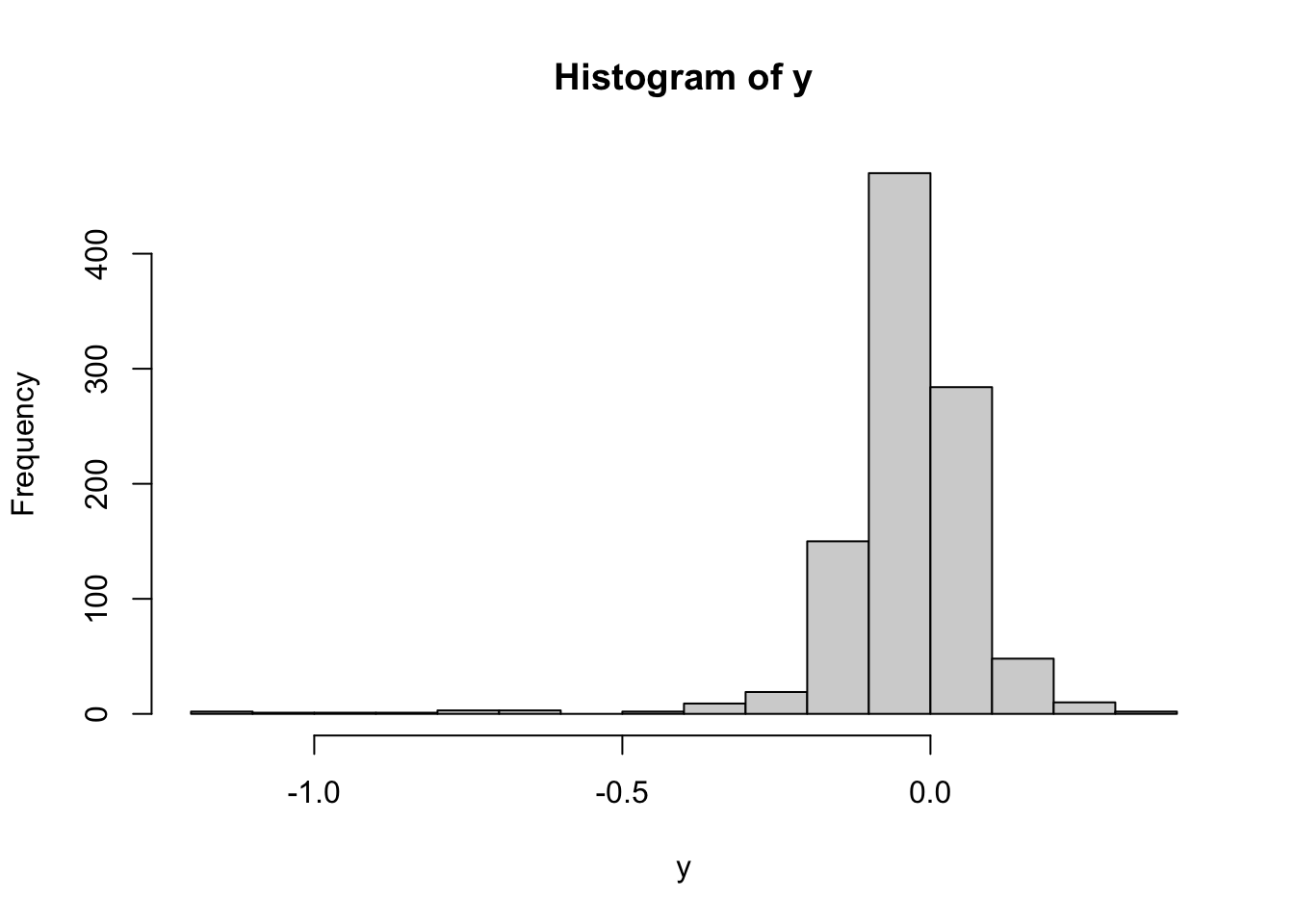
index_less_than_minus_point_five <- which (y< (-0.5))sample_records_less_than_minus_point_five <- cell_sampleinfo_in_the_same_order[which (y< (-0.5)),]sample_records_less_than_minus_point_five$Index_number <- index_less_than_minus_point_fivecolnames(sample_records_less_than_minus_point_five)## [1] "DepMap_ID" "cell_line_name"
## [3] "stripped_cell_line_name" "CCLE_Name"
## [5] "alias" "COSMICID"
## [7] "sex" "source"
## [9] "RRID" "WTSI_Master_Cell_ID"
## [11] "sample_collection_site" "primary_or_metastasis"
## [13] "primary_disease" "Subtype"
## [15] "age" "Sanger_Model_ID"
## [17] "depmap_public_comments" "lineage"
## [19] "lineage_subtype" "lineage_sub_subtype"
## [21] "lineage_molecular_subtype" "default_growth_pattern"
## [23] "model_manipulation" "model_manipulation_details"
## [25] "patient_id" "parent_depmap_id"
## [27] "Cellosaurus_NCIt_disease" "Cellosaurus_NCIt_id"
## [29] "Cellosaurus_issues" "Index_number"6.7 Let’s use local interpretatble machine learning to learn about what are the top predictors for the individual cell lines
6.7.1 Let’s examine the top CCLE cancer cell lines that are dependent on EST1 for survival
sample_records_less_than_minus_point_five## DepMap_ID cell_line_name stripped_cell_line_name
## ACH-000019 ACH-000019 MCF7 MCF7
## ACH-000330 ACH-000330 EFM-19 EFM19
## ACH-001630 ACH-001630 PEO1 PEO1
## ACH-000876 ACH-000876 MDA-MB-415 MDAMB415
## ACH-000028 ACH-000028 KPL-1 KPL1
## ACH-000277 ACH-000277 HCC1419 HCC1419
## ACH-000783 ACH-000783 CAMA-1 CAMA1
## ACH-000147 ACH-000147 T-47D T47D
## ACH-000352 ACH-000352 HCC1428 HCC1428
## ACH-001632 ACH-001632 PEO4 PEO4
## ACH-000934 ACH-000934 MDA-MB-361 MDAMB361
## CCLE_Name alias COSMICID sex source RRID
## ACH-000019 MCF7_BREAST 905946 Female ATCC CVCL_0031
## ACH-000330 EFM19_BREAST 906851 Female DSMZ CVCL_0253
## ACH-001630 PEO1_OVARY 1480372 Female Sigma-Aldrich CVCL_2686
## ACH-000876 MDAMB415_BREAST 924240 Female ATCC CVCL_0621
## ACH-000028 KPL1_BREAST NA Female DSMZ CVCL_2094
## ACH-000277 HCC1419_BREAST 907045 Female ATCC CVCL_1251
## ACH-000783 CAMA1_BREAST 946382 Female ATCC CVCL_1115
## ACH-000147 T47D_BREAST 905945 Female ATCC CVCL_0553
## ACH-000352 HCC1428_BREAST 1290905 Female ATCC CVCL_1252
## ACH-001632 PEO4_OVARY NA Female Sigma-Aldrich CVCL_2690
## ACH-000934 MDAMB361_BREAST 908121 Female ATCC CVCL_0620
## WTSI_Master_Cell_ID sample_collection_site primary_or_metastasis
## ACH-000019 588 pleural_effusion Metastasis
## ACH-000330 1786 pleural_effusion Metastasis
## ACH-001630 177 ascites Metastasis
## ACH-000876 344 pleural_effusion Metastasis
## ACH-000028 NA pleural_effusion Metastasis
## ACH-000277 1551 breast Primary
## ACH-000783 363 pleural_effusion Metastasis
## ACH-000147 1286 pleural_effusion Metastasis
## ACH-000352 1567 pleural_effusion Metastasis
## ACH-001632 NA ascites Metastasis
## ACH-000934 880 central_nervous_system Metastasis
## primary_disease Subtype age Sanger_Model_ID
## ACH-000019 Breast Cancer Carcinoma 69 SIDM00148
## ACH-000330 Breast Cancer Breast Ductal Carcinoma 50 SIDM01056
## ACH-001630 Ovarian Cancer Cystadenocarcinoma SIDM00472
## ACH-000876 Breast Cancer Carcinoma 38 SIDM00630
## ACH-000028 Breast Cancer Carcinoma 69 SIDM00147
## ACH-000277 Breast Cancer Breast Ductal Carcinoma 42 SIDM00882
## ACH-000783 Breast Cancer Carcinoma 51 SIDM00920
## ACH-000147 Breast Cancer Breast Ductal Carcinoma 54 SIDM00097
## ACH-000352 Breast Cancer Carcinoma 49 SIDM00881
## ACH-001632 Ovarian Cancer Cystadenocarcinoma SIDM00471
## ACH-000934 Breast Cancer Carcinoma 40 SIDM00528
## depmap_public_comments
## ACH-000019
## ACH-000330
## ACH-001630
## ACH-000876
## ACH-000028 SNP fingerprinting confirms that this line is an MCF-7 derivative. DepMap metadata has been updated (as of 22Q2) to reflect this.
## ACH-000277
## ACH-000783
## ACH-000147
## ACH-000352
## ACH-001632
## ACH-000934
## lineage lineage_subtype lineage_sub_subtype
## ACH-000019 breast breast_carcinoma ERpos_HER2neg
## ACH-000330 breast breast_ductal_carcinoma ERpos_HER2neg
## ACH-001630 ovary ovary_adenocarcinoma
## ACH-000876 breast breast_carcinoma ERpos_HER2neg
## ACH-000028 breast breast_carcinoma ERpos_HER2neg
## ACH-000277 breast breast_ductal_carcinoma ERpos_HER2pos
## ACH-000783 breast breast_carcinoma ERpos_HER2neg
## ACH-000147 breast breast_ductal_carcinoma ERpos_HER2neg
## ACH-000352 breast breast_carcinoma ERpos_HER2neg
## ACH-001632 ovary ovary_adenocarcinoma
## ACH-000934 breast breast_carcinoma ERpos_HER2pos
## lineage_molecular_subtype default_growth_pattern
## ACH-000019 luminal 2D: adherent
## ACH-000330 luminal
## ACH-001630 2D: adherent
## ACH-000876 HER2_amp 2D: adherent
## ACH-000028 luminal 2D: adherent
## ACH-000277 HER2_amp
## ACH-000783 luminal 2D: adherent
## ACH-000147 luminal 2D: adherent
## ACH-000352 luminal
## ACH-001632 2D: adherent
## ACH-000934 HER2_amp 2D: adherent
## model_manipulation model_manipulation_details patient_id
## ACH-000019 PT-viJKnw
## ACH-000330 PT-ykXL80
## ACH-001630 PT-FMp5RI
## ACH-000876 PT-FjJcwU
## ACH-000028 PT-viJKnw
## ACH-000277 PT-osxHUh
## ACH-000783 PT-dK1tBm
## ACH-000147 PT-9WkxWW
## ACH-000352 PT-jBpXJh
## ACH-001632 PT-FMp5RI
## ACH-000934 PT-0VWXIm
## parent_depmap_id Cellosaurus_NCIt_disease
## ACH-000019 Invasive breast carcinoma of no special type
## ACH-000330 Breast ductal carcinoma
## ACH-001630 BRCA2 syndrome
## ACH-000876 Breast adenocarcinoma
## ACH-000028 ACH-000019 Invasive breast carcinoma of no special type
## ACH-000277 Breast ductal carcinoma
## ACH-000783 Breast adenocarcinoma
## ACH-000147 Invasive breast carcinoma of no special type
## ACH-000352 Breast adenocarcinoma
## ACH-001632 BRCA2 syndrome
## ACH-000934 Breast adenocarcinoma
## Cellosaurus_NCIt_id
## ACH-000019 C4194
## ACH-000330 C4017
## ACH-001630 C36101
## ACH-000876 C5214
## ACH-000028 C4194
## ACH-000277 C4017
## ACH-000783 C5214
## ACH-000147 C4194
## ACH-000352 C5214
## ACH-001632 C36101
## ACH-000934 C5214
## Cellosaurus_issues
## ACH-000019
## ACH-000330
## ACH-001630
## ACH-000876
## ACH-000028 Contaminated. Shown to be a MCF-7 derivative (PubMed=18304946; PubMed=20143388).
## ACH-000277
## ACH-000783
## ACH-000147
## ACH-000352
## ACH-001632
## ACH-000934
## Index_number
## ACH-000019 104
## ACH-000330 106
## ACH-001630 278
## ACH-000876 290
## ACH-000028 423
## ACH-000277 432
## ACH-000783 596
## ACH-000147 696
## ACH-000352 744
## ACH-001632 967
## ACH-000934 987sample_records_less_than_minus_point_five[sample_records_less_than_minus_point_five$primary_disease %in% c("Breast Cancer"),]$Index_number## [1] 104 106 290 423 432 596 696 744 987sample_records_less_than_minus_point_five[sample_records_less_than_minus_point_five$Index_number %in% c(104,106,290,423,432,596,696,744,987),]## DepMap_ID cell_line_name stripped_cell_line_name
## ACH-000019 ACH-000019 MCF7 MCF7
## ACH-000330 ACH-000330 EFM-19 EFM19
## ACH-000876 ACH-000876 MDA-MB-415 MDAMB415
## ACH-000028 ACH-000028 KPL-1 KPL1
## ACH-000277 ACH-000277 HCC1419 HCC1419
## ACH-000783 ACH-000783 CAMA-1 CAMA1
## ACH-000147 ACH-000147 T-47D T47D
## ACH-000352 ACH-000352 HCC1428 HCC1428
## ACH-000934 ACH-000934 MDA-MB-361 MDAMB361
## CCLE_Name alias COSMICID sex source RRID
## ACH-000019 MCF7_BREAST 905946 Female ATCC CVCL_0031
## ACH-000330 EFM19_BREAST 906851 Female DSMZ CVCL_0253
## ACH-000876 MDAMB415_BREAST 924240 Female ATCC CVCL_0621
## ACH-000028 KPL1_BREAST NA Female DSMZ CVCL_2094
## ACH-000277 HCC1419_BREAST 907045 Female ATCC CVCL_1251
## ACH-000783 CAMA1_BREAST 946382 Female ATCC CVCL_1115
## ACH-000147 T47D_BREAST 905945 Female ATCC CVCL_0553
## ACH-000352 HCC1428_BREAST 1290905 Female ATCC CVCL_1252
## ACH-000934 MDAMB361_BREAST 908121 Female ATCC CVCL_0620
## WTSI_Master_Cell_ID sample_collection_site primary_or_metastasis
## ACH-000019 588 pleural_effusion Metastasis
## ACH-000330 1786 pleural_effusion Metastasis
## ACH-000876 344 pleural_effusion Metastasis
## ACH-000028 NA pleural_effusion Metastasis
## ACH-000277 1551 breast Primary
## ACH-000783 363 pleural_effusion Metastasis
## ACH-000147 1286 pleural_effusion Metastasis
## ACH-000352 1567 pleural_effusion Metastasis
## ACH-000934 880 central_nervous_system Metastasis
## primary_disease Subtype age Sanger_Model_ID
## ACH-000019 Breast Cancer Carcinoma 69 SIDM00148
## ACH-000330 Breast Cancer Breast Ductal Carcinoma 50 SIDM01056
## ACH-000876 Breast Cancer Carcinoma 38 SIDM00630
## ACH-000028 Breast Cancer Carcinoma 69 SIDM00147
## ACH-000277 Breast Cancer Breast Ductal Carcinoma 42 SIDM00882
## ACH-000783 Breast Cancer Carcinoma 51 SIDM00920
## ACH-000147 Breast Cancer Breast Ductal Carcinoma 54 SIDM00097
## ACH-000352 Breast Cancer Carcinoma 49 SIDM00881
## ACH-000934 Breast Cancer Carcinoma 40 SIDM00528
## depmap_public_comments
## ACH-000019
## ACH-000330
## ACH-000876
## ACH-000028 SNP fingerprinting confirms that this line is an MCF-7 derivative. DepMap metadata has been updated (as of 22Q2) to reflect this.
## ACH-000277
## ACH-000783
## ACH-000147
## ACH-000352
## ACH-000934
## lineage lineage_subtype lineage_sub_subtype
## ACH-000019 breast breast_carcinoma ERpos_HER2neg
## ACH-000330 breast breast_ductal_carcinoma ERpos_HER2neg
## ACH-000876 breast breast_carcinoma ERpos_HER2neg
## ACH-000028 breast breast_carcinoma ERpos_HER2neg
## ACH-000277 breast breast_ductal_carcinoma ERpos_HER2pos
## ACH-000783 breast breast_carcinoma ERpos_HER2neg
## ACH-000147 breast breast_ductal_carcinoma ERpos_HER2neg
## ACH-000352 breast breast_carcinoma ERpos_HER2neg
## ACH-000934 breast breast_carcinoma ERpos_HER2pos
## lineage_molecular_subtype default_growth_pattern
## ACH-000019 luminal 2D: adherent
## ACH-000330 luminal
## ACH-000876 HER2_amp 2D: adherent
## ACH-000028 luminal 2D: adherent
## ACH-000277 HER2_amp
## ACH-000783 luminal 2D: adherent
## ACH-000147 luminal 2D: adherent
## ACH-000352 luminal
## ACH-000934 HER2_amp 2D: adherent
## model_manipulation model_manipulation_details patient_id
## ACH-000019 PT-viJKnw
## ACH-000330 PT-ykXL80
## ACH-000876 PT-FjJcwU
## ACH-000028 PT-viJKnw
## ACH-000277 PT-osxHUh
## ACH-000783 PT-dK1tBm
## ACH-000147 PT-9WkxWW
## ACH-000352 PT-jBpXJh
## ACH-000934 PT-0VWXIm
## parent_depmap_id Cellosaurus_NCIt_disease
## ACH-000019 Invasive breast carcinoma of no special type
## ACH-000330 Breast ductal carcinoma
## ACH-000876 Breast adenocarcinoma
## ACH-000028 ACH-000019 Invasive breast carcinoma of no special type
## ACH-000277 Breast ductal carcinoma
## ACH-000783 Breast adenocarcinoma
## ACH-000147 Invasive breast carcinoma of no special type
## ACH-000352 Breast adenocarcinoma
## ACH-000934 Breast adenocarcinoma
## Cellosaurus_NCIt_id
## ACH-000019 C4194
## ACH-000330 C4017
## ACH-000876 C5214
## ACH-000028 C4194
## ACH-000277 C4017
## ACH-000783 C5214
## ACH-000147 C4194
## ACH-000352 C5214
## ACH-000934 C5214
## Cellosaurus_issues
## ACH-000019
## ACH-000330
## ACH-000876
## ACH-000028 Contaminated. Shown to be a MCF-7 derivative (PubMed=18304946; PubMed=20143388).
## ACH-000277
## ACH-000783
## ACH-000147
## ACH-000352
## ACH-000934
## Index_number
## ACH-000019 104
## ACH-000330 106
## ACH-000876 290
## ACH-000028 423
## ACH-000277 432
## ACH-000783 596
## ACH-000147 696
## ACH-000352 744
## ACH-000934 9876.7.2 Let’s zoom in on the top CCLE breast cancer cell lines that are dependent on EST1 for survival: we can see that they can be divided into two categories of “luminal” and “HER2_amp”. Let’s next use local interpretation to interpret each individual cell to see what the most important gene expression predictors are.
sample_records_less_than_minus_point_five[sample_records_less_than_minus_point_five$Index_number %in% c(104,106,290,423,432,596,696,744,987),c("cell_line_name","lineage_molecular_subtype","Index_number","CCLE_Name")]## cell_line_name lineage_molecular_subtype Index_number
## ACH-000019 MCF7 luminal 104
## ACH-000330 EFM-19 luminal 106
## ACH-000876 MDA-MB-415 HER2_amp 290
## ACH-000028 KPL-1 luminal 423
## ACH-000277 HCC1419 HER2_amp 432
## ACH-000783 CAMA-1 luminal 596
## ACH-000147 T-47D luminal 696
## ACH-000352 HCC1428 luminal 744
## ACH-000934 MDA-MB-361 HER2_amp 987
## CCLE_Name
## ACH-000019 MCF7_BREAST
## ACH-000330 EFM19_BREAST
## ACH-000876 MDAMB415_BREAST
## ACH-000028 KPL1_BREAST
## ACH-000277 HCC1419_BREAST
## ACH-000783 CAMA1_BREAST
## ACH-000147 T47D_BREAST
## ACH-000352 HCC1428_BREAST
## ACH-000934 MDAMB361_BREAST6.7.3 check the luminal ones in the most dependent cell lines (CRES score <-0.5)
For the example below, we can see that for luminal breast cancer cell lines that are dependent on ESR1 for survival. The top predictor for each individual cell’s ESR1 dependency are oftentimes PGR rather than ESR1 itself, with the two of them as the top two predictors. This suggest that PGR is probably a very good indicater of ESR1 pathway activity and potentially a good prognostic or predictive biomarkers in luminal breast cancers.
lime.explain26 <- LocalModel$new(iml_predictor, k=10,x.interest = data1[104, ])## Warning in private$aggregate(): Had to choose a smaller kplot(lime.explain26)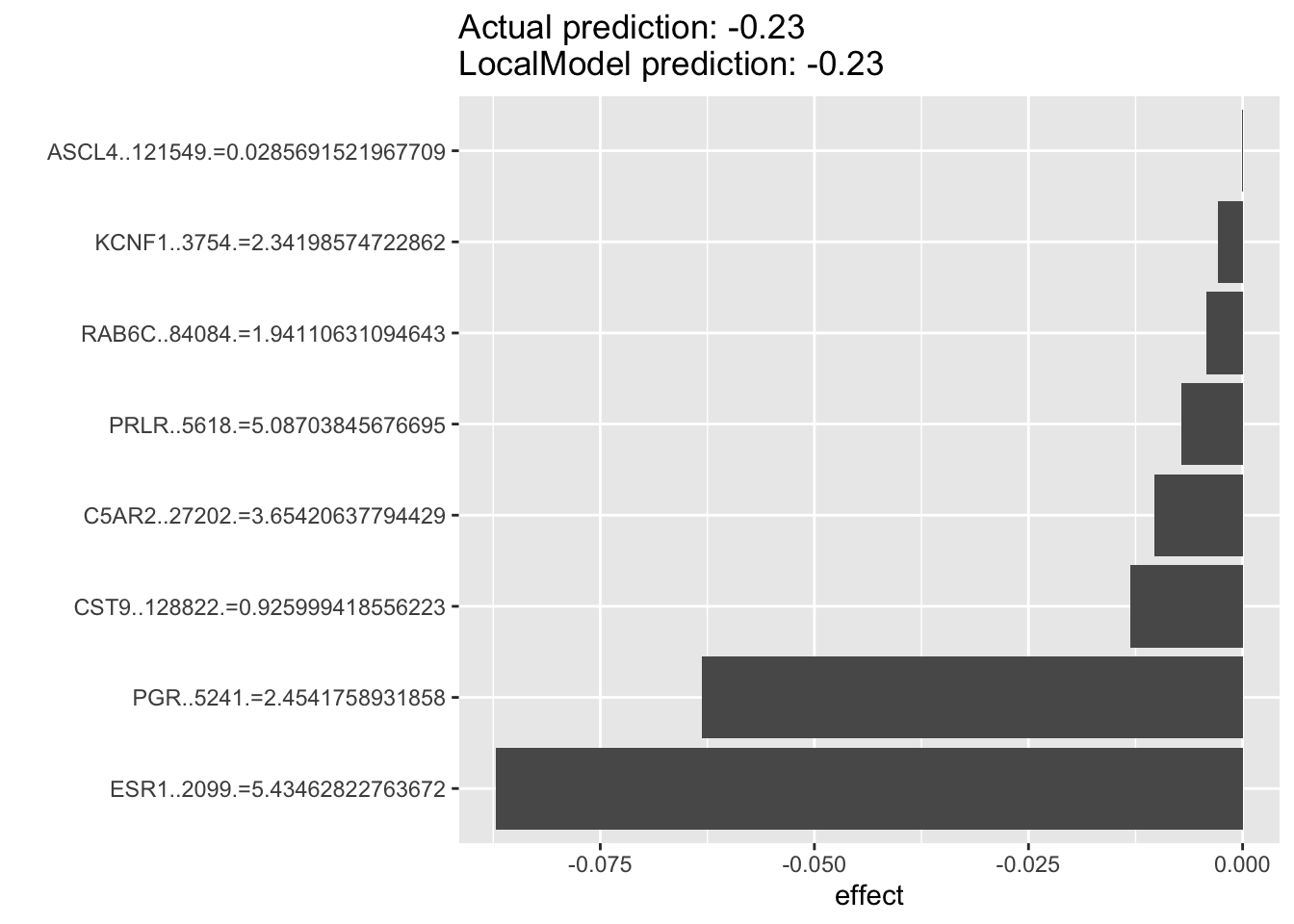
lime.explain209 <- LocalModel$new(iml_predictor, k=10,x.interest = data1[106, ])## Warning in private$aggregate(): Had to choose a smaller kplot(lime.explain209)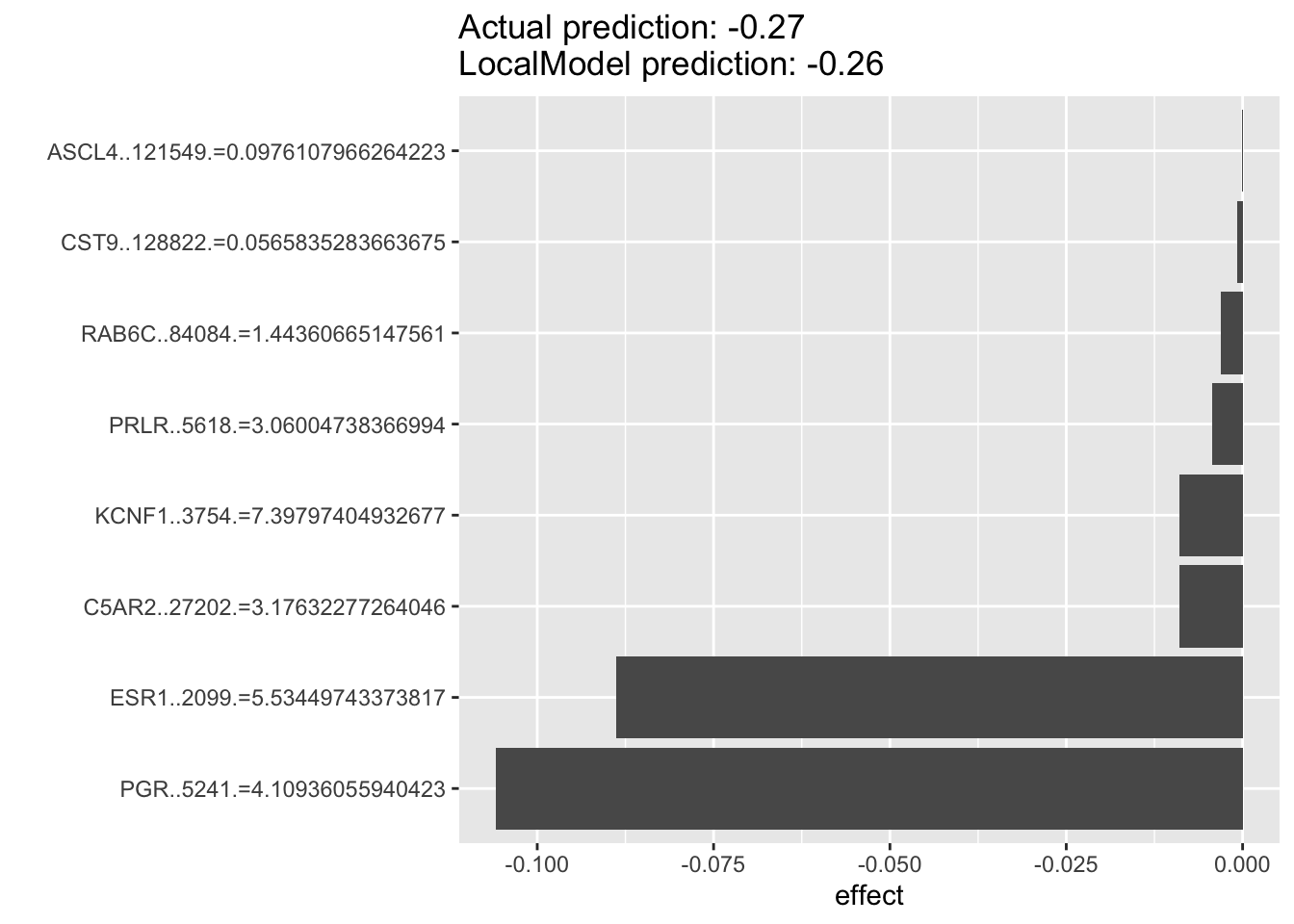
lime.explain209 <- LocalModel$new(iml_predictor, k=10,x.interest = data1[106, ])## Warning in private$aggregate(): Had to choose a smaller kplot(lime.explain209)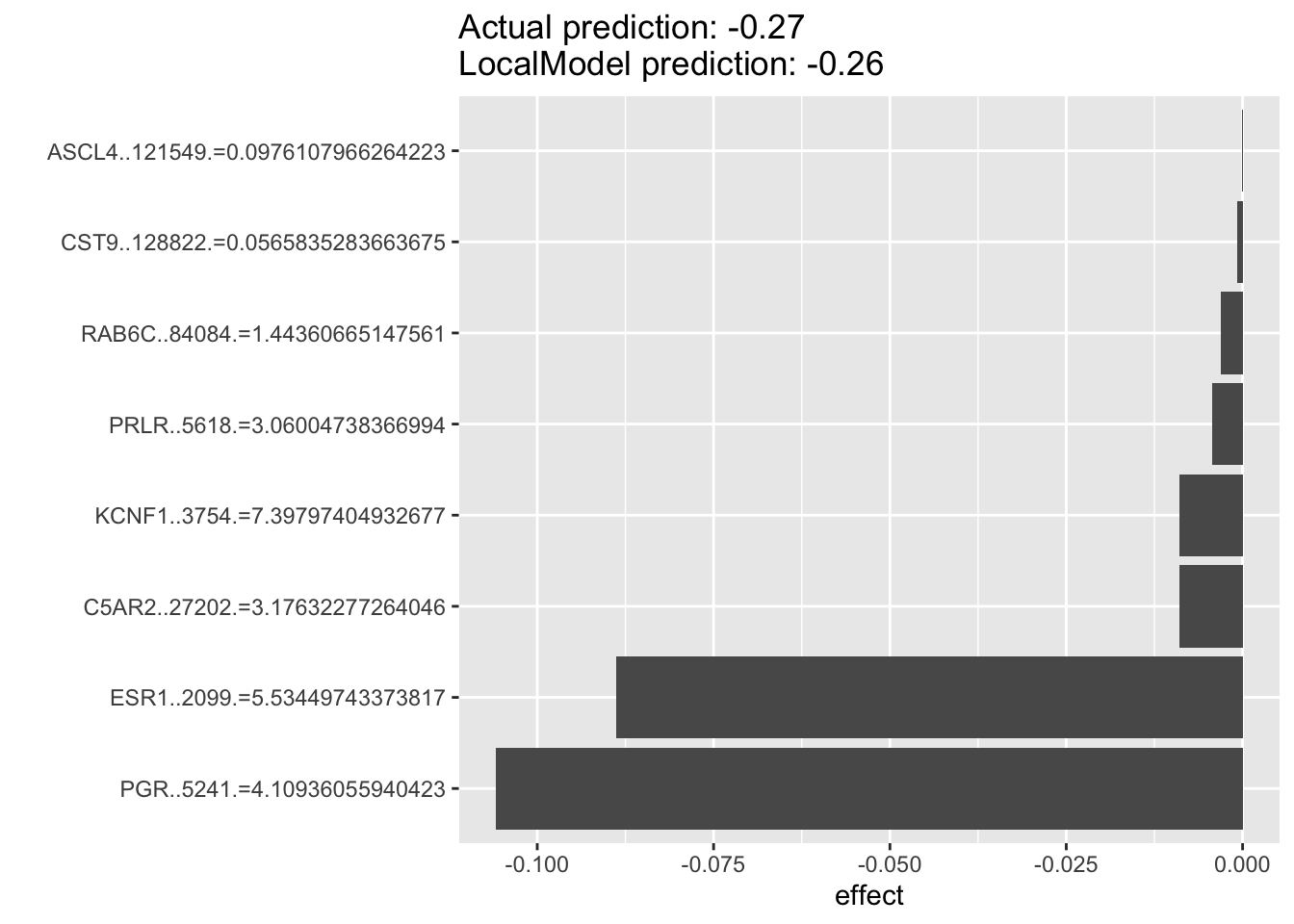
lime.explain744 <- LocalModel$new(iml_predictor, k=10,x.interest = data1[744, ])## Warning in private$aggregate(): Had to choose a smaller kplot(lime.explain744)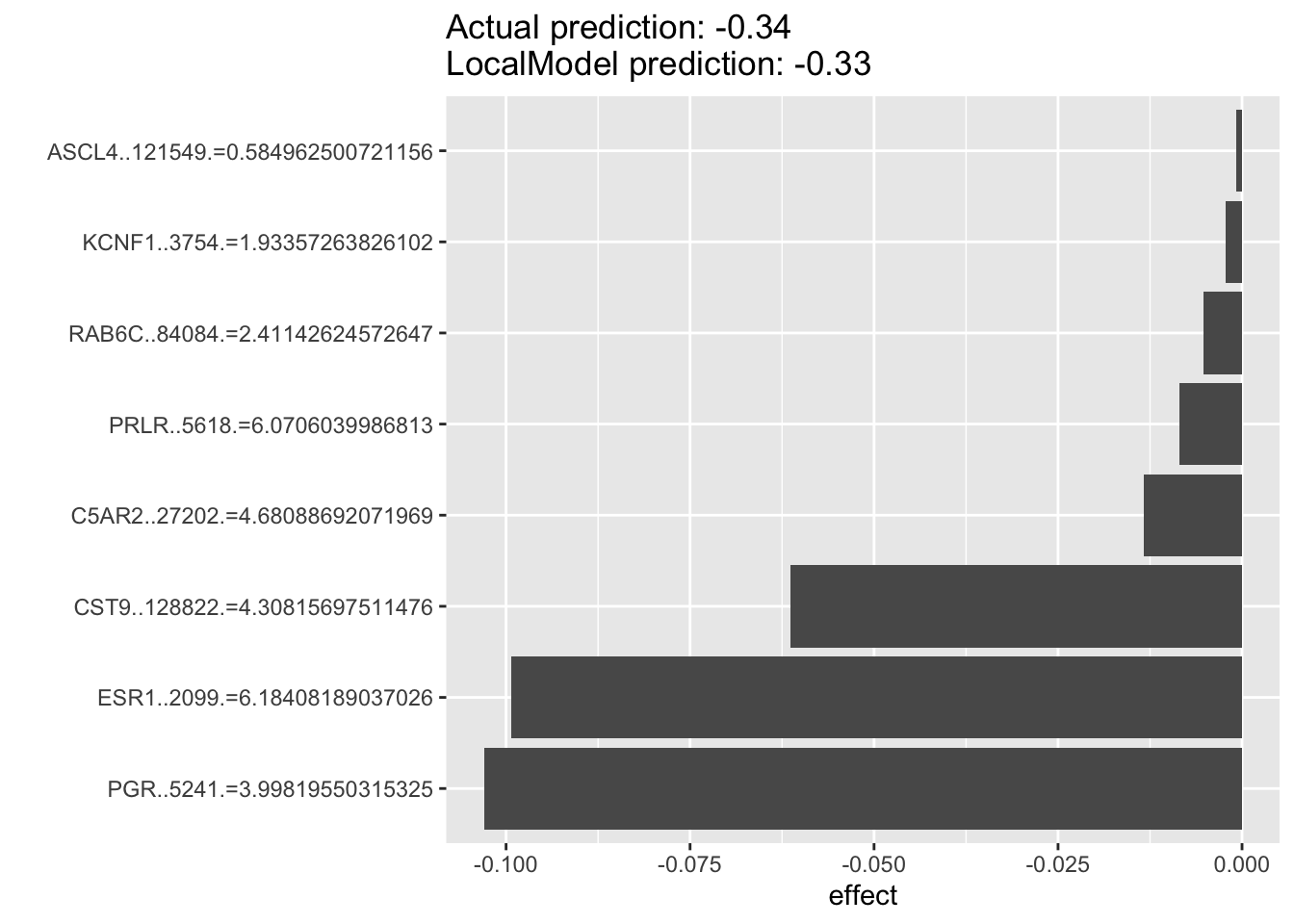
6.7.4 Let’s check the HER2 amplified cell lines in the most dependent cell lines (CRES score <-0.5)
For the example below, we can see that for HER2 amplified breast cancer cell lines that are dependent on ESR1 for survival. The top predictor for each individual cell’s ESR1 dependency are oftentimes ESR1 without PGR among the top predictors. If the previous results suggest that PGR is probably a very good indicater of ESR1 pathway activity and potentially a good prognostic or predictive biomarkers in luminal breast cancers, then it suggests to us that in the HER2-amplified cell lines, PGR will probably not be of much predictive value.
lime.explain142 <- LocalModel$new(iml_predictor, k=10,x.interest = data1[290, ])## Warning in private$aggregate(): Had to choose a smaller kplot(lime.explain142)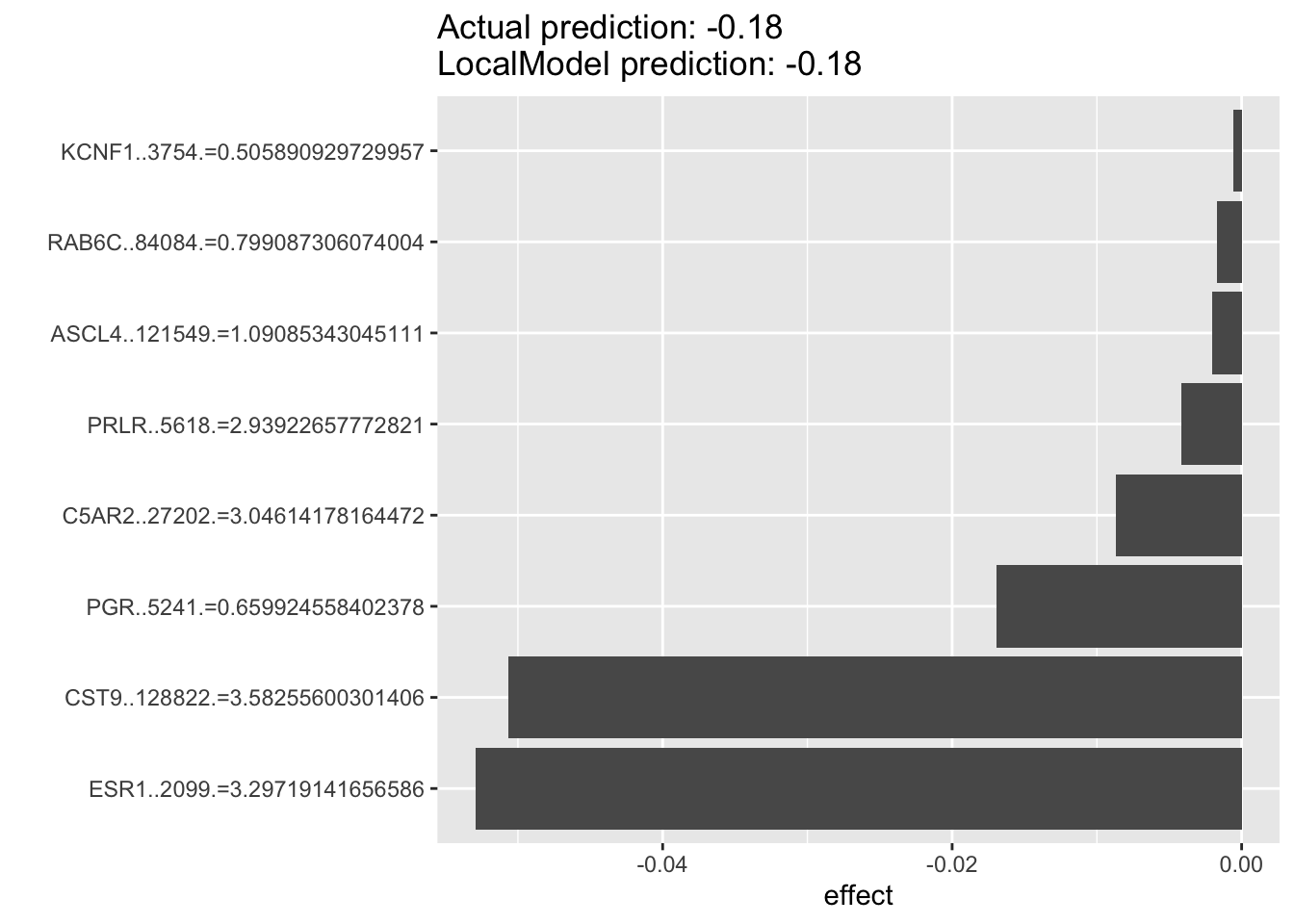
lime.explain502 <- LocalModel$new(iml_predictor, k=10,x.interest = data1[432, ])## Warning in private$aggregate(): Had to choose a smaller kplot(lime.explain502)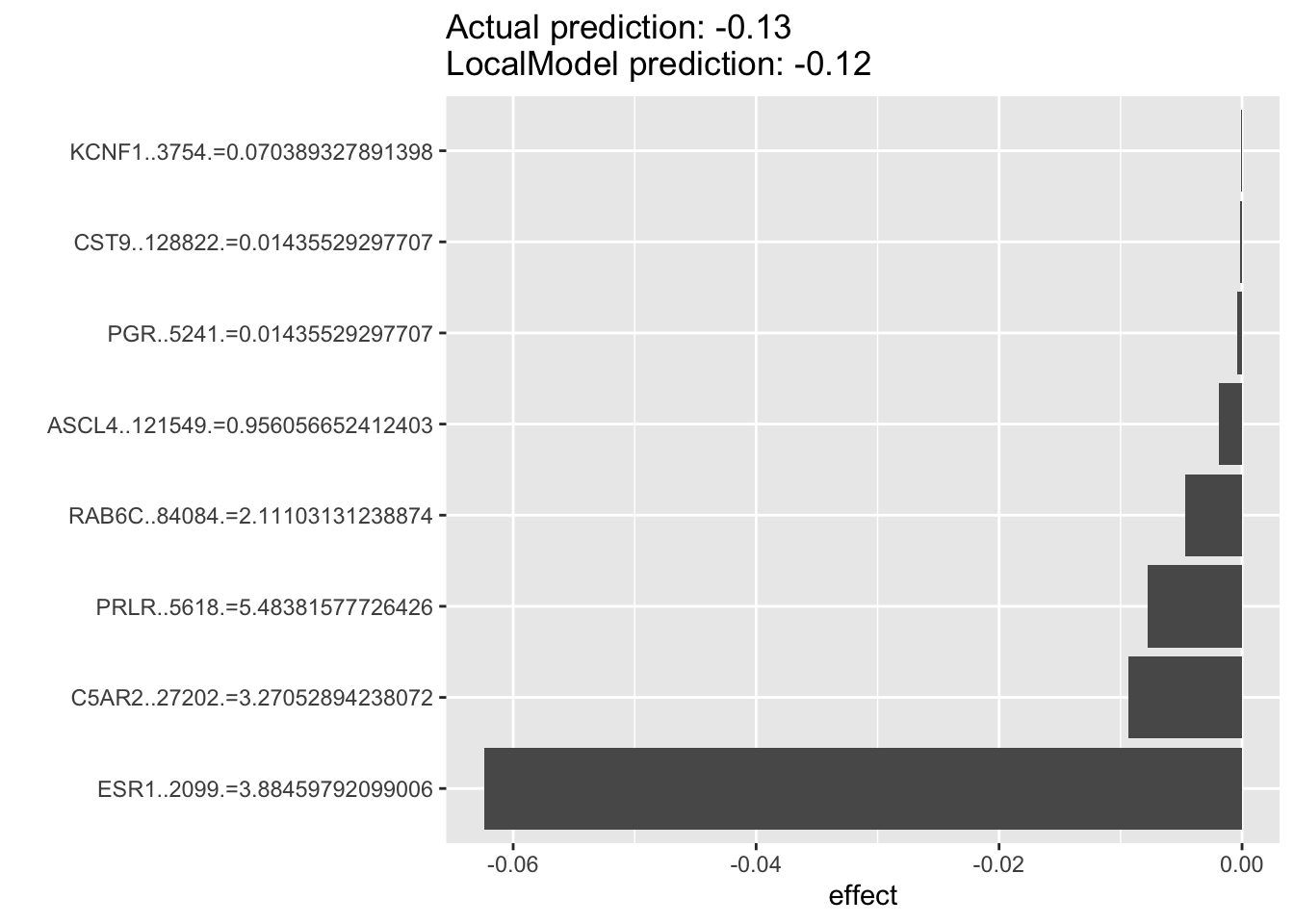
6.8 The overall summary of interpretative machine learning results above
Technical summary: (1) the feature importance based on permutation in iml results are similar to the coeficients of the glm model, although not always the same (2) The local models for the highly dependent ones are all similar to the global models in both the bladder and pancreatic cancer smaples
Scientific summary: (1) ESR1 and PGR expressions are top predictors for ESR1 dependencies in CCLE cell lines globally; (2) In Luminal breast cancers, PGR expression might be the top predictor for ESR1 dependencies than ESR1 expression itself.
6.9 Back to the question of predictive biomarker for anti-ER agents
For a good predictive biomarker of any given anti-ER agent, there are two additional key pieces of information needed: (1) The modeling above essentially is for the ER dependency using crisper in cancer cell lines. We know that different drugs, even if they have the same mechanism of action, may still act differently. Also, cancer cell line data may or may not be predictive of real outcomes in patients. So, we need to have “compound-specific” clinical trial data for PGR+ and PGR- patients. (2) For PGR to be predictive, it means that patients receiving a particular treatment should be getting more benefit in PGR+ patients vs PGR- patients than the control group if control group are present.
6.10 Use the publicly revealed clinical trial data and real-world evidence to support and validate the biomarker hypothesis
The following two publications provided the needed evidence to support and validate the previous biomarker hypothesis. Disclosure: I am a co-author of both publications.
- Hamilton E, Wang J, Pluard T, Johnson S, Morikawa A, Dees E, Jones R, Haley B, Armstrong A, Cohen A, Munster P, Wright G, Kayali F, Korpal M, Yu L, Cantagallo L, Destenaves B, Zhang Z, Gao L, Pipas M, Sahmoud T, Gualberto A and Juric D. Phase I/II trial of H3B-6545, a novel selective estrogen receptor covalent antagonist (SERCA), in estrogen receptor positive (ER+), human factor receptor 2 negative (HER2-) advanced breast cancer. San Antonio Breast Cancer Symposium, Dec 8-12, 2020
H3B-6545 is an anti-ER agent being developed for ER+ advanced or metastatic breast cancer. In the 2020 SABCS publication listed above, the median PFS (progression-free survival) is 5.5 months for ER+ PGR+ breast caner, and 2.1 months for the ER+ PGR- breast cancer. This is a single-arm study, so we do not know what a potential arm would look like.
- Chen T, Zhang Z, Gao L, Scholz C, Gualberto A, Yu L, Yu K. Using real-world data to evaluate the performance of endocrine therapies in ER+/Her2- metastatic breast cancer patients. AACR Annual Meeting, Apr 8-13, 2022.
The RWE (real-world evidence) data provide an opportunity to look at potential control arms using the Flatiron RWE data. From this 2022 AACR publication above, we know that the median TTD (time-to-treatment discontinuation, a measure that is similar to PFS and often used in the RWE data) is 3.7 months ER+ PGR+ metastatic breast caner, and 2.1 months for the ER+ PGR- metastatic breast cancer.
Together with these two publicly available clinical trial and RWE data, along with the modeling above, we can say with relatively strong confidence that, PGR is potentially a good predictive biomarker for ER+ metastatic breast cancer.
6.11 Why do we care about predictive biomarkers?
Let’s go back to the beginning: why do we need to identify predictive biomarkers? A good predictive biomarker can potentially help identify patients who will likely benefit from such a treatment, and also may help avoid exposing patients who might not benefit. Clinical development with a good predictive biomarker may be more ethical in this sense.
Also, with a patient population identified to be more likely responding to the treatment on interest, the success rate of the such a clinical program might be higher, leading to cost savings for the pharmaceutical company and the society as a whole.
6.12 What about the anti-ER therapies?
Breast cancer is the most prevalent malignancies in women, and there is a high unmet medical needs for new therapies, including new anti-ER therapies, because the previously approved anti-ER degrader, Fulvestrant, needs muscle ingection and has poor pharmacokinetics.
However, two promsing anti-ER agents fell short recently in late stage development. One from Roche: https://pharmaphorum.com/news/roches-oral-serd-giredestrant-fails-breast-cancer-trial/
And the other one from Sanofi: https://www.fiercebiotech.com/biotech/long-wait-pivotal-data-sanofis-oral-serd-ends-failure
As far as I know, there is no biomarker-driven patient selection or enrichment strategies in either trial. So one may ask: would these trial turn out differently if predictive biomarker strategies are applied? It is a big if. I am sure there will be more pre-clinical and clinical work trying to address this. And I hope a successful predictive biomarker strategy could be implemented in future anti-ER therapy trials to aid with clinical development.