Chapter 4 Simulation study: Gender and age restrictions
4.1 Gender
library(boot)
library(purrr)
dat_sim <- dat %>%
nest() %>%
mutate(area = "Overall") %>%
bind_rows(dat %>%
mutate(area = ifelse(area == "0_periurban",
"Periurban",
"Rural")) %>%
group_by(area) %>%
nest()) %>%
crossing(p_out = as.character(unique(dat$nm_sex))) %>%
mutate(data = map(.x = data,
.f = ~mutate(.x, comm = as.factor(as.character(comm)))),
boot = map2(.x = data, .y = p_out,
.f = ~boot(data = .x,
statistic = standardization_bin,
treatment = work_out,
outcome = SEROPOSITIVE,
var_sim = nm_sex,
cat_sim = .y,
formula1 = f,
R = R
)),
p_out_label = ifelse(p_out == "0_female",
"Female", "Male"),
boot_tab = map2(.x = boot, .y = p_out_label,
.f = ~tab_bin(.x,
cat_sim = .y)))
saveRDS(dat_sim, "./_out/_rds/result_05-sex.rds")dat_sim_res <- dat_sim %>%
select(area, p_out_label, boot_tab) %>%
mutate(p_out_label_s = ifelse(p_out_label == "Female",
"F", "M"),) %>%
unnest()4.2 Plots Gender
p_gender <- dat_sim_res %>%
filter(type != "NE - OBS") %>% #UPDATE
mutate(type = case_when( #UPDATE
type == "OBS" ~ "NC",
type == "FE" ~ "FE",
type == "NE" ~ "NE",
type == "FE - NE" ~ "RD [FE - NE]",
type == "FE - OBS" ~ "RD [FE - NC]")
) %>%
ggplot(aes(x = type, y = mean, col = p_out_label)) +
geom_hline(aes(yintercept = 0), col = "grey50") +
geom_rect(aes(xmin = 3.5, xmax = Inf, ymin = -Inf, ymax = Inf), #UPDATE
alpha = .025, col = NA) +
geom_linerange(aes(ymin = ll, ymax = ul), alpha = .6,
size = 1.5,
position = position_dodge(width=0.5)) +
geom_point(size = .7, position = position_dodge(width=0.5)) +
#guides(col = F) +
scale_color_npg() +
scale_x_discrete(limits = c("NC", "FE","NE",
"RD [FE - NE]", "RD [FE - NC]")) +
#scale_color_viridis_c(option = "B", direction = -1) +
facet_grid(.~area) +
#geom_smooth(method = "lm", se = F, size = .2) +
labs(y = "Standardized mean outcome", x = "",
col = "Gender") +
theme_bw() +
theme(legend.position = "top",
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
axis.text.x = element_text(angle = 90,
vjust = 0.5, hjust=1))4.3 Age
library(boot)
library(purrr)
dat <- dat %>%
mutate(age_bin = ifelse(edad <= 17, "17 and under",
"18 and older"))
dat_sim <- dat %>%
nest() %>%
mutate(area = "Overall") %>%
bind_rows(dat %>%
mutate(area = ifelse(area == "0_periurban",
"Periurban",
"Rural")) %>%
group_by(area) %>%
nest()) %>%
crossing(p_out = unique(dat$age_bin)) %>%
mutate(data = map(.x = data,
.f = ~mutate(.x, comm = as.factor(as.character(comm)))),
boot = map2(.x = data, .y = p_out,
.f = ~boot(data = .x,
statistic = standardization_bin,
treatment = work_out,
outcome = SEROPOSITIVE,
var_sim = age_bin,
cat_sim = .y,
formula1 = f,
R = R
)),
# p_out_label = ifelse(p_out == "0_female",
# "Female", "Male"),
boot_tab = map2(.x = boot, .y = p_out,
.f = ~tab_bin(.x,
cat_sim = .y)))
saveRDS(dat_sim, "./_out/_rds/result_05-age.rds")dat_sim_res <- dat_sim %>%
select(area, p_out, boot_tab) %>%
# mutate(p_out_label_s = ifelse(p_out_label == "Female",
# "F", "M"),) %>%
unnest()4.4 Plots Age
p_age <- dat_sim_res %>%
filter(type != "NE - OBS") %>% #UPDATE
mutate(type = case_when( #UPDATE
type == "OBS" ~ "NC",
type == "FE" ~ "FE",
type == "NE" ~ "NE",
type == "FE - NE" ~ "RD [FE - NE]",
type == "FE - OBS" ~ "RD [FE - NC]")
) %>%
ggplot(aes(x = type, y = mean, col = p_out)) +
geom_hline(aes(yintercept = 0), col = "grey50") +
geom_rect(aes(xmin = 3.5, xmax = Inf, ymin = -Inf, ymax = Inf), #UPDATE
alpha = .025, col = NA) +
geom_linerange(aes(ymin = ll, ymax = ul), alpha = .6,
size = 1.5,
position = position_dodge(width=0.5)) +
geom_point(size = .7, position = position_dodge(width=0.5)) +
#guides(col = F) +
scale_color_npg() +
scale_x_discrete(limits = c("NC", "FE","NE",
"RD [FE - NE]", "RD [FE - NC]")) +
#scale_color_viridis_c(option = "B", direction = -1) +
facet_grid(.~area) +
#geom_smooth(method = "lm", se = F, size = .2) +
labs(y = "Standardized mean outcome", x = "",
col = "Age category") +
theme_bw() +
theme(legend.position = "top",
panel.grid.major.x = element_blank(),
panel.grid.minor.x = element_blank(),
axis.text.x = element_text(angle = 90,
vjust = 0.5, hjust=1))4.5 Summary Plot
library(cowplot)
plot_grid(p_gender, p_age, labels = c("A)", "B)"), ncol = 1)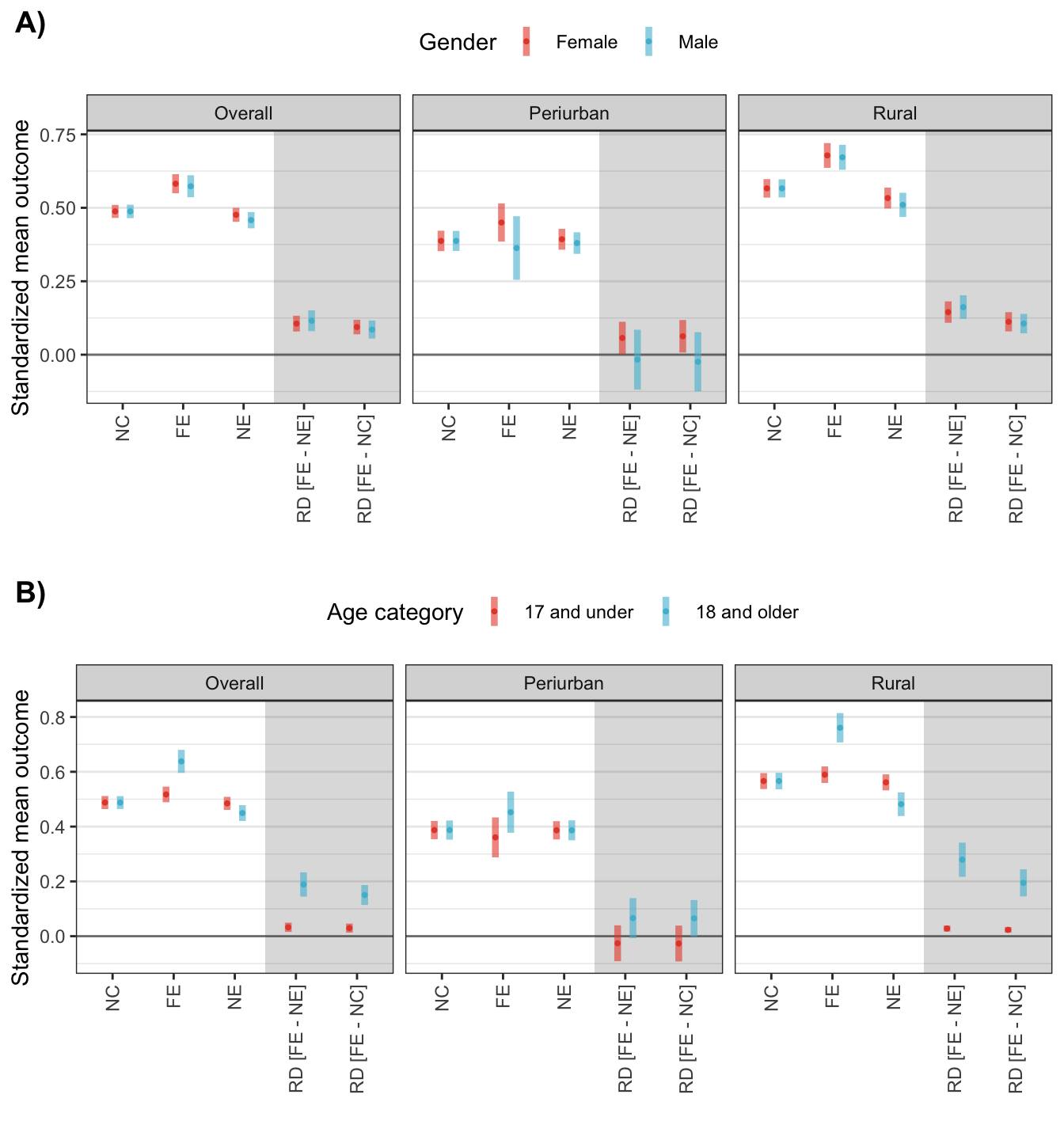
ggsave("./_out/fig5.png", width = 7, height = 7.5,
dpi = "retina", bg = "white")