Chapter 2 g-computation
Details about standarization function in APPENDIX.
2.1 Overall
Based on Hernan and Robins, 2020 Chapter 13
Using a modified Poisson Regression proposed by Zou, 2004
f <- SEROPOSITIVE ~ work_out*edad + work_out*nm_sex + edu_cat + hist_fever + comm
R <- 999 # N bootstraps - 5 for testing, chage upon production
library(boot)
# bootstrap
results <- boot(data = dat,
statistic = standardization,
treatment = work_out,
outcome = SEROPOSITIVE,
formula1 = f, #parameters from function
R = R)
saveRDS(results, "./_out/_rds/result_03-overall.rds")# generating confidence intervals
se <- c(sd(results$t[, 1]),
sd(results$t[, 2]),
sd(results$t[, 3]),
sd(results$t[, 4]))
mean <- results$t0
ll <- mean - qnorm(0.975) * se
ul <- mean + qnorm(0.975) * se
bootstrap <-data.frame(cbind(type=c("Observed", "No Exposed",
"Fully Exposed",
"Fully Exposed - No Exposed"),
mean, se, ll, ul)) %>%
mutate_at(.vars = c(2:5), ~as.numeric(as.character(.)))
kable(bootstrap)| type | mean | se | ll | ul |
|---|---|---|---|---|
| Observed | 0.4877095 | 0.0120416 | 0.4641084 | 0.5113106 |
| No Exposed | 0.4463283 | 0.0153259 | 0.4162900 | 0.4763666 |
| Fully Exposed | 0.6675274 | 0.0255392 | 0.6174716 | 0.7175832 |
| Fully Exposed - No Exposed | 0.2211991 | 0.0300898 | 0.1622242 | 0.2801741 |
2.2 Rural
dat_rural <- dat %>%
filter(area == "1_rural") %>%
mutate(comm = as.factor(as.character(comm)))
# bootstrap
results.r <- boot(data = dat_rural,
statistic = standardization,
treatment = work_out,
outcome = SEROPOSITIVE,
formula1 = f, #parameters from function
R = R)
saveRDS(results.r, "./_out/_rds/result_03-rural.rds")# generating confidence intervals
se.r <- c(sd(results.r$t[, 1]),
sd(results.r$t[, 2]),
sd(results.r$t[, 3]),
sd(results.r$t[, 4]))
mean.r <- results.r$t0
ll.r <- mean.r - qnorm(0.975) * se.r
ul.r <- mean.r + qnorm(0.975) * se.r
bootstrap.r <-data.frame(cbind(type=c("Observed", "No Exposed",
"Fully Exposed", "Fully Exposed - No Exposed"),
mean.r, se.r, ll.r, ul.r)) %>%
mutate_at(.vars = c(2:5), ~as.numeric(as.character(.)))
kable(bootstrap.r, digits = 5)| type | mean.r | se.r | ll.r | ul.r |
|---|---|---|---|---|
| Observed | 0.56617 | 0.01545 | 0.53588 | 0.59646 |
| No Exposed | 0.47695 | 0.02292 | 0.43203 | 0.52186 |
| Fully Exposed | 0.78395 | 0.02866 | 0.72777 | 0.84012 |
| Fully Exposed - No Exposed | 0.30700 | 0.03627 | 0.23591 | 0.37808 |
2.3 Periurban
dat_urban <- dat %>%
filter(area == "0_periurban") %>%
mutate(comm = as.factor(as.character(comm)))
# bootstrap
results.u <- boot(data = dat_urban,
statistic = standardization,
treatment = work_out,
outcome = SEROPOSITIVE,
formula1 = f, #parameters from function
R = R)
saveRDS(results.u, "./_out/_rds/result_03-urban.rds")# generating confidence intervals
se.u <- c(sd(results.u$t[, 1]),
sd(results.u$t[, 2]),
sd(results.u$t[, 3]),
sd(results.u$t[, 4]))
mean.u <- results.u$t0
ll.u <- mean.u - qnorm(0.975) * se.u
ul.u <- mean.u + qnorm(0.975) * se.u
bootstrap.u <-data.frame(cbind(type=c("Observed", "No Exposed",
"Fully Exposed", "Fully Exposed - No Exposed"),
mean.u, se.u, ll.u, ul.u)) %>%
mutate_at(.vars = c(2:5), ~as.numeric(as.character(.)))
kable(bootstrap.u, digits = 5)| type | mean.u | se.u | ll.u | ul.u |
|---|---|---|---|---|
| Observed | 0.38726 | 0.01750 | 0.35296 | 0.42157 |
| No Exposed | 0.38559 | 0.01869 | 0.34896 | 0.42221 |
| Fully Exposed | 0.42579 | 0.06533 | 0.29775 | 0.55382 |
| Fully Exposed - No Exposed | 0.04020 | 0.06745 | -0.09199 | 0.17239 |
2.4 Summary g-computation
library(ggsci)
bootstrap %>% mutate(model = "Overall") %>%
rbind(bootstrap.r %>%
set_names(colnames(bootstrap)) %>%
mutate(model = "Rural")) %>%
rbind(bootstrap.u %>%
set_names(colnames(bootstrap)) %>%
mutate(model = "Peri-urban")) %>%
mutate(type = case_when( #UPDATE
type == "Observed" ~ "Natural Course (NC)",
type == "No Exposed" ~ "No Exposed (NE)",
type == "Fully Exposed" ~ "Fully Exposed (FE)",
type == "Fully Exposed - No Exposed" ~ "Risk Difference (RD)\n[FE - NE]")) %>%
ggplot(aes(y = type, x = mean, col = type)) +
geom_rect(aes(ymin = -Inf, ymax = 1.5, xmin = -Inf, xmax = Inf), #UPDATE
alpha = .05, col = NA) +
geom_linerange(aes(xmin = ll, xmax = ul),
alpha = .6,
size = .5) +
geom_point(size = 1) +
#geom_point() +
geom_vline(xintercept = 0, linetype = "dashed") +
scale_color_npg(guide = F) +
scale_y_discrete(limits = rev(c("Natural Course (NC)",
"Fully Exposed (FE)",
"No Exposed (NE)",
"Risk Difference (RD)\n[FE - NE]"))) +
theme_bw() +
labs(y = "", x = "Standardized mean outcome", col = "") +
facet_grid(.~model) +
theme(legend.position = "top",
panel.grid.major.y = element_blank(),
panel.grid.minor.x = element_blank())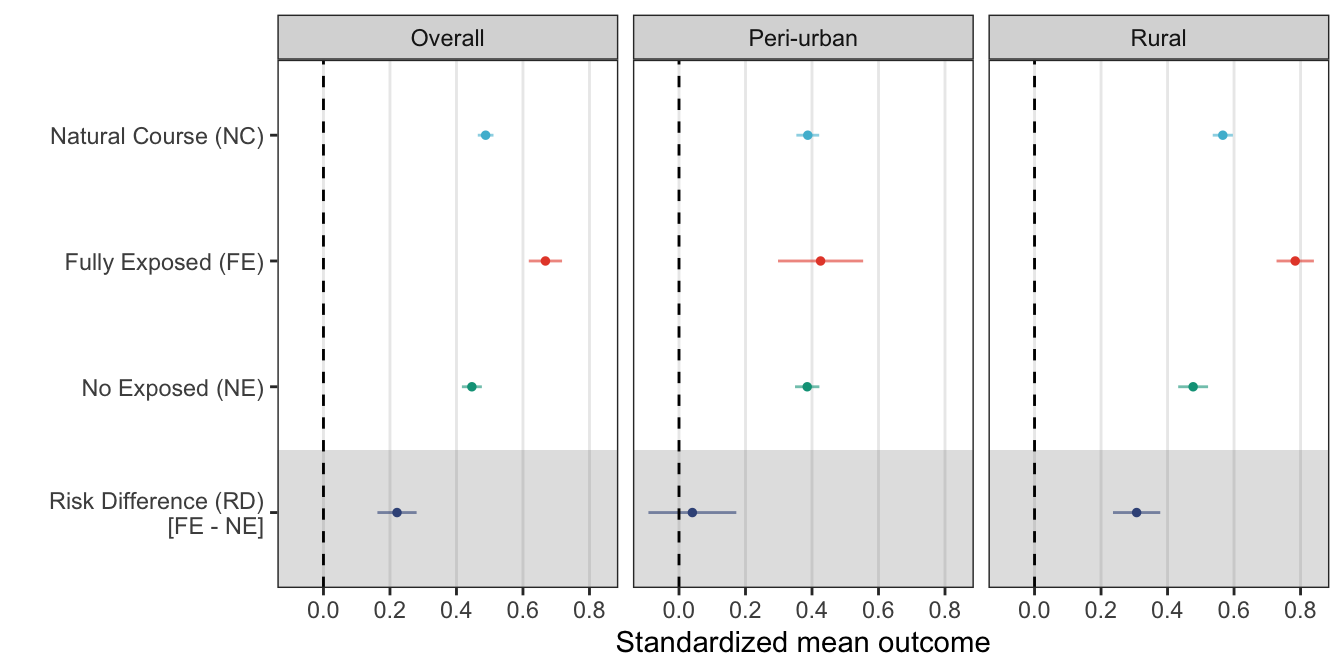
ggsave("./_out/fig3.png", width = 7, height = 3.5,
dpi = "retina", bg = "white")