Chapter 1 Descriptive Analysis
1.1 Table 1
t1 <- dat %>%
dplyr::select(area, comm, edad, age_cat, nm_sex, edu_cat,
work_out, travel_1m,TREATMENT,
resultado_micro, fever, temp_axilar,
hist_fever,SEROPOSITIVE)
library(furniture)
t1 %>%
table1(splitby = ~SEROPOSITIVE, na.rm = F,
NAkeep = T, total = T,
output = "html")| Total | Negative | Positive | |
|---|---|---|---|
| n = 1790 | n = 917 | n = 873 | |
| area | |||
| 0_periurban | 785 (43.9%) | 481 (52.5%) | 304 (34.8%) |
| 1_rural | 1005 (56.1%) | 436 (47.5%) | 569 (65.2%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| comm | |||
| 501_Rumococha | 250 (14%) | 165 (18%) | 85 (9.7%) |
| 502_Santo Tomás | 273 (15.3%) | 132 (14.4%) | 141 (16.2%) |
| 503_Quistococha | 262 (14.6%) | 184 (20.1%) | 78 (8.9%) |
| 901_Gamitanacocha | 47 (2.6%) | 6 (0.7%) | 41 (4.7%) |
| 902_Libertad | 179 (10%) | 45 (4.9%) | 134 (15.3%) |
| 903_Primero de Enero | 58 (3.2%) | 18 (2%) | 40 (4.6%) |
| 904_Salvador | 270 (15.1%) | 144 (15.7%) | 126 (14.4%) |
| 905_Lago Yuracyacu | 97 (5.4%) | 67 (7.3%) | 30 (3.4%) |
| 906_Puerto Alegre | 166 (9.3%) | 84 (9.2%) | 82 (9.4%) |
| 907_Urco Miraño | 188 (10.5%) | 72 (7.9%) | 116 (13.3%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| edad | |||
| 29.0 (22.0) | 20.6 (18.8) | 37.8 (21.8) | |
| age_cat | |||
| [0,5] | 178 (9.9%) | 147 (16%) | 31 (3.6%) |
| (5,15] | 568 (31.7%) | 405 (44.2%) | 163 (18.7%) |
| (15,30] | 299 (16.7%) | 148 (16.1%) | 151 (17.3%) |
| (30,50] | 388 (21.7%) | 126 (13.7%) | 262 (30%) |
| (50,117] | 357 (19.9%) | 91 (9.9%) | 266 (30.5%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| nm_sex | |||
| 0_female | 973 (54.4%) | 536 (58.5%) | 437 (50.1%) |
| 1_male | 817 (45.6%) | 381 (41.5%) | 436 (49.9%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| edu_cat | |||
| Primary or less | 1183 (66.1%) | 582 (63.5%) | 601 (68.8%) |
| Secondary or Superior | 607 (33.9%) | 335 (36.5%) | 272 (31.2%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| work_out | |||
| Inside | 1278 (71.4%) | 798 (87%) | 480 (55%) |
| Outside | 512 (28.6%) | 119 (13%) | 393 (45%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| travel_1m | |||
| No | 1436 (80.2%) | 794 (86.6%) | 642 (73.5%) |
| Yes | 349 (19.5%) | 120 (13.1%) | 229 (26.2%) |
| NA | 5 (0.3%) | 3 (0.3%) | 2 (0.2%) |
| TREATMENT | |||
| No treatment | 917 (51.2%) | 917 (100%) | 0 (0%) |
| Treatment | 873 (48.8%) | 0 (0%) | 873 (100%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| resultado_micro | |||
| Negative | 1729 (96.6%) | 894 (97.5%) | 835 (95.6%) |
| Positive | 38 (2.1%) | 12 (1.3%) | 26 (3%) |
| NA | 23 (1.3%) | 11 (1.2%) | 12 (1.4%) |
| fever | |||
| No | 1764 (98.5%) | 900 (98.1%) | 864 (99%) |
| Yes | 26 (1.5%) | 17 (1.9%) | 9 (1%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| temp_axilar | |||
| 36.2 (0.5) | 36.2 (0.5) | 36.2 (0.5) | |
| hist_fever | |||
| No | 1572 (87.8%) | 821 (89.5%) | 751 (86%) |
| Yes | 218 (12.2%) | 96 (10.5%) | 122 (14%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| SEROPOSITIVE | |||
| Negative | 917 (51.2%) | 917 (100%) | 0 (0%) |
| Positive | 873 (48.8%) | 0 (0%) | 873 (100%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
t1 %>%
table1(splitby = ~area, na.rm = F,
NAkeep = T, total = T,
output = "html")| Total | 0_periurban | 1_rural | |
|---|---|---|---|
| n = 1790 | n = 785 | n = 1005 | |
| area | |||
| 0_periurban | 785 (43.9%) | 785 (100%) | 0 (0%) |
| 1_rural | 1005 (56.1%) | 0 (0%) | 1005 (100%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| comm | |||
| 501_Rumococha | 250 (14%) | 250 (31.8%) | 0 (0%) |
| 502_Santo Tomás | 273 (15.3%) | 273 (34.8%) | 0 (0%) |
| 503_Quistococha | 262 (14.6%) | 262 (33.4%) | 0 (0%) |
| 901_Gamitanacocha | 47 (2.6%) | 0 (0%) | 47 (4.7%) |
| 902_Libertad | 179 (10%) | 0 (0%) | 179 (17.8%) |
| 903_Primero de Enero | 58 (3.2%) | 0 (0%) | 58 (5.8%) |
| 904_Salvador | 270 (15.1%) | 0 (0%) | 270 (26.9%) |
| 905_Lago Yuracyacu | 97 (5.4%) | 0 (0%) | 97 (9.7%) |
| 906_Puerto Alegre | 166 (9.3%) | 0 (0%) | 166 (16.5%) |
| 907_Urco Miraño | 188 (10.5%) | 0 (0%) | 188 (18.7%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| edad | |||
| 29.0 (22.0) | 30.6 (21.8) | 27.7 (22.2) | |
| age_cat | |||
| [0,5] | 178 (9.9%) | 52 (6.6%) | 126 (12.5%) |
| (5,15] | 568 (31.7%) | 227 (28.9%) | 341 (33.9%) |
| (15,30] | 299 (16.7%) | 169 (21.5%) | 130 (12.9%) |
| (30,50] | 388 (21.7%) | 176 (22.4%) | 212 (21.1%) |
| (50,117] | 357 (19.9%) | 161 (20.5%) | 196 (19.5%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| nm_sex | |||
| 0_female | 973 (54.4%) | 461 (58.7%) | 512 (50.9%) |
| 1_male | 817 (45.6%) | 324 (41.3%) | 493 (49.1%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| edu_cat | |||
| Primary or less | 1183 (66.1%) | 427 (54.4%) | 756 (75.2%) |
| Secondary or Superior | 607 (33.9%) | 358 (45.6%) | 249 (24.8%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| work_out | |||
| Inside | 1278 (71.4%) | 688 (87.6%) | 590 (58.7%) |
| Outside | 512 (28.6%) | 97 (12.4%) | 415 (41.3%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| travel_1m | |||
| No | 1436 (80.2%) | 768 (97.8%) | 668 (66.5%) |
| Yes | 349 (19.5%) | 17 (2.2%) | 332 (33%) |
| NA | 5 (0.3%) | 0 (0%) | 5 (0.5%) |
| TREATMENT | |||
| No treatment | 917 (51.2%) | 481 (61.3%) | 436 (43.4%) |
| Treatment | 873 (48.8%) | 304 (38.7%) | 569 (56.6%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| resultado_micro | |||
| Negative | 1729 (96.6%) | 742 (94.5%) | 987 (98.2%) |
| Positive | 38 (2.1%) | 20 (2.5%) | 18 (1.8%) |
| NA | 23 (1.3%) | 23 (2.9%) | 0 (0%) |
| fever | |||
| No | 1764 (98.5%) | 771 (98.2%) | 993 (98.8%) |
| Yes | 26 (1.5%) | 14 (1.8%) | 12 (1.2%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| temp_axilar | |||
| 36.2 (0.5) | 36.2 (0.5) | 36.1 (0.5) | |
| hist_fever | |||
| No | 1572 (87.8%) | 719 (91.6%) | 853 (84.9%) |
| Yes | 218 (12.2%) | 66 (8.4%) | 152 (15.1%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
| SEROPOSITIVE | |||
| Negative | 917 (51.2%) | 481 (61.3%) | 436 (43.4%) |
| Positive | 873 (48.8%) | 304 (38.7%) | 569 (56.6%) |
| NA | 0 (0%) | 0 (0%) | 0 (0%) |
1.2 Descriptives
1.3 Proportions
library(tidyverse)
library(ggmosaic)
library(innovar)
prop_bar <- function(cat_var, title = NULL, l.pos = "none", labx = "") {
title1 <- if (is.null(title)) {expression(cat_var)} else {title}
var1 <- enquo(cat_var)
t1 %>%
filter(!is.na(resultado_micro)) %>%
mutate(area = ifelse(area == "0_periurban", "Periurban", "Rural"),
nm_sex = ifelse(nm_sex == "0_female", "Female", "Male"),
var2 = !!var1,
mm = ifelse(resultado_micro=="Positive", "micro+", "micro-"),
ss = ifelse(SEROPOSITIVE=="Positive", "sero+", "sero-"),
out = paste0(mm, " | ",ss),
age_cat = factor(age_cat),
out = factor(out),
out = fct_rev(out)) %>%
filter(!is.na(var2)) %>%
ggplot() +
geom_mosaic(aes(x = product(out, age_cat), fill = out)#,
#offset = 0.1
) +
scale_fill_innova(palette = "mlobo") +
# scale_fill_discrete_sequential(palette="Heat 2",
# rev = F) +
# scale_fill_manual(values=c("#1b4332", "#b7e4c7",
# "#006d77", "#83c5be")) +
labs(y = "proportion", fill = "Outcome", x = labx) +
theme_minimal() +
facet_grid(~var2) +
ggtitle(title1) +
theme(panel.grid.major = element_blank(),
panel.grid.minor = element_blank(),
axis.text.y=element_blank(),
axis.ticks.y=element_blank(),
axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1),
legend.position = l.pos)
}
f1a <- prop_bar(area, "Area")
f1b <- prop_bar(nm_sex, "Sex")
f1c <- prop_bar(work_out, "Activities outside village")
f1d <- prop_bar(travel_1m, "Travel in the last month")
f1e <- prop_bar(edu_cat, "Education", labx = "Age category")
f1f <- prop_bar(hist_fever, "Fever in last week", labx = "Age category")
library(cowplot)
legend <- get_legend(
prop_bar(hist_fever, "Fever in last week", "bottom") +
theme(legend.box.margin = margin(0, 0, 0, 12))
)
lplot <- plot_grid(legend)
plots <- plot_grid(f1a, f1b, f1c, f1d, f1e, f1f, ncol = 2)
p2 <- plot_grid(plots, lplot, ncol = 1, rel_heights = c(9,1))
p2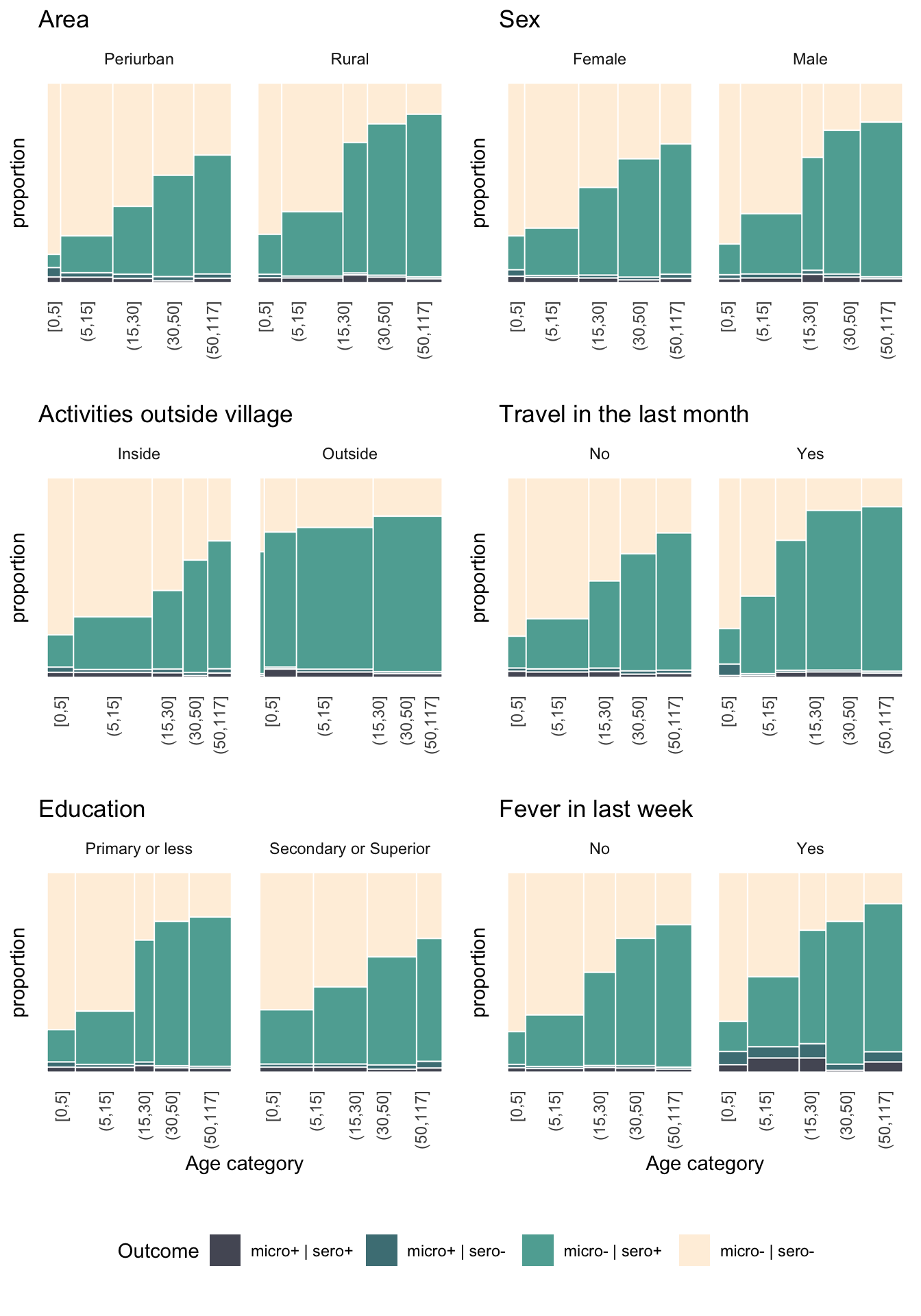
1.4 Maps
1.4.1 Static
library(maptools)
library(ggspatial)
# library(ggmap)
# map <- get_map(location = st_bbox(d2) %>% as.numeric(), maptype = "watercolor", source = "osm", zoom = 10)
#
# ggmap(map) +
# coord_sf(crs = st_crs(3857)) +
# geom_sf(data = d2 %>%
# group_by(id_house) %>%
# summarise(p = mean(sero)),
# aes(col = p), inherit.aes = FALSE, alpha=.4) +
# scale_color_viridis(option = "magma") +
# labs(col = "Household \nSeropositivity Rate")
library(basemaps)
# get_maptypes()
set_defaults(map_service = "carto",
map_type = "light_no_labels")
# basemap_ggplot(d2)
data1 = d2 %>%
group_by(id_house) %>%
summarise(p = mean(sero)) %>%
st_transform(crs = 3857)
buffer <- st_bbox(d2) %>%
st_as_sfc() %>%
st_buffer(10000)
p1 <- ggplot() +
basemap_gglayer(buffer) +
coord_sf(expand = F) +
scale_fill_identity() +
geom_sf(data = data1, aes(col = p)) +
coord_sf(expand = F) +
labs(col = "Household \nSeropositivity rate") +
scale_color_viridis_c(option = "magma") +
annotation_scale(location = "br") +
coord_sf(expand = F) +
annotation_north_arrow(location = "tl",
style = north_arrow_nautical) +
coord_sf(expand = F) +
#theme_void() +
theme(panel.background = element_rect(fill = "white",
colour = "black"),
legend.position = "bottom") +
guides(colour = guide_colourbar(title.position="top",
title.hjust = 0.5),
size = guide_legend(title.position="top",
title.hjust = 0.5))## Loading basemap 'light_no_labels' from map service 'carto'...plot_grid(p1, p2, nrow = 1, labels = c("A)", "B)"),
rel_widths = c(4,5))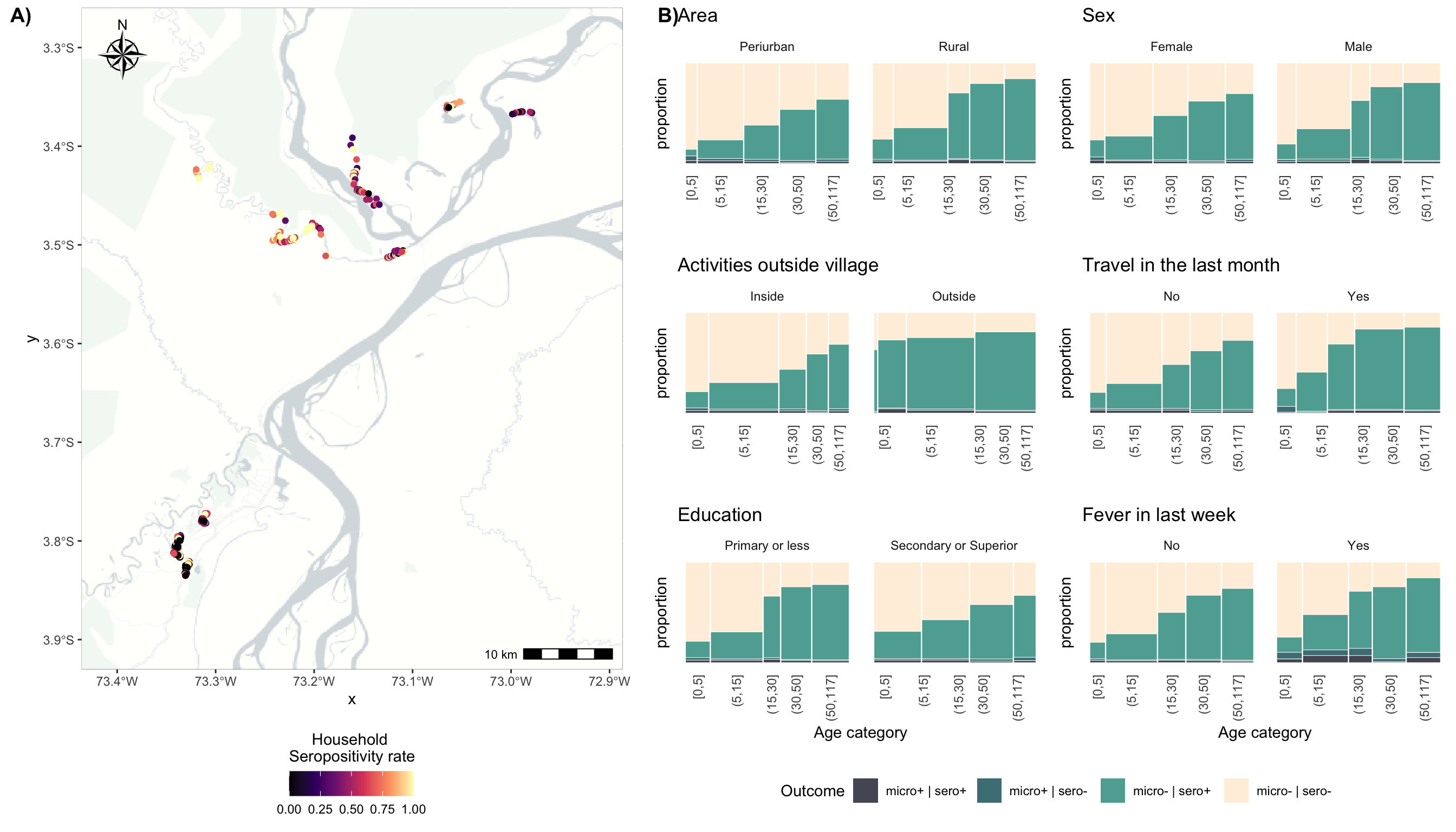
ggsave("./_out/fig1.png", width = 14, height = 8,
dpi = "retina", bg = "white")1.4.2 Dynamic
library(leaflet)
dat_ci_map <- d2 %>%
group_by(id_house) %>%
summarise(p = mean(sero, na.rm = T), area = first(area), age = mean(edad, na.rm = T)) %>%
filter(!is.na(p)) %>%
filter(!is.infinite(p)) %>%
dplyr::mutate(lat = sf::st_coordinates(.)[,2],
long = sf::st_coordinates(.)[,1])
pal <- colorBin("inferno", bins = seq(0,1,.1), reverse=T)
dat_ci_map %>%
leaflet() %>%
addTiles() %>%
addCircles(lng = ~long, lat = ~lat, color = ~ pal(p),
popup = paste("ID Household", dat_ci_map$id_house, "<br>",
"Area:", dat_ci_map$area, "<br>",
"Mean age:", dat_ci_map$age, "<br>",
"Prevalence:", dat_ci_map$p)) %>%
addLegend("bottomright",
pal = pal, values = ~p,
title = "Prevalence") %>%
addScaleBar(position = c("bottomleft"))