Chapter 3 Clustering
3.1 Number of Clusters
Clustering Methods
- C1: average silhouette width
- C2: gap statistics
- C3: Hierarchical (spatial) cluster analysis cutoff = 50m
- C4: Hierarchical (spatial) cluster analysis cutoff = 10km
- C5: Hierarchical (spatial) cluster analysis cutoff = 200km
dat.t <- dat.n %>%
#slice(1) %>%
mutate(c3 = map(data, ~n.clust.sp(dat = ., full=F)), #unique places (50m)
)## # A tibble: 6 x 4
## patientID n uniq c3
## <dbl> <int> <int> <int>
## 1 4 127 102 9
## 2 9 86 81 22
## 3 10 77 21 9
## 4 11 186 144 8
## 5 14 135 123 5
## 6 17 56 36 223.1.1 Distribution of Number of locations (within 50m) per participant
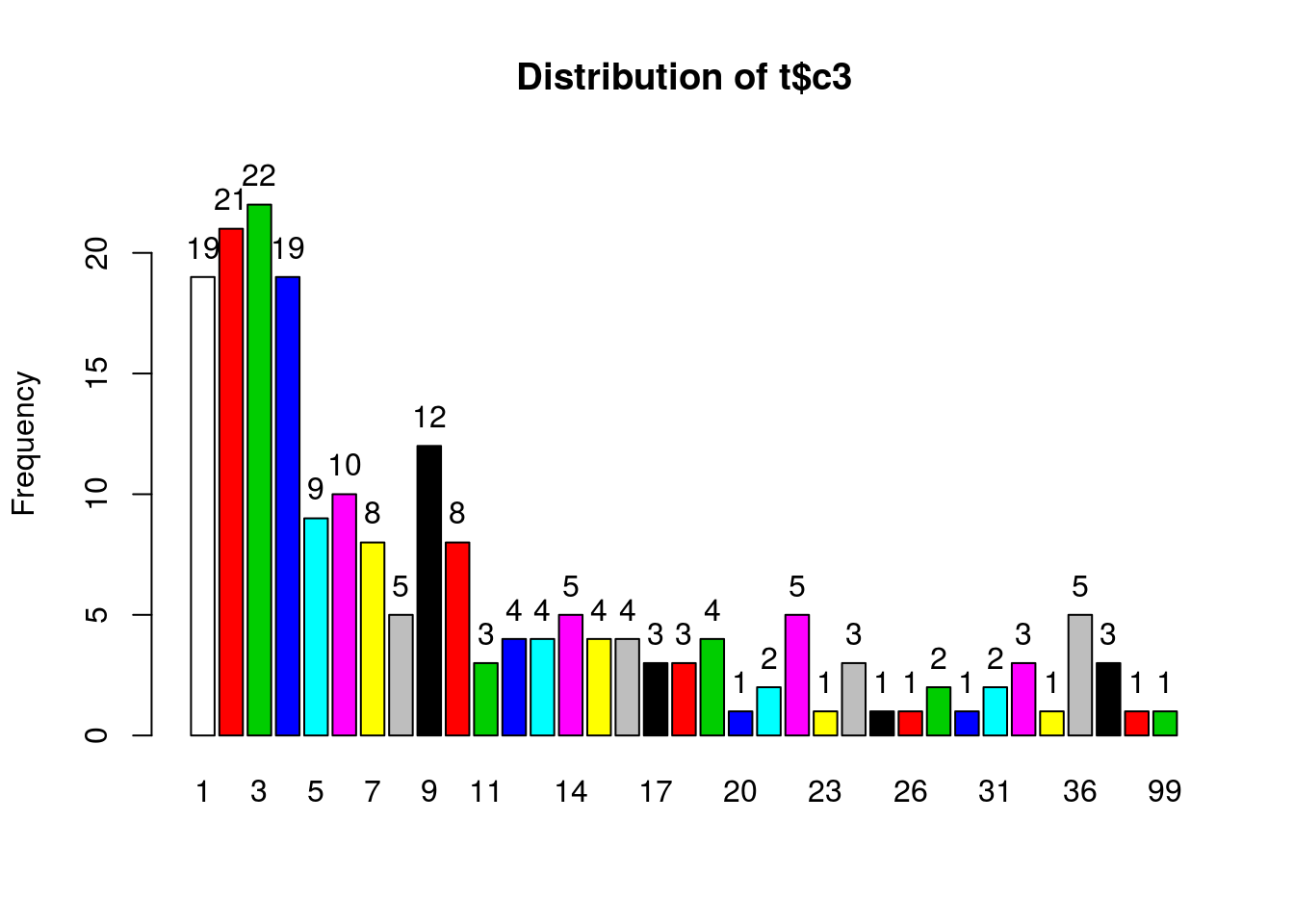
## t$c3 :
## Frequency Percent Cum. percent
## 1 19 9.5 9.5
## 2 21 10.5 20.0
## 3 22 11.0 31.0
## 4 19 9.5 40.5
## 5 9 4.5 45.0
## 6 10 5.0 50.0
## 7 8 4.0 54.0
## 8 5 2.5 56.5
## 9 12 6.0 62.5
## 10 8 4.0 66.5
## 11 3 1.5 68.0
## 12 4 2.0 70.0
## 13 4 2.0 72.0
## 14 5 2.5 74.5
## 15 4 2.0 76.5
## 16 4 2.0 78.5
## 17 3 1.5 80.0
## 18 3 1.5 81.5
## 19 4 2.0 83.5
## 20 1 0.5 84.0
## 21 2 1.0 85.0
## 22 5 2.5 87.5
## 23 1 0.5 88.0
## 24 3 1.5 89.5
## 25 1 0.5 90.0
## 26 1 0.5 90.5
## 28 2 1.0 91.5
## 29 1 0.5 92.0
## 31 2 1.0 93.0
## 32 3 1.5 94.5
## 33 1 0.5 95.0
## 36 5 2.5 97.5
## 41 3 1.5 99.0
## 43 1 0.5 99.5
## 99 1 0.5 100.0
## Total 200 100.0 100.0Only 9.5% of participants recorded videos in a single location.
3.2 Distance from base (most frequent location)
3.2.1 Defining most frequent location (mfl)
dat.t2 <- dat.t %>%
#slice(1) %>%
mutate(c3_df = map(data, ~n.clust.sp(dat = ., full=F, df = T)),
c3_mfl = map(c3_df, ~mfl(.))
)## # A tibble: 9 x 5
## clust x1 x2 loc mfl
## <int> <dbl> <dbl> <int> <dbl>
## 1 1 -117. 32.9 87 1
## 2 2 -117. 32.9 3 0
## 3 3 -117. 32.9 2 0
## 4 4 -117. 32.9 5 0
## 5 5 -122. 37.8 23 0
## 6 6 -117. 32.9 1 0
## 7 7 -122. 37.0 1 0
## 8 8 -117. 32.9 1 0
## 9 9 -117. 33.0 4 03.2.2 Geodesic distance from mfl
## to_cluster distance_m dist_cat GC_dist GC_cat loc
## 1 1 0.000 base 0.0000 base 87
## 2 2 1851.759 local 1851.2128 local 3
## 3 3 1767.785 local 1767.1582 local 2
## 4 4 1876.144 local 1875.2589 local 5
## 5 5 710341.286 distant 709687.8539 distant 23
## 6 6 330.870 local 330.1236 local 1
## 7 7 632069.323 distant 631383.2844 distant 1
## 8 8 1777.499 local 1776.6424 local 1
## 9 9 18830.318 local 18809.6115 local 4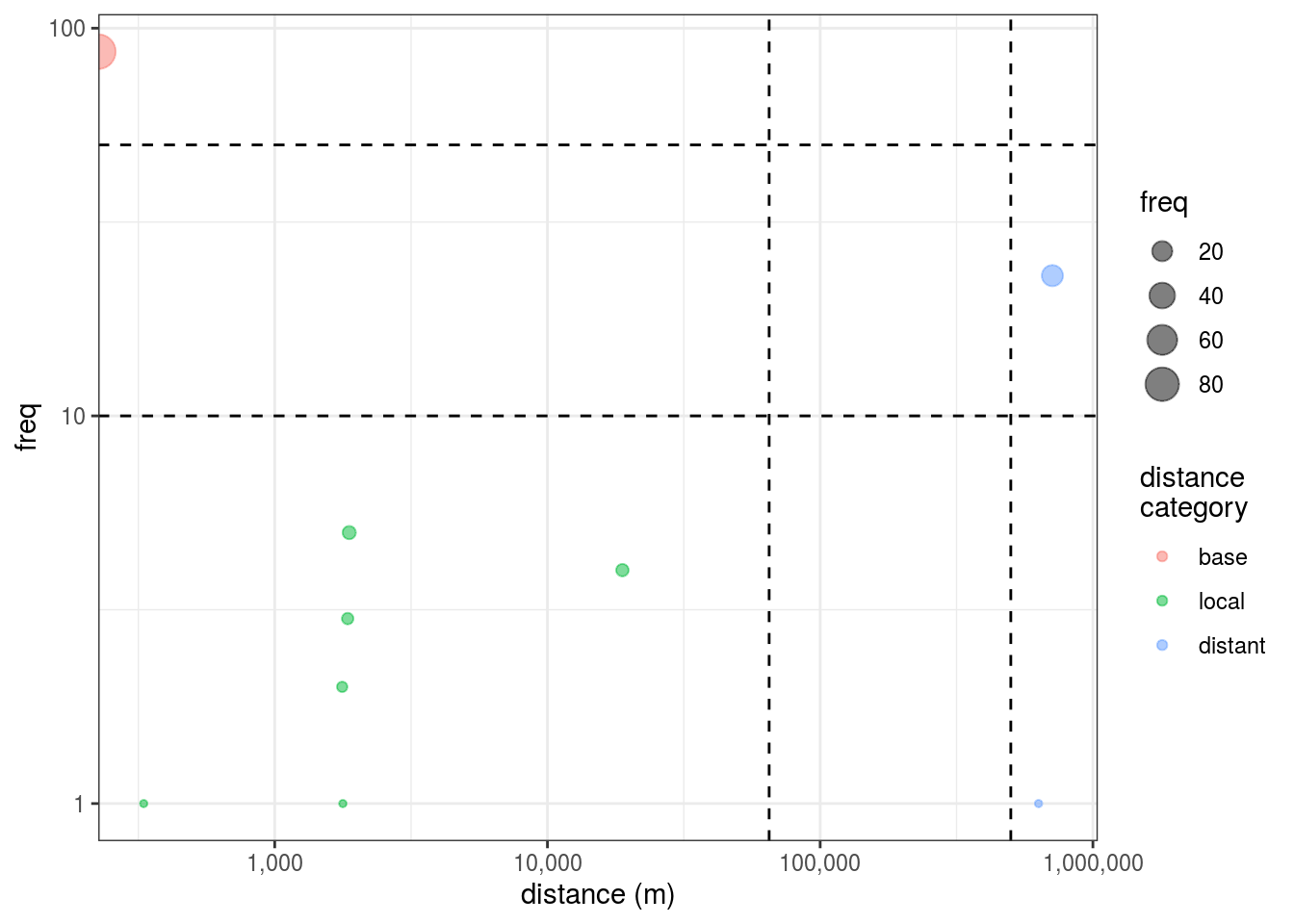
3.2.3 Distance summaries
## # A tibble: 3 x 11
## dist_cat avg_gd min_gd max_gd sd_gd avg_GC min_GC max_GC sd_GC freq n_clust
## <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <int> <int>
## 1 base 0. 0. 0. NA 0. 0. 0. NA 87 1
## 2 local 4.41e3 3.31e2 1.88e4 7092. 4.40e3 3.30e2 1.88e4 7084. 16 6
## 3 distant 6.71e5 6.32e5 7.10e5 55347. 6.71e5 6.31e5 7.10e5 55370. 24 23.3 Final dataset
dat.s <- dat.t4 %>%
dplyr::select(patientID, c3, c3_s) %>%
#unnest(cols = c(c1,c2,c3,c4, c5, c3_s, c4_s, c5_s))
unnest() %>%
mutate_if(is.numeric, .funs = ~ifelse(.==Inf | .==-Inf, NA, .)) %>%
rename(c3_avg_gd = avg_gd, c3_min_gd=min_gd, c3_max_gd=max_gd, c3_sd_gd=sd_gd,
c3_avg_GC=avg_GC, c3_min_GC=min_GC, c3_max_GC=max_GC, c3_sd_GC=sd_GC
)
# [c*_s = 1]: No summaries
# [c*_s = 2]: Only 1 distance to compute; avg=min=max; no sd## # A tibble: 6 x 13
## patientID c3 dist_cat c3_avg_gd c3_min_gd c3_max_gd c3_sd_gd c3_avg_GC
## <dbl> <int> <fct> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 4 9 base 0 0 0 NA 0
## 2 4 9 local 4406. 331. 18830. 7092. 4402.
## 3 4 9 distant 671205. 632069. 710341. 55347. 670536.
## 4 9 22 base 0 0 0 NA 0
## 5 9 22 local 8479. 228. 24159. 8299. 8475.
## 6 10 9 base 0 0 0 NA 0
## # … with 5 more variables: c3_min_GC <dbl>, c3_max_GC <dbl>, c3_sd_GC <dbl>,
## # freq <int>, n_clust <int>