Chapter 2 Analysis
2.1 Dataset
library(tidyverse)
library(readxl)
library(lubridate)
dat <- read_excel("./_data/VDOT_GIS_v3.xlsx") %>%
filter(latitude>0) %>%
mutate(month = month(dtRecorded),
year = year(dtRecorded),
semester = semester(dtRecorded),
quater = quarter(dtRecorded),
index = (((year-2013)*12) + month)-9,
index_sem = (((year-2013)*2) + semester)-1,
index_q = (((year-2013)*4) + quater)-3,
out_bound = ifelse(latitude>48|latitude<20|longitude<(-123)|longitude>(-80),1,0))
covars <- read.csv("./_data/VDOT_PatientLevelData_2_27_18.csv", stringsAsFactors=FALSE) %>%
dplyr::select(incomea, incommo, dpndntb, marij, drugs, age, latino, race, sex, cborn, edu, ADH_A, ADH_AS, ADH_ASP, ADHR_A, patientID, C_endstat, PI_Hisp, PI_IDU, PI_IDUyr)
dat.n <- dat %>%
dplyr::select(patientID, longitude, latitude) %>%
nest(-patientID) %>%
mutate(n = map(data, ~nrow(.)),
uniq = map(data, ~nrow(unique(.)))) %>%
filter(n>2 & uniq>2) # individuals with enought sample size2.2 Example single Participant
2.2.1 Hierarchical Clustering
library(factoextra)
source("./_data/vdot_helpers.R")
a <- dat.n$data[dat.n$patientID==100][[1]]
a %>%
ggplot() +
geom_point(aes(longitude, latitude), size = 3, alpha = .5) +
theme_bw()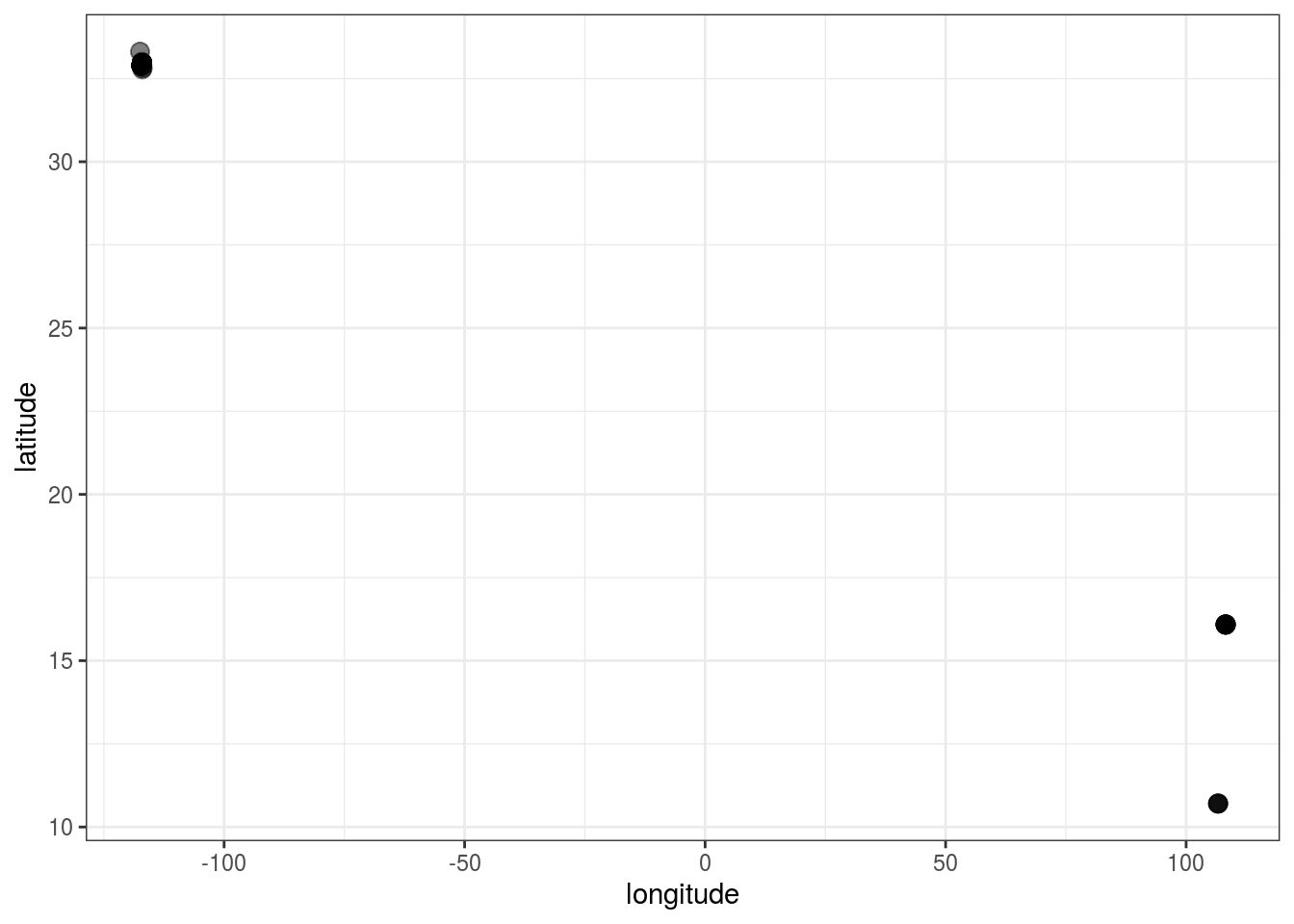
## $`Optimal k`
## [1] 2
##
## $`Plot optimization`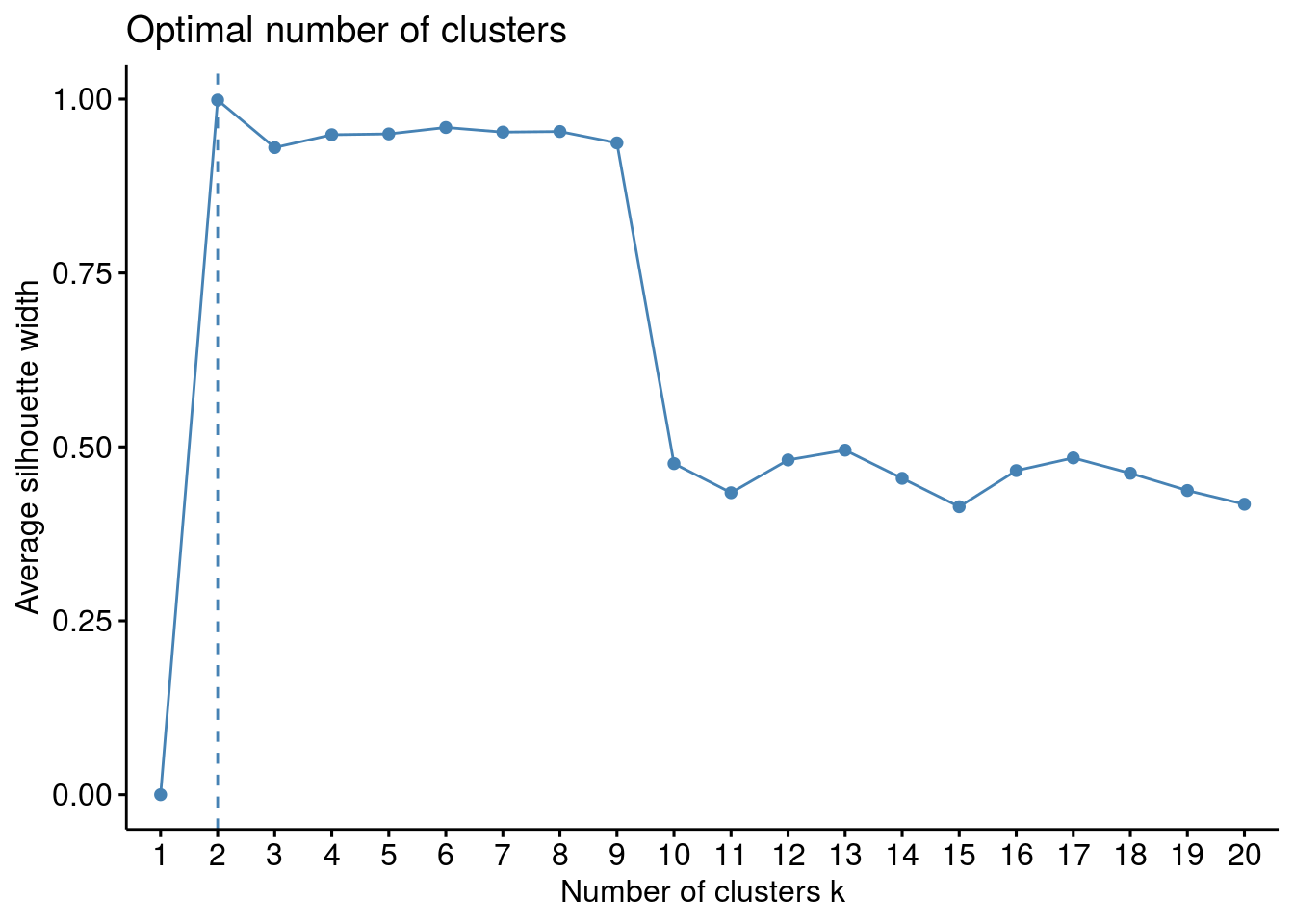
##
## $`Cluster Plot`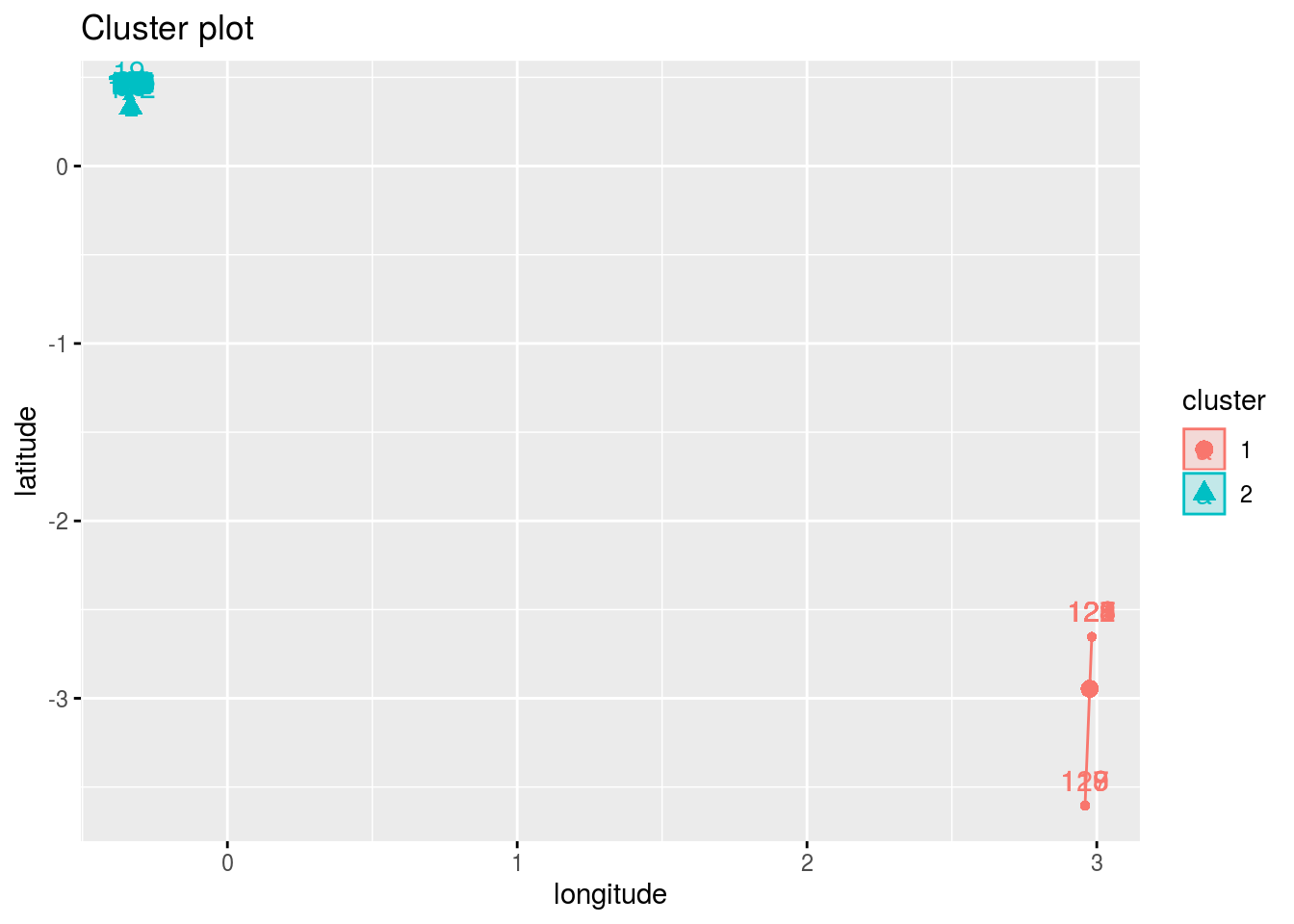
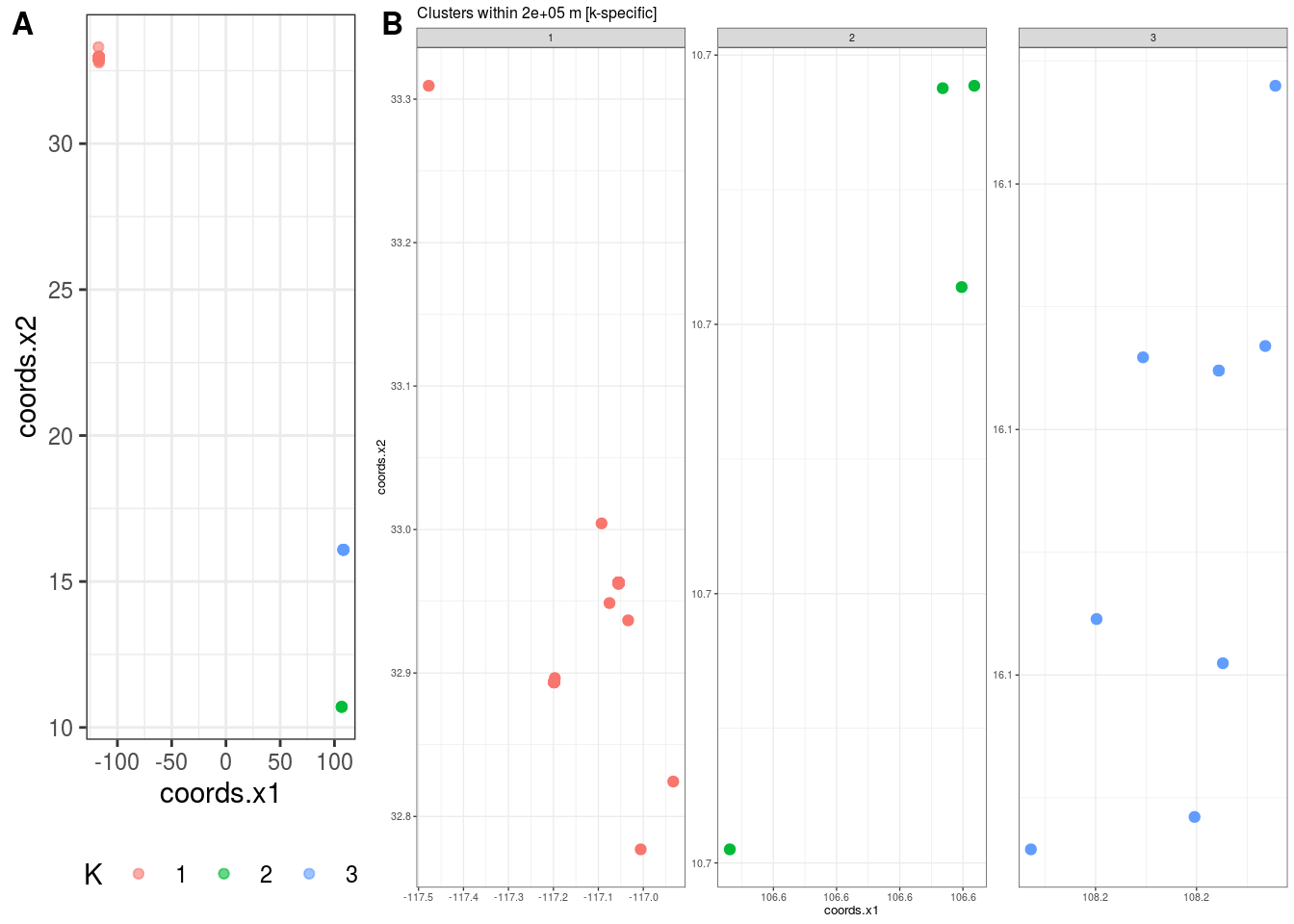
2.2.2 Spatial Clustering
## $K
## [1] 13
##
## $`Cluster Plot`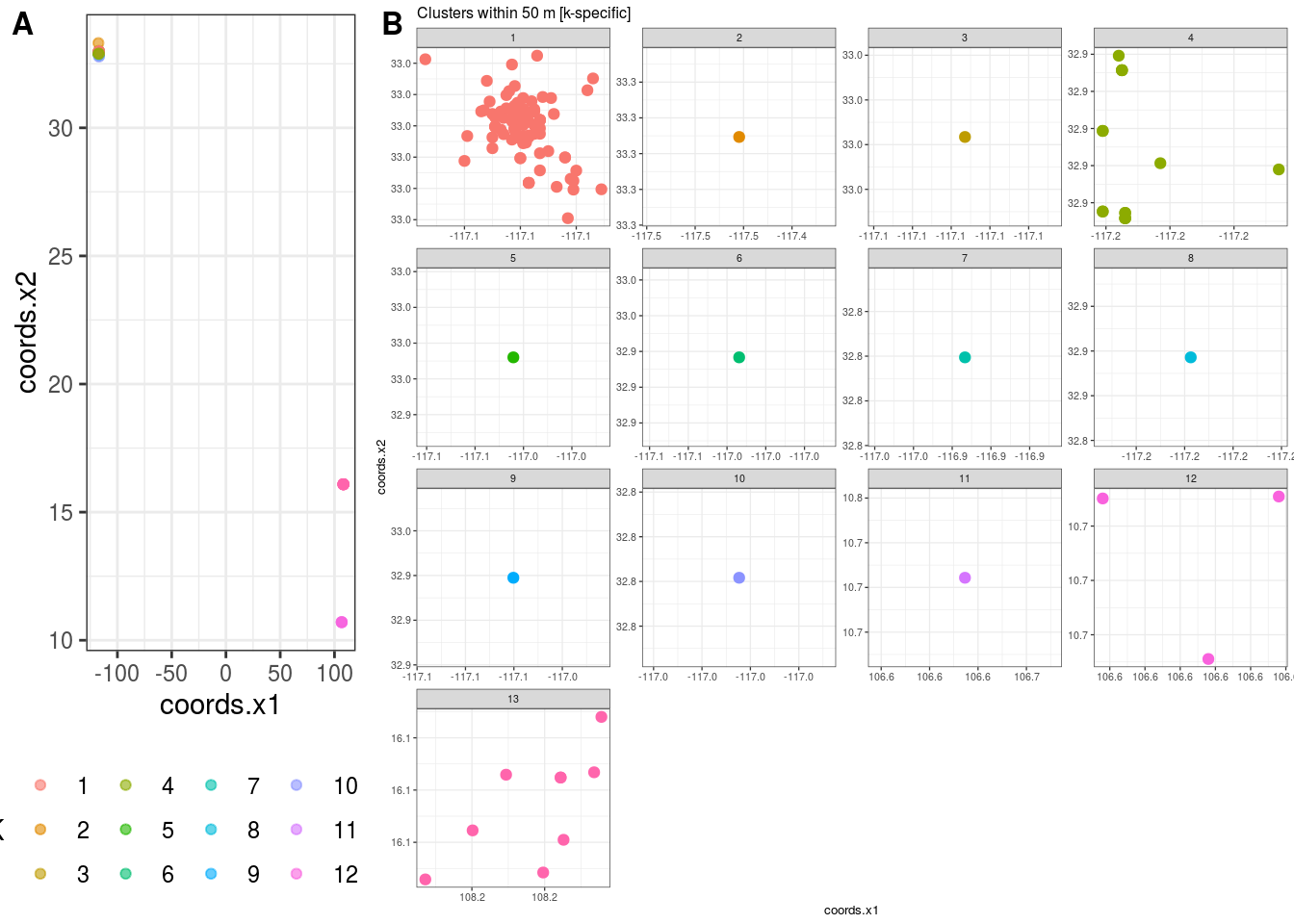
## $K
## [1] 6
##
## $`Cluster Plot`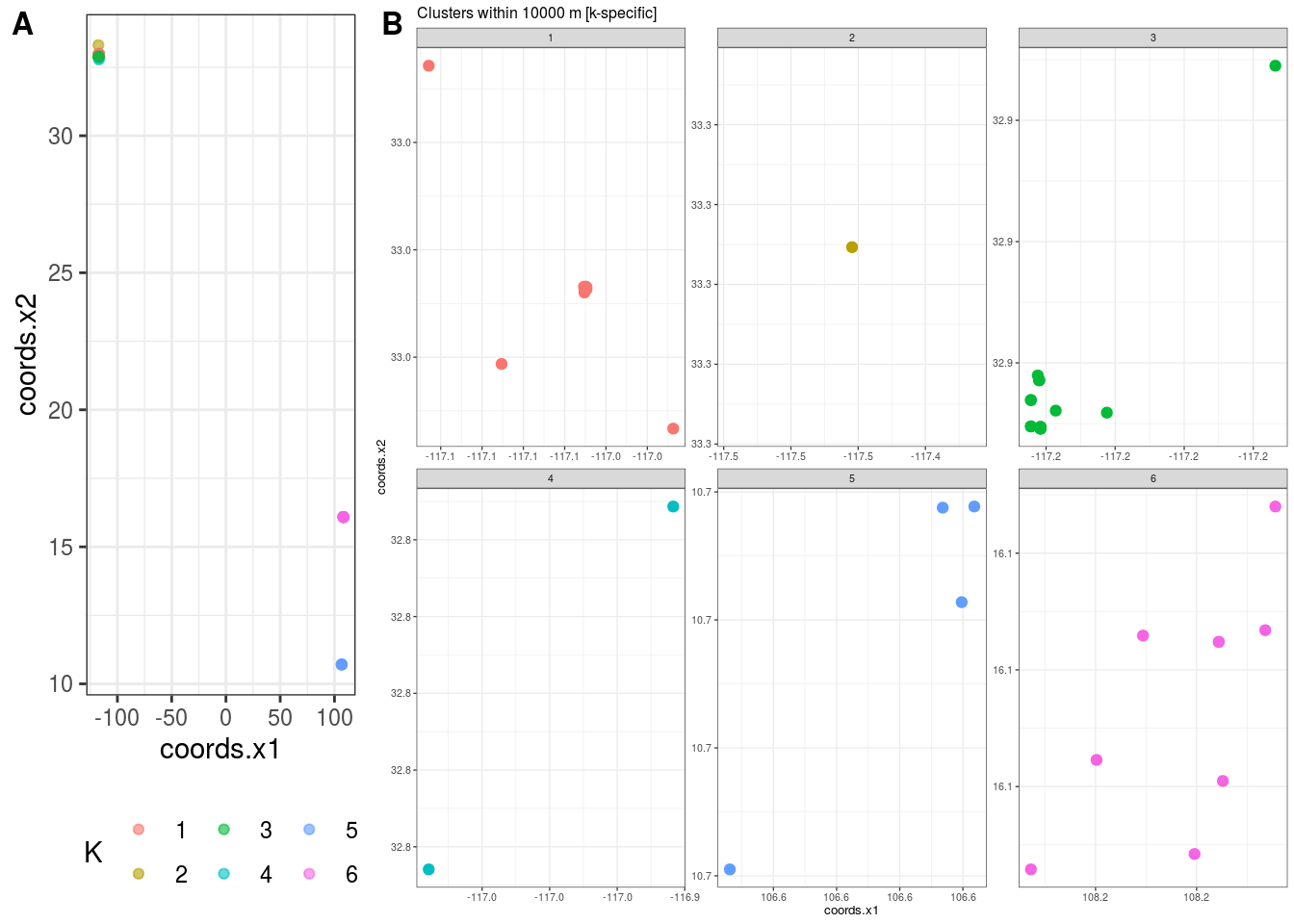
## $K
## [1] 3
##
## $`Cluster Plot`