Chapter 4 DENGUE DATA
4.1 Init
library(tidyverse)
library(lubridate)
library(rEDM)
library(ISOweek)
library(stringr)
library(cowplot)
library(purrr)
library(naniar)
library(ggsci)4.2 Data
rm(list = ls())
load("./_data/climate_vars/Data.RData")
den <- read_csv("_data/LOR_dengue.csv")
nl <- nl %>%
select(-ntl2014)
nl2 <- readRDS("./_data/climate_vars/NightTimeLight.rds")
ubi_den <- unique(den$ubigeo)
ubi_clim <- unique(temp$UBIGEO) %>% as.numeric()
unique(ubi_clim[! ubi_clim %in% ubi_den]) # Rosa Panduro creado 2014## [1] 1608024.3 Assembly Climate
data_frames <- do.call("list",mget(ls())) %>%
enframe() %>%
#filter(name == "nl2") %>%
filter(!name %in% c("den", "ubi_clim","ubi_den", "data_frames")) %>%
mutate(dat_edit = map(.x = value,
.f = ~gather(.x, var_date, value, 5:last_col()) %>%
separate(var_date, into = c("var", "date"),
sep = "(?<=[A-Za-z])(?=[_0-9])") %>%
mutate(date = str_sub(date, -7,-1)) %>%
separate(date, into = c("year", "month"),
sep = "-", convert = T) %>%
mutate(month = ifelse(is.na(month), 1, month)))) %>%
select(name, dat_edit) %>%
unnest(cols = "dat_edit") %>%
select(-name) %>%
spread(var, value) %>%
filter(year<2021) %>%
mutate(UBIGEO = as.numeric(UBIGEO))
# 53 districts * 12 months * 21 years = 13,356
# a <- nl2 %>%
# gather(var_date, value, 5:last_col()) %>%
# separate(var_date,
# into = c("var", "date"),
# sep = "(?<=[A-Za-z])(?=[_0-9])"
# ) %>%
# mutate(date = str_sub(date, -7,-1)) %>%
# separate(date,
# into = c("year", "month"),
# sep = "-",
# convert = T) %>%
# mutate(month = ifelse(is.na(month), 1, month))
vis_miss(data_frames %>%
arrange(year, month))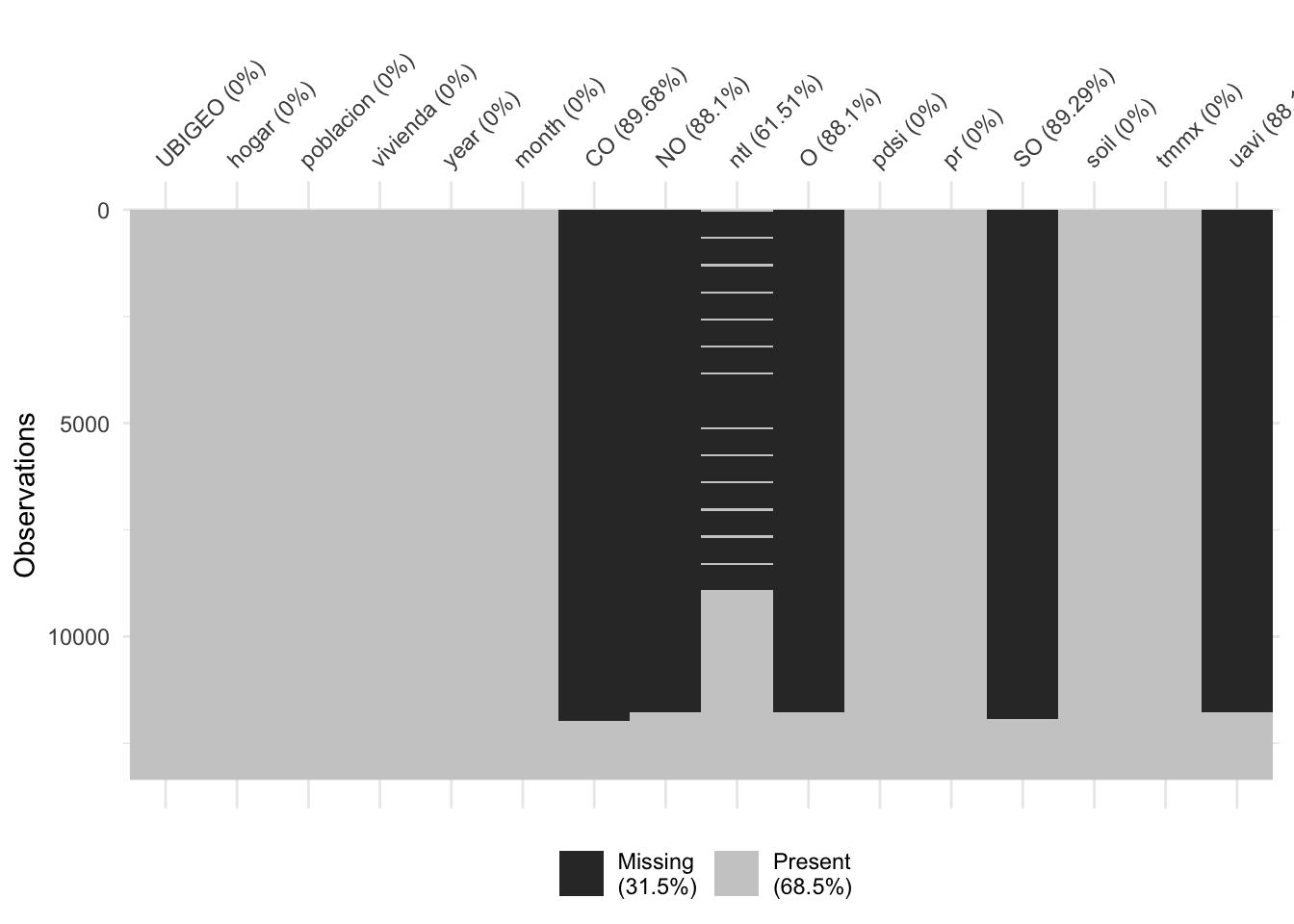
4.4 Assembly Cases
den_proto <- den %>%
filter(complete.cases(.),
yy<2021) %>%
expand(ubigeo, yy, mm) %>% # 52 districts * 12 months * 21 years = 13,104
full_join(den %>%
select(ubigeo, yy, mm, tot_casos) %>%
filter(yy<2021), by = c("ubigeo", "yy", "mm")) %>%
mutate(tot_casos = replace_na(tot_casos, 0)) %>%
full_join(den %>%
distinct(ubigeo, departam, provincia, distrito), by = "ubigeo") %>%
rename(UBIGEO = ubigeo,
year = yy,
month = mm,
department = departam,
province = provincia,
district = distrito,
den_cases = tot_casos)
dat <- den_proto %>%
inner_join(data_frames, by = c("UBIGEO", "year", "month")) %>%
mutate(date = ymd(paste(year, month, "01", sep = "-")))
vis_miss(dat %>%
arrange(year, month))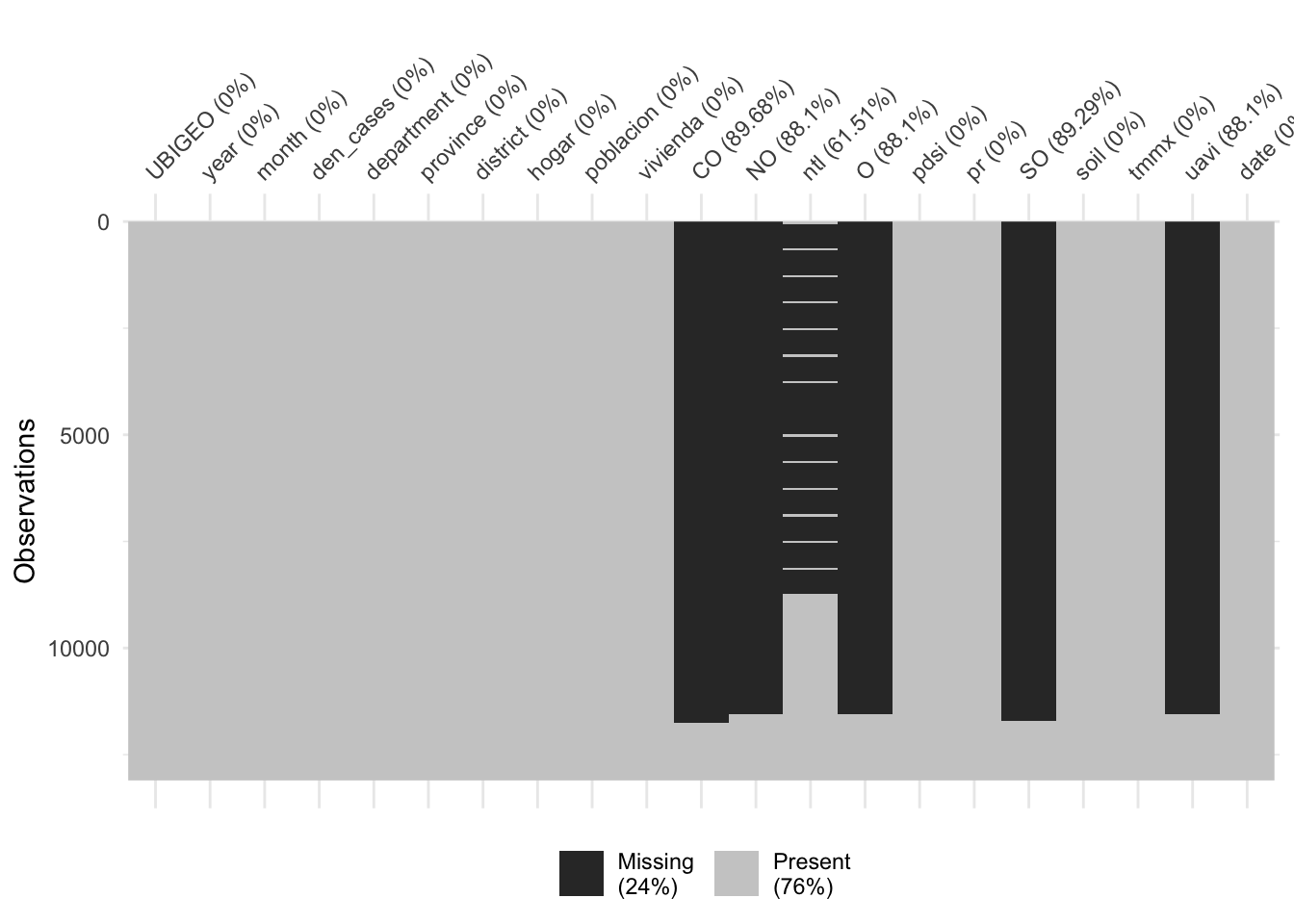
write_csv(dat, "_data/LOR_dengue_clim.csv", na = "")4.5 EDA
top8 <- dat %>%
group_by(district) %>%
summarise(cases = sum(den_cases)) %>%
arrange(-cases) %>%
slice(1:8) %>%
select(district) %>%
unlist()
capitals <- c("IQUITOS","YURIMAGUAS", "NAUTA", "RAMON CASTILLA",
"REQUENA", "CONTAMANA", "BARRANCA", "PUTUMAYO")
plot <- dat %>%
mutate_at(.vars = c(4, 11:20),
.funs = ~scale_this(.)) %>%
select(7, 21, 4, 11:20) %>%
gather(var, val, 3:13) %>%
filter(district %in% top8) %>%
#filter(district %in% capitals) %>%
filter(var %in% c("pdsi", "pr", "soil", "tmmx", "den_cases"))
plot_cases <- plot %>%
filter(var != "den_cases") %>%
inner_join(plot %>%
filter(var == "den_cases") %>%
select(-var), by = c("district", "date"))
plot %>%
filter(var != "den_cases") %>%
ggplot(aes(x = date, col = var, group = var)) +
geom_line(aes(y = val), size =.3) +
geom_line(dat = plot_cases, aes(y = val.y), col = "black", size =.3) +
facet_wrap(factor(district, levels = top8)~var,
scales = "free_y", ncol = 4) +
scale_x_date(date_breaks = "5 year", date_labels = "%Y")+
theme_bw() +
scale_color_npg() +
theme(legend.position = "bottom") 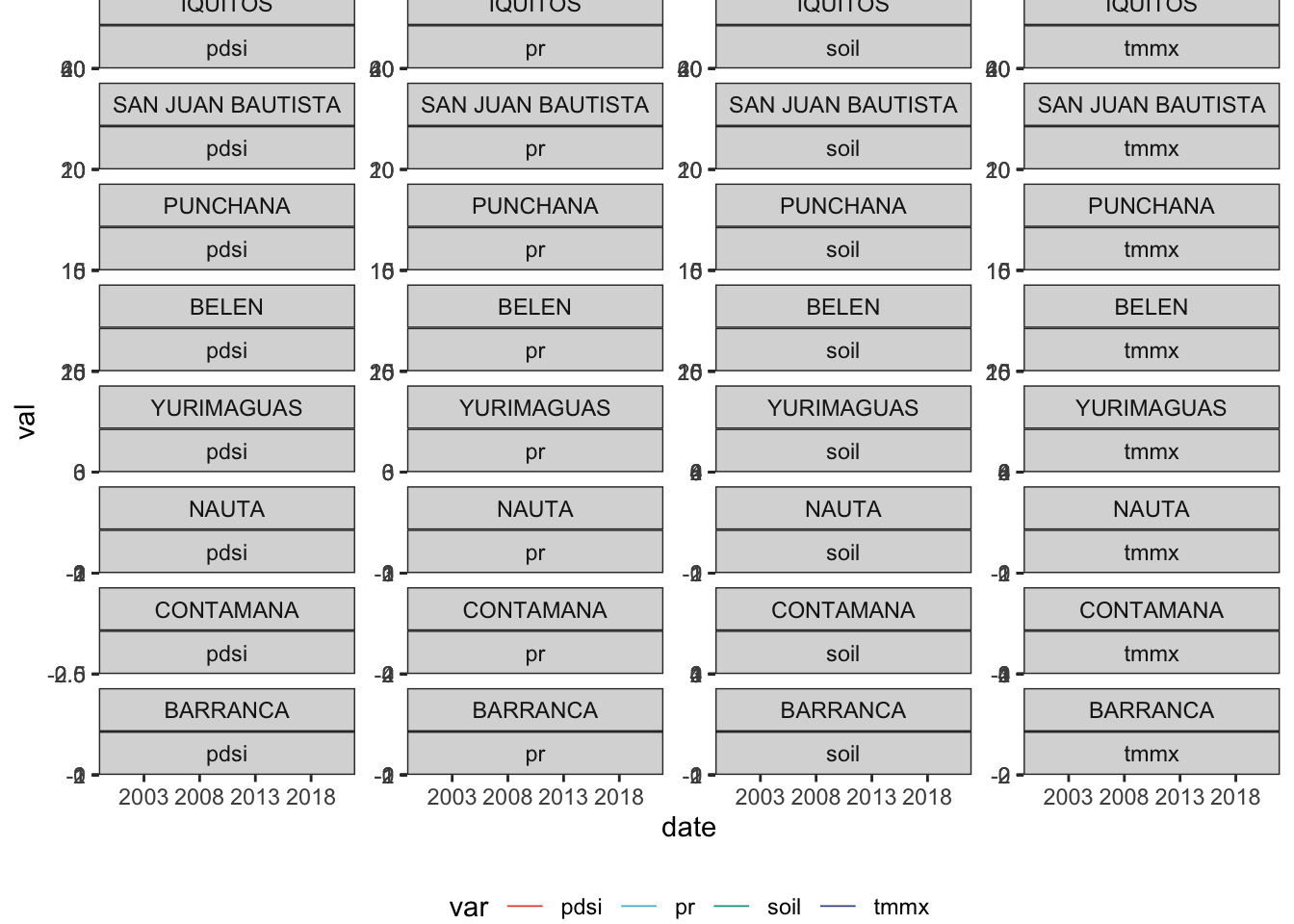
ggsave("./_out/time_series.png", width = 7, height = 10)