Chapter 2 Digital trace data
One elementary skill for Computational social scientists is the analysis of “found” data from the internet. There are two basic approaches to acquire those data: scraping web pages and sending API requests.
Figure 1 shows an overview of different technologies that are used to store and disseminate data (Munzert et al. 2014: 10).
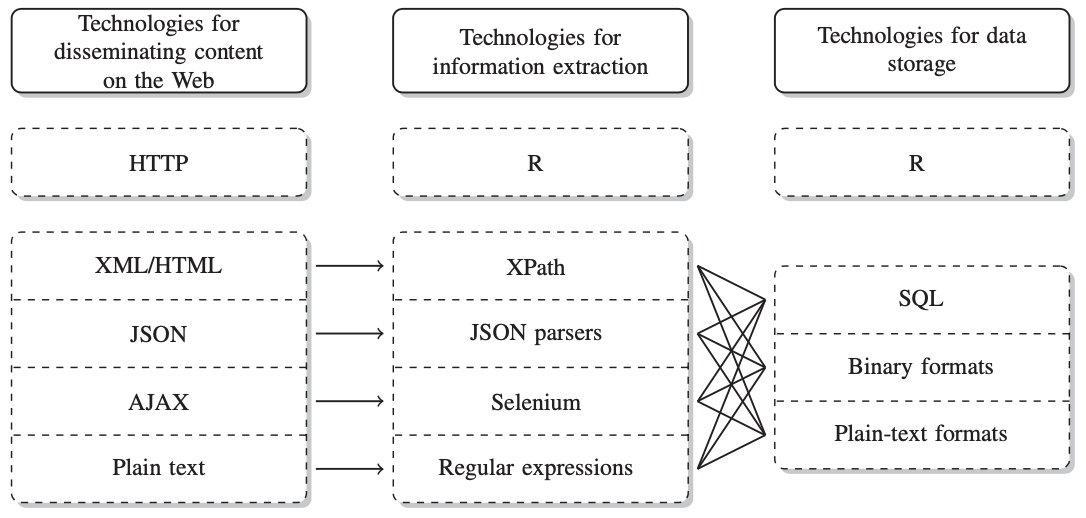
Figure 1: technologies for putting data online and how to extract and store them
HTML is usually used for contents on more basic websites, AJAX on fancier ones which change appearance etc. using JavaScript. We use rvest (Wickham 2019a) for web scraping. AJAX, however, requires more elaborate software (e.g., RSelenium). I will link an extensive tutorial for that later. JSON is the format many application programming interfaces (APIs) provide data in and I will, therefore, dwell a bit more on it in the chapter on APIs.
A problem with data from the web is their messiness. There might be some special characters in there that you want to get rid of or unnecessary text that you need to remove. The most common way to do this is by using regular expressions (regexes) which are introduced in the first part of this chapter. Then, I will introduce the actual scraping. I will also give a brief introduction to HTML, as it will enable you to pre-select relevant parts from the particular web page. rvest has some handy functions to extract certain kinds of content. Those will be introduced, too. Of course, one big advantage of doing scraping with R is the fact that we can automate the process. For instance, we can tell the machine to first scrape a list of links and then following those links and extract information from there (e.g., if you want to want to collect data on housing prices). However, many web pages will not want you to extract their entire page or only do so at a certain rate limit and we definitely need to respect that. This is what the polite (Perepolkin 2019) package is for which works well in connection with rvest. In the final part, you will learn more on APIs and how you can communicate with them. For this, I will also introduce you to JSON, the data format most APIs work with.
2.1 String manipulation
When working with data, a significant number of variables will be in some sort of text format. When you want to manipulate those variables, an easy approach would be exporting the data to MS Excel and then just performing those manipulations by hand. This is very time-consuming, though, and, hence, I rather recommend the R way which scales well and works fast for data sets of varying sizes.
Quick reminder: a string is an element of a character vector and can be created by simply wrapping some text in back ticks:
library(tidyverse)
string <- "Hi, how are you doing?"
vector_of_strings <- c("Hi, how are you doing?", "I'm doing well, HBY?", "Me too, thanks for asking.")The stringr package (Wickham 2019b) contains a multitude of commands (49 in total) which can be used to achieve a couple of things: manipulating character vectors; operations which are sensitive to different locales; matching patterns. Basically, those goals can also be achieved with base R functions, but stringr’s advantage is its consistency. The makers of stringr describe it as
A consistent, simple and easy to use set of wrappers around the fantastic ‘stringi’ package. All function and argument names (and positions) are consistent, all functions deal with “NA”’s and zero length vectors in the same way, and the output from one function is easy to feed into the input of another.
Every stringr function starts with str_ – which facilitates finding the proper command: just type str_ and RStudio’s auto-suggest function should take care of the rest (if it doesn’t pop up by itself, you can trigger it by hitting the tab-key). Also, they take a vector of strings as their first argument, which facilitates using them in a %>%-pipeline and adding them to a mutate()-call.
One important component of stringr functions is regular expressions which will be introduced later as well.
2.1.1 Basic manipulations
In the following, I will introduce you to a number of different operations that can be performed on strings.
2.1.1.1 Changing the case of the words
A basic operation is changing words’ case.
library(tidyverse) #stringr is part of the core tidyverse
str_to_lower(vector_of_strings)## [1] "hi, how are you doing?" "i'm doing well, hby?"
## [3] "me too, thanks for asking."str_to_upper(vector_of_strings)## [1] "HI, HOW ARE YOU DOING?" "I'M DOING WELL, HBY?"
## [3] "ME TOO, THANKS FOR ASKING."str_to_title(vector_of_strings)## [1] "Hi, How Are You Doing?" "I'm Doing Well, Hby?"
## [3] "Me Too, Thanks For Asking."str_to_sentence(vector_of_strings)## [1] "Hi, how are you doing?" "I'm doing well, hby?"
## [3] "Me too, thanks for asking."2.1.1.2 Determining a string’s length
Determining the string’s number of characters goes as follows:
str_length(vector_of_strings)## [1] 22 20 262.1.1.3 Extracting particular characters
Characters can be extracted (by position) using str_sub
str_sub(vector_of_strings, start = 1, end = 5) # extracting first to fifth character## [1] "Hi, h" "I'm d" "Me to"str_sub(vector_of_strings, start = -5, end = -1) # extracting fifth-to-last to last character## [1] "oing?" " HBY?" "king."You can also use str_sub() to replace strings. E.g., to replace the last character by a full stop, you can do the following:
str_sub(vector_of_strings, start = -1) <- "."
vector_of_strings## [1] "Hi, how are you doing." "I'm doing well, HBY."
## [3] "Me too, thanks for asking."However, in everyday use you would probably go with str_replace() and regular expressions.
2.1.1.4 Concatenating strings
Similar to how c() puts together different elements (or vectors of length 1) into a single vector, str_c() can be used to concatenate several strings into a single string. This can, for instance, be used to write some birthday invitations.
names <- c("Inger", "Peter", "Kalle", "Ingrid")
str_c("Hi", names, "I hope you're doing well. As per this letter, I invite you to my birthday party.")## [1] "HiIngerI hope you're doing well. As per this letter, I invite you to my birthday party."
## [2] "HiPeterI hope you're doing well. As per this letter, I invite you to my birthday party."
## [3] "HiKalleI hope you're doing well. As per this letter, I invite you to my birthday party."
## [4] "HiIngridI hope you're doing well. As per this letter, I invite you to my birthday party."Well, this looks kind of ugly, as there are no spaces and commas are lacking as well. You can fix that by determining a separator using the sep argument.
str_c("Hi", names, "I hope you're doing well. As per this letter, I invite you to my birthday party.", sep = ", ")## [1] "Hi, Inger, I hope you're doing well. As per this letter, I invite you to my birthday party."
## [2] "Hi, Peter, I hope you're doing well. As per this letter, I invite you to my birthday party."
## [3] "Hi, Kalle, I hope you're doing well. As per this letter, I invite you to my birthday party."
## [4] "Hi, Ingrid, I hope you're doing well. As per this letter, I invite you to my birthday party."You could also collapse the strings contained in a vector together into one single string using the collapse argument.
str_c(names, collapse = ", ")## [1] "Inger, Peter, Kalle, Ingrid"This can also be achieved using the str_flatten() function.
str_flatten(names, collapse = ", ")## [1] "Inger, Peter, Kalle, Ingrid"2.1.1.5 Repetition
Repeating (or duplicating) strings is performed using str_dup(). The function takes two arguments: the string to be duplicated and the number of times.
str_dup("felix", 2)## [1] "felixfelix"str_dup("felix", 1:3)## [1] "felix" "felixfelix" "felixfelixfelix"str_dup(names, 2)## [1] "IngerInger" "PeterPeter" "KalleKalle" "IngridIngrid"str_dup(names, 1:4)## [1] "Inger" "PeterPeter"
## [3] "KalleKalleKalle" "IngridIngridIngridIngrid"2.1.1.6 Removing unnecessary whitespaces
Often text contains unnecessary whitespaces.
unnecessary_whitespaces <- c(" on the left", "on the right ", " on both sides ", " literally everywhere ")Removing the ones at the beginning or the end of a string can be accomplished using str_trim().
str_trim(unnecessary_whitespaces, side = "left")## [1] "on the left" "on the right "
## [3] "on both sides " "literally everywhere "str_trim(unnecessary_whitespaces, side = "right")## [1] " on the left" "on the right"
## [3] " on both sides" " literally everywhere"str_trim(unnecessary_whitespaces, side = "both") # the default option## [1] "on the left" "on the right"
## [3] "on both sides" "literally everywhere"str_trim() could not fix the last string though, where unnecessary whitespaces were also present in between words. Here, str_squish is more appropriate. It removes leading or trailing whitespaces as well as duplicated ones in between words.
str_squish(unnecessary_whitespaces)## [1] "on the left" "on the right" "on both sides"
## [4] "literally everywhere"2.1.2 Regular expressions
Up to now, you have been introduced to the more basic functions of the stringr package. Those are useful, for sure, yet limited. However, to make use of the full potential of stringr, you will first have to get acquainted to regular expressions (also often abbreviated as “regex” with plural “regexes”).
Those regular expressions are patterns that can be used to describe certain strings. Hence, if you want to replace certain words with another one, you can write the proper regex and it will identify the strings you want to replace and the stringr function (i.e., str_replace()) will take care of the rest. Exemplary use cases of regexes are the identification of phone numbers, email addresses, or whether a password you choose on a web page consists of enough characters, an upper-case character, and at least one special character.
Before you dive into regexes, beware that they are quite complicated in the beginning (honestly, I was quite overwhelmed when I encountered them first). Yet, mastering them is very rewarding and will definitely pay off in the future.
2.1.2.1 Literal characters
The most basic regex patterns consist of literal characters only. str_view() tells you which parts of a string match a pattern is present in the element.
five_largest_cities <- c("Stockholm", "Göteborg", "Malmö", "Uppsala", "Västerås")Note that regexes are case-sensitive.
str_view(five_largest_cities, "stockholm")str_view(five_largest_cities, "Stockholm")## [1] │ <Stockholm>They also match parts of words:
str_view(five_largest_cities, "borg")## [2] │ Göte<borg>Moreover, they are “greedy,” they only match the first occurrence (in “Stockholm”):
str_view(five_largest_cities, "o")## [1] │ St<o>ckh<o>lm
## [2] │ Göteb<o>rgThis can be addressed in the stringr package by using str_._all() function – but more on that later.
If you want to match multiple literal characters (or words, for that sake), you can connect them using the | meta character (more on meta characters later).
str_view(five_largest_cities, "Stockholm|Göteborg")## [1] │ <Stockholm>
## [2] │ <Göteborg>Every letter of the English alphabet (or number/or combination of those) can serve as a literal character. Those literal characters match themselves. This is, however, not the case with the other sort of characters, so-called meta characters.
2.1.2.2 Metacharacters
When using regexes, the following characters are considered meta characters and have a special meaning:
. \ | ( ) { } [ ] ^ $ - * + ?
2.1.2.2.1 The wildcard
Did you notice how I used the dot to refer to the entirety of the str_._all() functions? This is basically what the . meta-character does: it matches every character except for a new line. The first call extracts all function names from the stringr package, the second one shows the matches (i.e., the elements of the vector where it can find the pattern).
stringr_functions <- ls("package:stringr")
str_detect(stringr_functions, "str_._all")## [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [13] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [25] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [49] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [61] FALSE FALSEWell, as you can see, there are none. This is due to the fact that the . can only replace one character. We need some sort of multiplier to find them. The ones available are:
?– zero or one*– zero or more+– one or more{n}– exactly n{n,}– n or more{n,m}– between n and m
In our case, the appropriate one is +:
str_detect(stringr_functions, "str_.+_all")## [1] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE
## [13] FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE FALSE FALSE FALSE
## [25] FALSE FALSE TRUE FALSE TRUE FALSE FALSE FALSE FALSE TRUE FALSE TRUE
## [37] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE
## [49] FALSE FALSE FALSE FALSE FALSE FALSE FALSE FALSE TRUE FALSE FALSE FALSE
## [61] FALSE FALSEHowever, if you want to match the character dot? This problem may arise when searching for clock time. A naive regex might look like this:
vectors_with_time <- c("13500", "13M00", "13.00")
str_detect(vectors_with_time, "13.00")## [1] TRUE TRUE TRUEYet, it matches everything. We need some sort of literal dot. Here, the meta character \ comes in handy. By putting it in front of the meta character, it does no longer has its special meaning, and is interpreted as a literal character. This procedure is referred to as “escaping.” Hence, \ is also referred to as the “escape character.” Note that you will need to escape \ as well, and therefore it will look like this: \\..
str_detect(vectors_with_time, "13\\.00")## [1] FALSE FALSE TRUE2.1.2.3 Sets of characters
You can also define sets of multiple characters using the [ ] meta characters. This can be used to define multiple possible characters that can appear in the same place.
sp_ce <- c("spice", "space")
str_view(sp_ce, "sp[ai]ce")## [1] │ <spice>
## [2] │ <space>You can also define certain ranges of characters using the - meta character:
Same holds for numbers:
american_phone_number <- "(555) 555-1234"
str_view(american_phone_number, "\\([:digit:]{3}\\) [0-9]{3}-[0-9]{4}")## [1] │ <(555) 555-1234>There are also predefined sets of characters, for instance digits or letters, which are called character classes. You can find them on the stringr cheatsheet.
Furthermore, you can put almost every meta character inside the square brackets without escaping them. This does not apply to the the caret (^) in first position, the dash -, the closing square bracket ], and the backslash \.
str_view(vector_of_strings, "[.]")## [1] │ Hi, how are you doing<.>
## [2] │ I'm doing well, HBY<.>
## [3] │ Me too, thanks for asking<.>2.1.2.4 Anchors
There is also a way to define whether you want the pattern to be present in the beginning ^ or at the end $ of a string. sentences are a couple of (i.e., 720) predefined example sentences. If I were now interested in the number of sentences that begin with a “the,” I could write the following regex:
shortened_sentences <- sentences[1:10]
str_view(shortened_sentences, "^The") ## [1] │ <The> birch canoe slid on the smooth planks.
## [4] │ <The>se days a chicken leg is a rare dish.
## [6] │ <The> juice of lemons makes fine punch.
## [7] │ <The> box was thrown beside the parked truck.
## [8] │ <The> hogs were fed chopped corn and garbage.If I wanted to know how many start with a “The” and end with a full stop, I could do this one:
str_view(shortened_sentences, "^The.+\\.$") ## [1] │ <The birch canoe slid on the smooth planks.>
## [4] │ <These days a chicken leg is a rare dish.>
## [6] │ <The juice of lemons makes fine punch.>
## [7] │ <The box was thrown beside the parked truck.>
## [8] │ <The hogs were fed chopped corn and garbage.>2.1.2.4.1 Boundaries
Note that right now, the regex also matches the sentence which starts with a “These.” In order to address this, I need to tell the machine that it should only accept a “The” if there starts a new word thereafter. In regex syntax, this is done using so-called boundaries. Those are defined as \b as a word boundary and \B as no word boundary. (Note that you will need an additional escape character as you will have to escape the escape character itself.)
In my example, I would include the former if I were to search for sentences that begin with a single “The” and the latter if I were to search for sentences that begin with a word that starts with a “The” but are not “The” – such as “These.”
str_view(shortened_sentences, "^The\\b.+\\.$") ## [1] │ <The birch canoe slid on the smooth planks.>
## [6] │ <The juice of lemons makes fine punch.>
## [7] │ <The box was thrown beside the parked truck.>
## [8] │ <The hogs were fed chopped corn and garbage.>str_view(shortened_sentences, "^The\\B.+\\.$") ## [4] │ <These days a chicken leg is a rare dish.>2.1.2.4.2 Lookarounds
A final common task is to extract certain words or values based on what comes before or after them. Look at the following example:
heights <- c("1m30cm", "2m01cm", "3m10cm")Here, in order to identify the height in meters, the first task is to identify all the numbers that are followed by an “m”. The regex syntax for this looks like this: A(?=pattern) with A being the entity that is supposed to be found (hence, in this case, [0-9]+).
str_view(heights, "[0-9]+(?=m)")## [1] │ <1>m30cm
## [2] │ <2>m01cm
## [3] │ <3>m10cmThe second step now is to identify the centimeters. This could of course be achieved using the same regex and replacing m by cm. However, we can also harness a so-called negative look ahead A(?!pattern), a so-called look behind (?<=pattern)A. The negative counterpart, the negative look behind (?<!pattern)A could be used to extract the meters.
The negative look ahead basically returns everything that is not followed by the defined pattern. The look behind returns everything that is preceded by the pattern, the negative look behind returns everything that is not preceded by the pattern.
In the following, I demonstrate how you could extract the centimeters using negative look ahead and look behind.
str_view(heights, "[0-9]+(?!m)") # negative look ahead## [1] │ 1m<30>cm
## [2] │ 2m<01>cm
## [3] │ 3m<10>cmstr_view(heights, "(?<=m)[0-9]+") # look behind## [1] │ 1m<30>cm
## [2] │ 2m<01>cm
## [3] │ 3m<10>cm2.1.3 More advanced string manipulation
Now that you have learned about regexes, you can unleash the full power of stringr.
The basic syntax of a stringr function looks as follows: str_.*(string, regex("")). Some stringr functions also have the suffix _all which implies that they perform the operation not only on the first match (“greedy”) but on every match.
In order to demonstrate the different functions, I will again rely on the subset of example sentences.
2.1.3.1 Detect matches
str_detect can be used to determine whether a certain pattern is present in the string.
str_detect(shortened_sentences, "The\\b")## [1] TRUE FALSE FALSE FALSE FALSE TRUE TRUE TRUE FALSE FALSEThis also works very well in a dplyr::filter() call. Finding all action movies in the IMDB data set can be solved like this:
imdb_raw <- read_csv("https://www.dropbox.com/s/81o3zzdkw737vt0/imdb2006-2016.csv?dl=1")## Rows: 1000 Columns: 12
## ── Column specification ──────────────────
## Delimiter: ","
## chr (5): Title, Genre, Description, Director, Actors
## dbl (7): Rank, Year, Runtime (Minutes), Rating, Votes, Revenue (Millions), M...
##
## ℹ Use `spec()` to retrieve the full column specification for this data.
## ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.imdb_raw %>%
filter(str_detect(Genre, "Action"))## # A tibble: 303 × 12
## Rank Title Genre Descr…¹ Direc…² Actors Year Runti…³ Rating Votes Reven…⁴
## <dbl> <chr> <chr> <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 1 Guard… Acti… A grou… James … Chris… 2014 121 8.1 757074 333.
## 2 5 Suici… Acti… A secr… David … Will … 2016 123 6.2 393727 325.
## 3 6 The G… Acti… Europe… Yimou … Matt … 2016 103 6.1 56036 45.1
## 4 9 The L… Acti… A true… James … Charl… 2016 141 7.1 7188 8.01
## 5 13 Rogue… Acti… The Re… Gareth… Felic… 2016 133 7.9 323118 532.
## 6 15 Colos… Acti… Gloria… Nacho … Anne … 2016 109 6.4 8612 2.87
## 7 18 Jason… Acti… The CI… Paul G… Matt … 2016 123 6.7 150823 162.
## 8 25 Indep… Acti… Two de… Roland… Liam … 2016 120 5.3 127553 103.
## 9 27 Bahub… Acti… In anc… S.S. R… Prabh… 2015 159 8.3 76193 6.5
## 10 30 Assas… Acti… When C… Justin… Micha… 2016 115 5.9 112813 54.6
## # … with 293 more rows, 1 more variable: Metascore <dbl>, and abbreviated
## # variable names ¹Description, ²Director, ³`Runtime (Minutes)`,
## # ⁴`Revenue (Millions)`If you want to know whether there are multiple matches present in each string, you can use str_count. Here, it might by advisable to set the ignore_case option to TRUE:
str_count(shortened_sentences, regex("the\\b", ignore_case = TRUE))## [1] 2 2 1 0 0 1 2 1 0 0If you want to locate the match in the string, use str_locate. This returns a matrix, which is basically a vector of multiple dimensions.
str_locate(shortened_sentences, regex("The\\b", ignore_case = TRUE))## start end
## [1,] 1 3
## [2,] 6 8
## [3,] 19 21
## [4,] NA NA
## [5,] NA NA
## [6,] 1 3
## [7,] 1 3
## [8,] 1 3
## [9,] NA NA
## [10,] NA NAMoreover, this is a good example for the greediness of stringr functions. Hence, it is advisable to use str_locate_all which returns a list with one matrix for each element of the original vector:
str_locate_all(shortened_sentences, regex("The\\b", ignore_case = TRUE))## [[1]]
## start end
## [1,] 1 3
## [2,] 25 27
##
## [[2]]
## start end
## [1,] 6 8
## [2,] 19 21
##
## [[3]]
## start end
## [1,] 19 21
##
## [[4]]
## start end
##
## [[5]]
## start end
##
## [[6]]
## start end
## [1,] 1 3
##
## [[7]]
## start end
## [1,] 1 3
## [2,] 27 29
##
## [[8]]
## start end
## [1,] 1 3
##
## [[9]]
## start end
##
## [[10]]
## start end2.1.3.2 Mutating strings
Mutating strings usually implies the replacement of certain elements (e.g., words) with other elements (or removing them, which is basically a special case of replacing them). In stringr this is performed using str_replace(string, pattern, replacement) and str_replace_all(string, pattern, replacement).
If I wanted, for instance, replace the first occurrence of “m” letters by “meters,” I would go about this the following way:
str_replace(heights, "m", "meters")## [1] "1meters30cm" "2meters01cm" "3meters10cm"Note that str_replace_all would have lead to the following outcome:
str_replace_all(heights, "m", "meters")## [1] "1meters30cmeters" "2meters01cmeters" "3meters10cmeters"However, I also want to replace the “cm” with “centimeters,” hence, I can harness another feature of str_replace_all():
str_replace_all(heights, c("m" = "meters", "cm" = "centimeters"))## [1] "1meters30centimeterseters" "2meters01centimeterseters"
## [3] "3meters10centimeterseters"What becomes obvious is that a “simple” regex containing just literal characters more often than not does not suffice. It will be your task to fix this. And while on it, you can also address the meter/meters problem – a “1” needs meter instead of meters. Another feature is that the replacements are performed in order. You can harness this for solving the problem.
Solution. Click to expand!
Solution:
str_replace_all(heights, c("(?<=[2-9]{1})m" = "meters", "(?<=[0-9]{2})m" = "meters", "(?<=1)m" = "meter", "(?<=01)cm$" = "centimeter", "cm$" = "centimeters"))## [1] "1meter30centimeters" "2meters01centimeter" "3meters10centimeters"2.1.3.3 Extracting text
str_extract(_all)() can be used to extract matching strings. In the mtcars data set, the first word describes the car brand. Here, I harness another regexp, the \\w which stands for any word character. Its opponent is \\W for any non-word character.
mtcars %>%
rownames_to_column(var = "car_model") %>%
transmute(manufacturer = str_extract(car_model, "^\\w+\\b"))## manufacturer
## 1 Mazda
## 2 Mazda
## 3 Datsun
## 4 Hornet
## 5 Hornet
## 6 Valiant
## 7 Duster
## 8 Merc
## 9 Merc
## 10 Merc
## 11 Merc
## 12 Merc
## 13 Merc
## 14 Merc
## 15 Cadillac
## 16 Lincoln
## 17 Chrysler
## 18 Fiat
## 19 Honda
## 20 Toyota
## 21 Toyota
## 22 Dodge
## 23 AMC
## 24 Camaro
## 25 Pontiac
## 26 Fiat
## 27 Porsche
## 28 Lotus
## 29 Ford
## 30 Ferrari
## 31 Maserati
## 32 Volvo2.1.3.4 Split vectors
Another use case here would have been to split it into two columns: manufacturer and model. One approach would be to use str_split(). This function splits the string at every occurrence of the predefined pattern. In this example, I use a word boundary as the pattern:
manufacturer_model <- rownames(mtcars)
str_split(manufacturer_model, "\\b") %>%
head()## [[1]]
## [1] "" "Mazda" " " "RX4" ""
##
## [[2]]
## [1] "" "Mazda" " " "RX4" " " "Wag" ""
##
## [[3]]
## [1] "" "Datsun" " " "710" ""
##
## [[4]]
## [1] "" "Hornet" " " "4" " " "Drive" ""
##
## [[5]]
## [1] "" "Hornet" " " "Sportabout" ""
##
## [[6]]
## [1] "" "Valiant" ""This outputs a list containing the different singular words/special characters. This doesn’t make sense in this case. Here, however, the structure of the string is always roughly the same: “\[manufacturer\]\[ \]\[model description\]”. Moreover, the manufacturer is only one word. Hence, the task can be fixed by splitting the string after the first word, which should indicate the manufacturer. This can be accomplished using str_split_fixed(). Fixed means that the number of splits is predefined. This returns a matrix that can easily become a tibble.
str_split_fixed(manufacturer_model, "(?<=\\w)\\b", n = 2) %>%
as_tibble() %>%
rename(manufacturer = V1,
model = V2) %>%
mutate(model = str_squish(model))## Warning: The `x` argument of `as_tibble.matrix()`
## must have unique column names if
## `.name_repair` is omitted as of tibble
## 2.0.0.
## ℹ Using compatibility `.name_repair`.## # A tibble: 32 × 2
## manufacturer model
## <chr> <chr>
## 1 Mazda "RX4"
## 2 Mazda "RX4 Wag"
## 3 Datsun "710"
## 4 Hornet "4 Drive"
## 5 Hornet "Sportabout"
## 6 Valiant ""
## 7 Duster "360"
## 8 Merc "240D"
## 9 Merc "230"
## 10 Merc "280"
## # … with 22 more rows2.2 Web scraping
Today’s session will be dedicated to getting data from the web. This process is also called scraping since we scrape data off from the surface and remodel it for our inferences. The following picture shows you the web scraping cheat sheet that outlines the process of scraping the web. On the left side, you can see the first step in scraping the web which is requesting the information from the server. This is basically what is going under the hood when you make requests using a browser. The response is the website, usually stored in an XML document, which is then the starting point for your subsequent queries and data extraction.
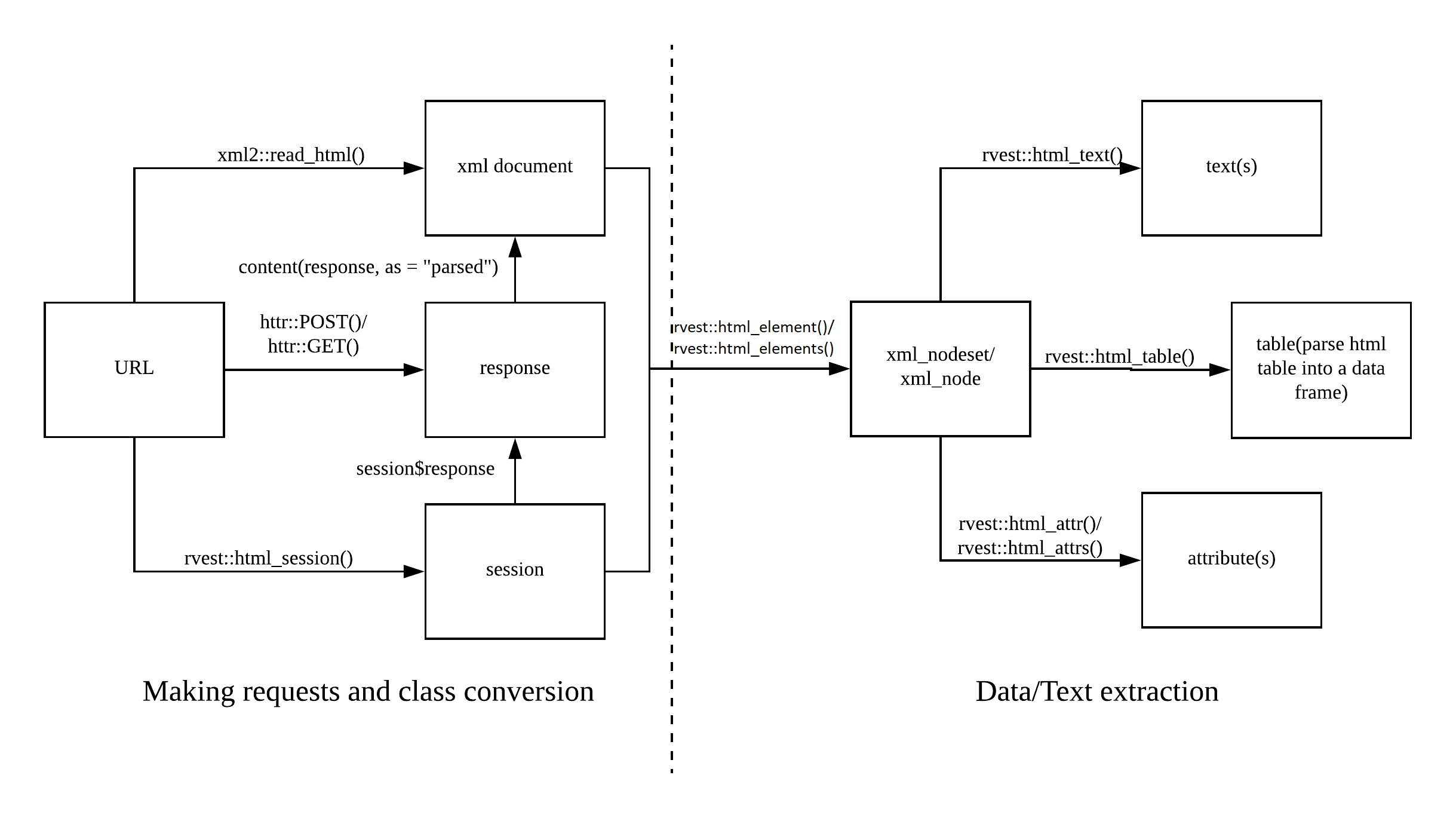
Web scraping cheat sheet
In today’s session, you will learn different techniques to get your hands on data. In particular, this will encompass making simple URL requests with read_html(), using session()s to navigate around on a web page, submitting html_form()s to fill in forms on a web page, and making structured requests to APIs.
2.2.1 Making requests
The most basic form of making a request is by using read_html() from the xml2 package.
#install.packages("rvest")
library(rvest) # this also loads the xml2 package
library(tidyverse)
page <- read_html("https://en.wikipedia.org/wiki/Tidyverse")
page %>% str()## List of 2
## $ node:<externalptr>
## $ doc :<externalptr>
## - attr(*, "class")= chr [1:2] "xml_document" "xml_node"#page %>% as.character() %>% write_lines("wiki.html")This is perfectly fine for making requests to static pages where you do not need to take any further action. Sometimes, however, this is not enough, and you want to accept cookies or move on the page.
2.2.1.1 session()
However, the slickest way to do this is by using a session(). In a session, R behaves like a normal browser, stores cookies, allows you to navigate between pages, by going session_forward() or session_back(), session_follow_link()s on the page itself or session_jump_to() a different URL, or submit form()s with session_submit().
First, you start the session by simply calling session().
my_session <- session("https://scrapethissite.com/")Some servers may not want robots to make requests and block you for this reason. To circumnavigate this, we can set a “user agent” in a session. The user agent contains data that the server receives from us when we make the request. Hence, by adapting it we can trick the server into thinking that we are humans instead of robots. Let’s check the current user agent first:
my_session$response$request$options$useragent## [1] "libcurl/7.85.0 r-curl/4.3.3 httr/1.4.4"Not very human. We can set it to a common one using the httr package (which powers rvest).
user_a <- httr::user_agent("Mozilla/5.0 (Macintosh; Intel Mac OS X 12_0_1) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/95.0.4638.69 Safari/537.36")
session_with_ua <- session("https://scrapethissite.com/", user_a)
session_with_ua$response$request$options$useragent## [1] "Mozilla/5.0 (Macintosh; Intel Mac OS X 12_0_1) AppleWebKit/537.36 (KHTML, like Gecko) Chrome/95.0.4638.69 Safari/537.36"You can check the response using session$response$status_code – 200 is good.
my_session$response$status_code## [1] 200When you want to save a page from the session, do so using read_html().
page <- read_html(session_with_ua)If you want to open a new URL, hit session_jump_to().
library(magrittr)
session_with_ua %<>% session_jump_to("https://www.scrapethissite.com/pages/")
session_with_ua## <session> https://www.scrapethissite.com/pages/
## Status: 200
## Type: text/html; charset=utf-8
## Size: 10603Once you are familiar with CSS selectors – which will be introduced tomorrow – you can also click buttons on the page:
session_with_ua %<>% session_jump_to("https://www.scrapethissite.com/") %>%
session_follow_link(css = ".btn-default")## Navigating to /pages/session_with_ua## <session> https://www.scrapethissite.com/pages/
## Status: 200
## Type: text/html; charset=utf-8
## Size: 10603Wanna go back – session_back(); thereafter you can go session_forward(), too.
session_with_ua %<>%
session_back()
session_with_ua## <session> https://www.scrapethissite.com/
## Status: 200
## Type: text/html; charset=utf-8
## Size: 8117session_with_ua %<>%
session_forward()
session_with_ua## <session> https://www.scrapethissite.com/pages/
## Status: 200
## Type: text/html; charset=utf-8
## Size: 10603You can look at what your scraper has done with session_history().
session_with_ua %>% session_history()## https://www.scrapethissite.com/
## https://www.scrapethissite.com/pages/
## https://www.scrapethissite.com/
## - https://www.scrapethissite.com/pages/2.2.1.2 Forms
Sometimes we also want to provide certain input, e.g., to provide login credentials or to scrape a website more systematically. That information is usually provided using so-called forms. A <form> element can contain different other elements such as text fields or checkboxes. Basically, we use html_form() to extract the form, html_form_set() to define what we want to submit, and html_form_submit() to finally submit it. For a basic example, wesearch for something on Google.
google <- read_html("http://www.google.com")
search <- html_form(google) %>% pluck(1)
search %>% str()## List of 5
## $ name : chr "f"
## $ method : chr "GET"
## $ action : chr "http://www.google.com/search"
## $ enctype: chr "form"
## $ fields :List of 10
## ..$ ie :List of 4
## .. ..$ type : chr "hidden"
## .. ..$ name : chr "ie"
## .. ..$ value: chr "ISO-8859-1"
## .. ..$ attr :List of 3
## .. .. ..$ name : chr "ie"
## .. .. ..$ value: chr "ISO-8859-1"
## .. .. ..$ type : chr "hidden"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ hl :List of 4
## .. ..$ type : chr "hidden"
## .. ..$ name : chr "hl"
## .. ..$ value: chr "fr"
## .. ..$ attr :List of 3
## .. .. ..$ value: chr "fr"
## .. .. ..$ name : chr "hl"
## .. .. ..$ type : chr "hidden"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ source:List of 4
## .. ..$ type : chr "hidden"
## .. ..$ name : chr "source"
## .. ..$ value: chr "hp"
## .. ..$ attr :List of 3
## .. .. ..$ name : chr "source"
## .. .. ..$ type : chr "hidden"
## .. .. ..$ value: chr "hp"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ biw :List of 4
## .. ..$ type : chr "hidden"
## .. ..$ name : chr "biw"
## .. ..$ value: NULL
## .. ..$ attr :List of 2
## .. .. ..$ name: chr "biw"
## .. .. ..$ type: chr "hidden"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ bih :List of 4
## .. ..$ type : chr "hidden"
## .. ..$ name : chr "bih"
## .. ..$ value: NULL
## .. ..$ attr :List of 2
## .. .. ..$ name: chr "bih"
## .. .. ..$ type: chr "hidden"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ q :List of 4
## .. ..$ type : chr "text"
## .. ..$ name : chr "q"
## .. ..$ value: chr ""
## .. ..$ attr :List of 8
## .. .. ..$ class : chr "lst"
## .. .. ..$ style : chr "margin:0;padding:5px 8px 0 6px;vertical-align:top;color:#000"
## .. .. ..$ autocomplete: chr "off"
## .. .. ..$ value : chr ""
## .. .. ..$ title : chr "Recherche Google"
## .. .. ..$ maxlength : chr "2048"
## .. .. ..$ name : chr "q"
## .. .. ..$ size : chr "57"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ btnG :List of 4
## .. ..$ type : chr "submit"
## .. ..$ name : chr "btnG"
## .. ..$ value: chr "Recherche Google"
## .. ..$ attr :List of 4
## .. .. ..$ class: chr "lsb"
## .. .. ..$ value: chr "Recherche Google"
## .. .. ..$ name : chr "btnG"
## .. .. ..$ type : chr "submit"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ btnI :List of 4
## .. ..$ type : chr "submit"
## .. ..$ name : chr "btnI"
## .. ..$ value: chr "J'ai de la chance"
## .. ..$ attr :List of 5
## .. .. ..$ class: chr "lsb"
## .. .. ..$ id : chr "tsuid_1"
## .. .. ..$ value: chr "J'ai de la chance"
## .. .. ..$ name : chr "btnI"
## .. .. ..$ type : chr "submit"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ iflsig:List of 4
## .. ..$ type : chr "hidden"
## .. ..$ name : chr "iflsig"
## .. ..$ value: chr "AK50M_UAAAAAY8Vj4sbCxc2sMzWuEW653JADeqw5D70q"
## .. ..$ attr :List of 3
## .. .. ..$ value: chr "AK50M_UAAAAAY8Vj4sbCxc2sMzWuEW653JADeqw5D70q"
## .. .. ..$ name : chr "iflsig"
## .. .. ..$ type : chr "hidden"
## .. ..- attr(*, "class")= chr "rvest_field"
## ..$ gbv :List of 4
## .. ..$ type : chr "hidden"
## .. ..$ name : chr "gbv"
## .. ..$ value: chr "1"
## .. ..$ attr :List of 4
## .. .. ..$ id : chr "gbv"
## .. .. ..$ name : chr "gbv"
## .. .. ..$ type : chr "hidden"
## .. .. ..$ value: chr "1"
## .. ..- attr(*, "class")= chr "rvest_field"
## - attr(*, "class")= chr "rvest_form"search_something <- search %>% html_form_set(q = "something")
resp <- html_form_submit(search_something, submit = "btnG")
read_html(resp)## {html_document}
## <html lang="fr">
## [1] <head>\n<meta http-equiv="Content-Type" content="text/html; charset=UTF-8 ...
## [2] <body jsmodel="hspDDf">\n<header id="hdr"><script nonce="ucJld5BfOeOfHXT_ ...vals <- list(q = "web scraping", hl = "en")
search <- search %>% html_form_set(!!!vals)
resp <- html_form_submit(search)
read_html(resp)## {html_document}
## <html lang="en-FR">
## [1] <head>\n<meta http-equiv="Content-Type" content="text/html; charset=UTF-8 ...
## [2] <body jsmodel="hspDDf">\n<header id="hdr"><script nonce="VRuYic_pVjYumWvi ...If you are working with a session, the workflow is as follows:
- Extract the form.
- Set it.
- Start your session on the page with the form.
- Submit the form using
session_submit().
google_form <- read_html("http://www.google.com") %>%
html_form() %>%
pluck(1) #another way to do [[1]]
search_something <- google_form %>% html_form_set(q = "something")
google_session <- session("http://www.google.com") %>%
session_submit(search_something, submit = "btnG")
google_session %>%
read_html()## {html_document}
## <html lang="fr">
## [1] <head>\n<meta http-equiv="Content-Type" content="text/html; charset=UTF-8 ...
## [2] <body jsmodel="hspDDf">\n<header id="hdr"><script nonce="T-AeKAjCymYu2Wdr ...2.2.1.3 Scraping hacks
Some web pages are a bit fancier than the ones we have looked at so far (i.e., they use JavaScript). rvest works nicely for static web pages, but for more advanced ones you need different tools such as RSelenium. This, however, goes beyond the scope of this tutorial.
A web page may sometimes give you time-outs (i.e., it doesn’t respond within a given time). This can break your loop. Wrapping your code in safely() or insistently() from the purrr package might help. The former moves on and notes down what has gone wrong, the latter keeps sending requests until it has been successful. They both work easiest if you put your scraping code in functions and wrap those with either insistently() or safely().
Sometimes a web page keeps blocking you. Consider using a proxy server.
my_proxy <- httr::use_proxy(url = "http://example.com",
user_name = "myusername",
password = "mypassword",
auth = "one of basic, digest, digest_ie, gssnegotiate, ntlm, any")
my_session <- session("https://scrapethissite.com/", my_proxy)Find more useful information – including the stuff we just described – and links on this GitHub page.
2.2.2 Application Programming Interfaces (APIs)
While web scraping (or screen scraping, as you extract the stuff that appears on your screen) is certainly fun, it should be seen as a last resort. More and more web platforms provide so-called Application Programming Interfaces (APIs).
“An application programming interface (API) is a connection between computers or between computer programs.” (Wikipedia)
There are a bunch of different sorts of APIs, but the most common one is the REST API. REST stands for “Representational State Transfer” and describes a set of rules the API designers are supposed to obey when developing their particular interface. You can make different requests, such as GET content, POST a file to a server – PUT is similar, or request to DELETE a file. We will only focus on the GET part.
APIs offer you a structured way to communicate with the platform via your machine. In our use case, this means that you can get the data you want in a usually well-structured format and without all the “dirt” that you need to scrape off tediously (enough web scraping metaphors for today). With APIs, you can generally quite clearly define what you want and how you want it. In R, we achieve this by using the httr (Wickham 2020) package. Moreover, using APIs does not bear the risk of acquiring the information you are not supposed to access and you also do not need to worry about the server not being able to handle the load of your requests (usually, there are rate limits in place to address this particular issue). However, it’s not all fun and games with APIs: they might give you their data in a special format, both XML and JSON are common. The former is the one rvest uses as well, the latter can be tamed using jsonlite (Ooms, Temple Lang, and Hilaiel 2020) which is to be introduced as well. Moreover, you usually have to ask the platform for permission and perhaps pay to get it. Once you have received the keys you need, you can tell R to fill them automatically, similar to how your browser knows your Amazon password, etc.; usethis (Wickham et al. 2021) can help you with such tasks. An overview of currently existing APIs can be found on The Programmable Web
The best thing that can happen with APIs: some of them are so popular that people have already written specific R packages for working with them – an overview can be found on the ROpenSci website. One example of this is Twitter and the rtweet package (Kearney 2019) which will be introduced in the end. Less work for us, great.
2.2.2.1 Obtaining their data
API requests are performed using URLs. Those start with the basic address of the API (e.g., https://api.nytimes.com), followed by the endpoint that you want to use (e.g., /lists). They also contain so-called headers which are provided as key-value pairs. Those headers can contain for instance authentification tokens or different search parameters. A request to the New York Times API to obtain articles for January 2019 would then look like this: https://api.nytimes.com/svc/archive/v1/2019/1.json?api-key=yourkey.
At most APIs, you will have to register first. As we will play with the New York Times API, do this here.
2.2.2.1.1 Making queries
A basic query is performed using the GET() function. However, first, you need to define the call you want to make. The different keys and values they can take can be found in the API documentation. Of course, there is also a neater way to deal with the key problem. We will show it later.
library(httr)
key <- "xxx"
nyt_headlines <- modify_url(
url = "https://api.nytimes.com/",
path = "svc/news/v3/content/nyt/business.json",
query = list(`api-key` = key))
response <- GET(nyt_headlines)When it comes to the NYT news API, there is the problem that the type of section is specified not in the query but in the endpoint path itself. Hence, if we were to scrape the different sections, we would have to change the path itself, e.g., through str_c().
The Status: code you want to see here is 200 which stands for success. If you want to put it inside a function, you might want to break the function once you get a non-successful query. http_error() or http_status() are your friends here.
response %>% http_error()## [1] TRUEresponse %>% http_status()## $category
## [1] "Client error"
##
## $reason
## [1] "Unauthorized"
##
## $message
## [1] "Client error: (401) Unauthorized"content() will give you the content of the request.
response %>% content() %>% glimpse()What you see is also the content of the call – which is what we want. It is in a format that we cannot work with right away, though, it is in JSON.
2.2.2.1.2 JSON
The following unordered list is stolen from this blog entry:
- The data are in name/value pairs
- Commas separate data objects
- Curly braces {} hold objects
- Square brackets [] hold arrays
- Each data element is enclosed with quotes “” if it is a character, or without quotes if it is a numeric value
writeLines(rawToChar(response$content))jsonlite helps us to bring this output into a data frame.
library(jsonlite)
response %>%
content(as = "text") %>%
fromJSON()## $fault
## $fault$faultstring
## [1] "Invalid ApiKey"
##
## $fault$detail
## $fault$detail$errorcode
## [1] "oauth.v2.InvalidApiKey"2.2.2.1.3 Dealing with authentification
Well, as we saw before, we would have to put our official NYT API key publicly visible in this script. This is bad practice and should be avoided, especially if you work on a joint project (where everybody uses their code) or if you put your scripts in public places (such as GitHub). The usethis package can help you here.
#usethis::edit_r_environ() # save key there
Sys.getenv("nyt_key")Hence, if we now search for articles – find the proper parameters here, we provide the key by using the Sys.getenv function. So, if somebody wants to work with your code and their own key, all they need to make sure is that they have the API key stored in the environment with the same name.
modify_url(
url = "http://api.nytimes.com/svc/search/v2/articlesearch.json",
query = list(q = "Trump",
pub = "20161101",
end_date = "20161110",
`api-key` = Sys.getenv("nyt_key"))
) %>%
GET()## Response [http://api.nytimes.com/svc/search/v2/articlesearch.json?q=Trump&pub=20161101&end_date=20161110&api-key=qekEhoGTXqjsZnXpqHns0Vfa2U6T7ABf]
## Date: 2023-01-16 13:49
## Status: 200
## Content-Type: application/json
## Size: 190 kB2.2.2.2 rtweet
Twitter is quite popular among social scientists. The main reason for this is arguably its data accessibility. For R users, the package you want to use is rtweet by Michael Kearney (find an overview of the different packages and their capabilities here). There is a great and quite complete presentation demonstrating its capabilities. This presentation is a bit outdated. The main difference to the package these days is that you will not need to register an app upfront anymore. All you need is a Twitter account. When you make your first request, a browser window will open where you log on to Twitter, authorize the app, and then you can just go for it. There are certain rate limits, too, which you will need to be aware of when you try to acquire data. Rate limits and the parameters you need to specify can be found in the extensive documentation.
In the following, we will just link to the respective vignettes. Please, feel free to play around with the functions yourself. As a starting point, we provide you with the list of all British MPs and some tasks in exercise 3. A first introduction gives this vignette.
library(rtweet)
auth_as('create_token')
uk_mps <- lists_members(
list_id = "217199644"
)2.2.3 Extracting data from unstructured pages
So far, you have been shown how to make calls to web pages and get responses. Moreover, you were introduced to making calls to APIs which (usually) give you content in a nice and structured manner. In this chapter, the guiding topic will be how you can extract content from web pages that give you unstructured content in a structured way. The (in our opinion) easiest way to achieve that is by harnessing the way the web is written.
2.2.3.1 HTML 101
Web content is usually written in HTML (Hyper Text Markup Language). An HTML document is comprised of elements that are letting its content appear in a certain way.
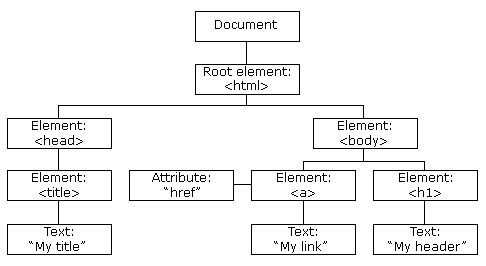
The tree-like structure of an HTML document
The way these elements look is defined by so-called tags.

The opening tag is the name of the element (p in this case) in angle brackets, and the closing tag is the same with a forward slash before the name. p stands for a paragraph element and would look like this (since RMarkdown can handle HTML tags, the second line will showcase how it would appear on a web page:
<p> My cat is very grumpy. <p/>
My cat is very grumpy.
The <p> tag makes sure that the text is standing by itself and that a line break is included thereafter:
<p>My cat is very grumpy</p>. And so is my dog. would look like this:
My cat is very grumpy
. And so is my dog.
There do exist many types of tags indicating different kinds of elements (about 100). Every page must be in an <html> element with two children <head> and <body>. The former contains the page title and some metadata, the latter the contents you are seeing in your browser. So-called block tags, e.g., <h1> (heading 1), <p> (paragraph), or <ol> (ordered list), structure the page. Inline tags (<b> – bold, <a> – link) format text inside block tags.
You can nest elements, e.g., if you want to make certain things bold, you can wrap text in <b>:
My cat is very grumpy
Then, the <b> element is considered the child of the <p> element.
Elements can also bear attributes:
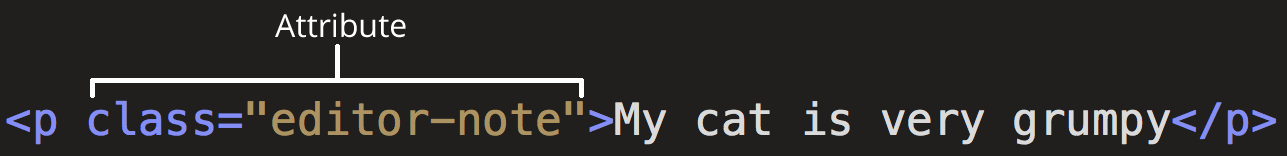
Those attributes will not appear in the actual content. Moreover, they are super-handy for us as scrapers. Here, class is the attribute name and "editor-note" the value. Another important attribute is id. Combined with CSS, they control the appearance of the element on the actual page. A class can be used by multiple HTML elements whereas an id is unique.
2.2.3.2 Extracting content in rvest
To scrape the web, the first step is to simply read in the web page. rvest then stores it in the XML format – just another format to store information. For this, we use rvest’s read_html() function.
To demonstrate the usage of CSS selectors, I create my own, basic web page using the rvest function minimal_html():
library(rvest)
library(tidyverse)
basic_html <- minimal_html('
<html>
<head>
<title>Page title</title>
</head>
<body>
<h1 id="first">A heading</h1>
<p class="paragraph">Some text & <b>some bold text.</b></p>
<a> Some more <i> italicized text which is not in a paragraph. </i> </a>
<a class="paragraph">even more text & <i>some italicized text.</i></p>
<a id="link" href="www.nyt.com"> The New York Times </a>
</body>
')
basic_html## {html_document}
## <html>
## [1] <head>\n<meta http-equiv="Content-Type" content="text/html; charset=UTF-8 ...
## [2] <body>\n <h1 id="first">A heading</h1>\n <p class="paragraph">Some ...#https://htmledit.squarefree.comCSS is the abbreviation for cascading style sheets and is used to define the visual styling of HTML documents. CSS selectors map elements in the HTML code to the relevant styles in the CSS. Hence, they define patterns that allow us to easily select certain elements on the page. CSS selectors can be used in conjunction with the rvest function html_elements() which takes as arguments the read-in page and a CSS selector. Alternatively, you can also provide an XPath which is usually a bit more complicated and will not be covered in this tutorial.
pselects all<p>elements.
basic_html %>% html_elements(css = "h1")## {xml_nodeset (1)}
## [1] <h1 id="first">A heading</h1>.titleselects all elements that are ofclass“title”
basic_html %>% html_elements(css = ".title")## {xml_nodeset (0)}There are no elements of class “title”. But some of class “paragraph”.
basic_html %>% html_elements(css = ".paragraph")## {xml_nodeset (2)}
## [1] <p class="paragraph">Some text & <b>some bold text.</b></p>
## [2] <a class="paragraph">even more text & <i>some italicized text.</i>\n ...p.paragraphanalogously takes every<p>element which is ofclass“paragraph”.
basic_html %>% html_elements(css = "p.paragraph")## {xml_nodeset (1)}
## [1] <p class="paragraph">Some text & <b>some bold text.</b></p>#linkscrapes elements that are ofid“link”
basic_html %>% html_elements(css = "#link")## {xml_nodeset (1)}
## [1] <a id="link" href="www.nyt.com"> The New York Times </a>You can also connect children with their parents by using the combinator. For instance, to extract the italicized text from “a.paragraph,” I can do “a.paragraph i”.
basic_html %>% html_elements(css = "a.paragraph i")## {xml_nodeset (1)}
## [1] <i>some italicized text.</i>You can also look at the children by using html_children():
basic_html %>% html_elements(css = "a.paragraph") %>% html_children()## {xml_nodeset (1)}
## [1] <i>some italicized text.</i>read_html("https://rvest.tidyverse.org") %>%
html_elements("#installation , p")## {xml_nodeset (8)}
## [1] <p>rvest helps you scrape (or harvest) data from web pages. It is designe ...
## [2] <p>If you’re scraping multiple pages, I highly recommend using rvest in c ...
## [3] <h2 id="installation">Installation<a class="anchor" aria-label="anchor" h ...
## [4] <p>If the page contains tabular data you can convert it directly to a dat ...
## [5] <p></p>
## [6] <p>Developed by <a href="http://hadley.nz" class="external-link">Hadley W ...
## [7] <p></p>
## [8] <p>Site built with <a href="https://pkgdown.r-lib.org/" class="external-l ...Unfortunately, web pages in the wild are usually not as easily readable as the small example one I came up with. Hence, I would recommend you to use the SelectorGadget – just drag it into your bookmarks list.
Its usage could hardly be simpler:
- Activate it – i.e., click on the bookmark.
- Click on the content you want to scrape – the things the CSS selector selects will appear green.
- Click on the green things that you don’t want – they will turn red; click on what’s not green yet but what you want – it will turn green.
- copy the CSS selector the gadget provides you with and paste it into the
html_elements()function.
2.2.3.3 Scraping HTML pages with rvest
So far, I have shown you how HTML is written and how to select elements. However, what we want to achieve is extracting the data the elements contained in a proper format and storing it in some sort of tibble. Therefore, we need functions that allow us to grab the data.
The following overview taken from the web scraping cheatsheet shows you the basic “flow” of scraping web pages plus the corresponding functions. In this tutorial, I will limit myself to rvest functions. Those are of course perfectly compatible with things, for instance, RSelenium, as long as you feed the content in XML format (i.e., by using read_html()).

In the first part, I will introduce you to scraping singular pages and extracting their contents. rvest also allows for proper sessions where you navigate on the web pages and fill out forms. This is to be introduced in the second part.
2.2.3.3.1 html_text() and html_text2()
Extracting text from HTML is easy. You use html_text() or html_text2(). The former is faster but will give you not-so-nice results. The latter will give you the text like it would be returned in a web browser.
The following example is taken from the documentation
# To understand the difference between html_text() and html_text2()
# take the following html:
html <- minimal_html(
"<p>This is a paragraph.
This is another sentence.<br>This should start on a new line"
)# html_text() returns the raw underlying text, which includes white space
# that would be ignored by a browser, and ignores the <br>
html %>% html_element("p") %>% html_text() %>% writeLines()## This is a paragraph.
## This is another sentence.This should start on a new line# html_text2() simulates what a browser would display. Non-significant
# white space is collapsed, and <br> is turned into a line break
html %>% html_element("p") %>% html_text2() %>% writeLines()## This is a paragraph. This is another sentence.
## This should start on a new lineA “real example” would then look like this:
us_senators <- read_html("https://en.wikipedia.org/wiki/List_of_current_United_States_senators")
text <- us_senators %>%
html_elements(css = "p:nth-child(6)") %>%
html_text2()2.2.3.3.2 Extracting tables
The general output format we strive for is a tibble. Oftentimes, data is already stored online in a table format, basically ready for us to analyze them. In the next example, I want to get a table from the Wikipedia page that contains the senators of different States in the United States I have used before. For this first, basic example, I do not use selectors for extracting the right table. You can use rvest::html_table(). It will give you a list containing all tables on this particular page. We can inspect it using str() which returns an overview of the list and the tibbles it contains.
tables <- us_senators %>%
html_table()
str(tables)## List of 26
## $ : tibble [4 × 3] (S3: tbl_df/tbl/data.frame)
## ..$ Affiliation: chr [1:4] "" "" "" "Total"
## ..$ Affiliation: chr [1:4] "Republican Party" "Democratic Party" "Independent" "Total"
## ..$ Members : int [1:4] 49 48 3 100
## $ : tibble [11 × 1] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:11] "This article is part of a series on the" "United States Senate" "" "History of the United States Senate" ...
## $ : tibble [2 × 5] (S3: tbl_df/tbl/data.frame)
## ..$ Office : chr [1:2] "President of the Senate[a]" "President pro tempore"
## ..$ Party : chr [1:2] "Democratic" "Democratic"
## ..$ Officer: chr [1:2] "Kamala Harris" "Patty Murray"
## ..$ State : chr [1:2] "CA[b]" "WA"
## ..$ Since : chr [1:2] "January 20, 2021" "January 3, 2023"
## $ : tibble [14 × 4] (S3: tbl_df/tbl/data.frame)
## ..$ Office : chr [1:14] "Senate Majority LeaderChair, Senate Democratic Caucus" "Senate Majority Whip" "Chair, Senate Democratic Policy and Communications Committee" "Chair, Senate Democratic Steering Committee" ...
## ..$ Officer: chr [1:14] "Chuck Schumer" "Dick Durbin" "Debbie Stabenow" "Amy Klobuchar" ...
## ..$ State : chr [1:14] "NY" "IL" "MI" "MN" ...
## ..$ Since : chr [1:14] "January 20, 2021Party leader since January 3, 2017" "January 20, 2021Party whip since January 3, 2005" "January 3, 2017" "January 3, 2017" ...
## $ : tibble [9 × 4] (S3: tbl_df/tbl/data.frame)
## ..$ Office : chr [1:9] "Senate Minority Leader" "Senate Minority Whip" "Chair, Senate Republican Conference" "Chair, Senate Republican Policy Committee" ...
## ..$ Officer: chr [1:9] "Mitch McConnell" "John Thune" "John Barrasso" "Joni Ernst" ...
## ..$ State : chr [1:9] "KY" "SD" "WY" "IA" ...
## ..$ Since : chr [1:9] "January 20, 2021Party leader since January 3, 2007" "January 20, 2021Party whip since January 3, 2019" "January 3, 2019" "January 3, 2023" ...
## $ : tibble [100 × 12] (S3: tbl_df/tbl/data.frame)
## ..$ State : chr [1:100] "Alabama" "Alabama" "Alaska" "Alaska" ...
## ..$ Portrait : logi [1:100] NA NA NA NA NA NA ...
## ..$ Senator : chr [1:100] "Tommy Tuberville" "Katie Britt" "Lisa Murkowski" "Dan Sullivan" ...
## ..$ Party : logi [1:100] NA NA NA NA NA NA ...
## ..$ Party : chr [1:100] "Republican" "Republican" "Republican" "Republican" ...
## ..$ Born : chr [1:100] "(1954-09-18) September 18, 1954 (age 68)" "(1982-02-02) February 2, 1982 (age 40)" "(1957-05-22) May 22, 1957 (age 65)" "(1964-11-13) November 13, 1964 (age 58)" ...
## ..$ Occupation(s) : chr [1:100] "College football coachInvestment management firm partner" "Senate stafferUniversity administratorLawyer Campaign managerBusiness Council of Alabama President and CEO Alab"| __truncated__ "Lawyer" "U.S. Marine Corps officerLawyerAssistant Secretary of State for Economic and Business Affairs" ...
## ..$ Previous electiveoffice(s): chr [1:100] "None" "None" "Alaska House of Representatives" "Alaska Attorney General" ...
## ..$ Education : chr [1:100] "Southern Arkansas University (BS)" "University of Alabama (BS, JD)" "Georgetown University (AB)\nWillamette University (JD)" "Harvard University (AB)\nGeorgetown University (MS, JD)" ...
## ..$ Assumed office : chr [1:100] "January 3, 2021" "January 3, 2023" "December 20, 2002[c]" "January 3, 2015" ...
## ..$ Class : chr [1:100] "2026Class 2" "2028Class 3" "2028Class 3" "2026Class 2" ...
## ..$ Residence[1] : chr [1:100] "Auburn[2]" "Montgomery" "Girdwood" "Anchorage" ...
## $ : tibble [3 × 3] (S3: tbl_df/tbl/data.frame)
## ..$ vteCurrent United States senators: chr [1:3] "President: ▌ Kamala Harris (D) ‧ President pro tempore: ▌ Patty Murray (D)" "AL: ▌ Tuberville (R)⎣ ▌ Britt (R)\nAK: ▌ Murkowski (R)⎣ ▌ Sullivan (R)\nAZ: ▌ Sinema (I)⎣ ▌ Kelly (D)\nAR: ▌ Bo"| __truncated__ "▌ Republican: 49\n▌ Democratic: 48\n▌ Independent: 3"
## ..$ vteCurrent United States senators: chr [1:3] "President: ▌ Kamala Harris (D) ‧ President pro tempore: ▌ Patty Murray (D)" "AL: ▌ Tuberville (R)⎣ ▌ Britt (R)\nAK: ▌ Murkowski (R)⎣ ▌ Sullivan (R)\nAZ: ▌ Sinema (I)⎣ ▌ Kelly (D)\nAR: ▌ Bo"| __truncated__ "▌ Republican: 49\n▌ Democratic: 48\n▌ Independent: 3"
## ..$ vteCurrent United States senators: chr [1:3] "President: ▌ Kamala Harris (D) ‧ President pro tempore: ▌ Patty Murray (D)" "" "▌ Republican: 49\n▌ Democratic: 48\n▌ Independent: 3"
## $ : tibble [4 × 6] (S3: tbl_df/tbl/data.frame)
## ..$ vteLeadership of the United States Senate: chr [1:4] "President: Kamala Harris (D)President pro tempore: Patty Murray (D)" "Majority (Democratic)Minority (Republican)\nChuck Schumer (Leader and Caucus Chair)\nDick Durbin (Whip)\nDebbie"| __truncated__ "Majority (Democratic)" "Chuck Schumer (Leader and Caucus Chair)\nDick Durbin (Whip)\nDebbie Stabenow (Policy/Communications Committee C"| __truncated__
## ..$ vteLeadership of the United States Senate: chr [1:4] "President: Kamala Harris (D)President pro tempore: Patty Murray (D)" "Majority (Democratic)Minority (Republican)\nChuck Schumer (Leader and Caucus Chair)\nDick Durbin (Whip)\nDebbie"| __truncated__ "Minority (Republican)" "Mitch McConnell (Leader)\nJohn Thune (Whip)\nJohn Barrasso (Conference Chair)\nJoni Ernst (Policy Committee Cha"| __truncated__
## ..$ : chr [1:4] NA "Majority (Democratic)" NA NA
## ..$ : chr [1:4] NA "Minority (Republican)" NA NA
## ..$ : chr [1:4] NA "Chuck Schumer (Leader and Caucus Chair)\nDick Durbin (Whip)\nDebbie Stabenow (Policy/Communications Committee C"| __truncated__ NA NA
## ..$ : chr [1:4] NA "Mitch McConnell (Leader)\nJohn Thune (Whip)\nJohn Barrasso (Conference Chair)\nJoni Ernst (Policy Committee Cha"| __truncated__ NA NA
## $ : tibble [2 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:2] "Majority (Democratic)" "Chuck Schumer (Leader and Caucus Chair)\nDick Durbin (Whip)\nDebbie Stabenow (Policy/Communications Committee C"| __truncated__
## ..$ X2: chr [1:2] "Minority (Republican)" "Mitch McConnell (Leader)\nJohn Thune (Whip)\nJohn Barrasso (Conference Chair)\nJoni Ernst (Policy Committee Cha"| __truncated__
## $ : tibble [3 × 6] (S3: tbl_df/tbl/data.frame)
## ..$ vteChairs and ranking members of United States Senate committees: chr [1:3] "Chairs (Democratic)Ranking Members (Republican)\nAging (Special): Bob Casey\nAgriculture, Nutrition and Forestr"| __truncated__ "Chairs (Democratic)" "Aging (Special): Bob Casey\nAgriculture, Nutrition and Forestry: Debbie Stabenow\nAppropriations: Patty Murray\"| __truncated__
## ..$ vteChairs and ranking members of United States Senate committees: chr [1:3] "Chairs (Democratic)Ranking Members (Republican)\nAging (Special): Bob Casey\nAgriculture, Nutrition and Forestr"| __truncated__ "Ranking Members (Republican)" "Aging (Special): Tim Scott\nAgriculture, Nutrition and Forestry: John Boozman\nAppropriations: TBD\nArmed Servi"| __truncated__
## ..$ : chr [1:3] "Chairs (Democratic)" NA NA
## ..$ : chr [1:3] "Ranking Members (Republican)" NA NA
## ..$ : chr [1:3] "Aging (Special): Bob Casey\nAgriculture, Nutrition and Forestry: Debbie Stabenow\nAppropriations: Patty Murray\"| __truncated__ NA NA
## ..$ : chr [1:3] "Aging (Special): Tim Scott\nAgriculture, Nutrition and Forestry: John Boozman\nAppropriations: TBD\nArmed Servi"| __truncated__ NA NA
## $ : tibble [2 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:2] "Chairs (Democratic)" "Aging (Special): Bob Casey\nAgriculture, Nutrition and Forestry: Debbie Stabenow\nAppropriations: Patty Murray\"| __truncated__
## ..$ X2: chr [1:2] "Ranking Members (Republican)" "Aging (Special): Tim Scott\nAgriculture, Nutrition and Forestry: John Boozman\nAppropriations: TBD\nArmed Servi"| __truncated__
## $ : tibble [49 × 36] (S3: tbl_df/tbl/data.frame)
## ..$ vteUnited States Congress: chr [1:49] "House of Representatives\nSenate\nJoint session\n(117th ← 118th → 119th)\nLists of United States Congress" "Members and leadersMembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members"| __truncated__ "Members and leaders" "MembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting member"| __truncated__ ...
## ..$ vteUnited States Congress: chr [1:49] "House of Representatives\nSenate\nJoint session\n(117th ← 118th → 119th)\nLists of United States Congress" "Members and leadersMembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members"| __truncated__ "Members and leaders" "MembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting member"| __truncated__ ...
## ..$ : chr [1:49] NA "Members and leaders" NA "Membership" ...
## ..$ : chr [1:49] NA "Members and leaders" NA "Members\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseate"| __truncated__ ...
## ..$ : chr [1:49] NA "MembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting member"| __truncated__ NA "Members" ...
## ..$ : chr [1:49] NA "MembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting member"| __truncated__ NA "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" ...
## ..$ : chr [1:49] NA "Membership" NA "Senate" ...
## ..$ : chr [1:49] NA "Members\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseate"| __truncated__ NA "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" ...
## ..$ : chr [1:49] NA "Members" NA "House" ...
## ..$ : chr [1:49] NA "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" NA "Members\nseniority\nDean\nFormer\nExpelled, censured, and reprimanded\nServed a single term\nSwitched parties\n"| __truncated__ ...
## ..$ : chr [1:49] NA "Senate" NA "Leaders" ...
## ..$ : chr [1:49] NA "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" NA "Senate\nPresident\nlist\nPresident pro tempore\nlist\nLeaders\nDemocratic Caucus\nChair\nSecretary\nPolicy Comm"| __truncated__ ...
## ..$ : chr [1:49] NA "House" NA "Senate" ...
## ..$ : chr [1:49] NA "Members\nseniority\nDean\nFormer\nExpelled, censured, and reprimanded\nServed a single term\nSwitched parties\n"| __truncated__ NA "President\nlist\nPresident pro tempore\nlist\nLeaders\nDemocratic Caucus\nChair\nSecretary\nPolicy Committee Ch"| __truncated__ ...
## ..$ : chr [1:49] NA "Leaders" NA "House" ...
## ..$ : chr [1:49] NA "Senate\nPresident\nlist\nPresident pro tempore\nlist\nLeaders\nDemocratic Caucus\nChair\nSecretary\nPolicy Comm"| __truncated__ NA "Speaker\nlist\nLeaders\nBipartisan Legal Advisory Group\nDemocratic Caucus\nRepublican Conference" ...
## ..$ : chr [1:49] NA "Senate" NA "Districts" ...
## ..$ : chr [1:49] NA "President\nlist\nPresident pro tempore\nlist\nLeaders\nDemocratic Caucus\nChair\nSecretary\nPolicy Committee Ch"| __truncated__ NA "List\nApportionment\nGerrymandering" ...
## ..$ : chr [1:49] NA "House" NA "Groups" ...
## ..$ : chr [1:49] NA "Speaker\nlist\nLeaders\nBipartisan Legal Advisory Group\nDemocratic Caucus\nRepublican Conference" NA "Congressional caucus\nCaucuses of the United States CongressEthnic and racial\nAfrican-American members\nSenate"| __truncated__ ...
## ..$ : chr [1:49] NA "Districts" NA "Congressional caucus" ...
## ..$ : chr [1:49] NA "List\nApportionment\nGerrymandering" NA "Caucuses of the United States Congress" ...
## ..$ : chr [1:49] NA "Groups" NA "Ethnic and racial" ...
## ..$ : chr [1:49] NA "Congressional caucus\nCaucuses of the United States CongressEthnic and racial\nAfrican-American members\nSenate"| __truncated__ NA "African-American members\nSenate\nHouse\nBlack Caucus\nArab and Middle Eastern members\nAsian Pacific American "| __truncated__ ...
## ..$ : chr [1:49] NA "Congressional caucus" NA "Gender and sexual identity" ...
## ..$ : chr [1:49] NA "Caucuses of the United States Congress" NA "LGBT members\nLGBT Equality Caucus\nWomen\nSenate\nHouse\nIssues Caucus\nCurrent House" ...
## ..$ : chr [1:49] NA "Ethnic and racial" NA "Occupation" ...
## ..$ : chr [1:49] NA "African-American members\nSenate\nHouse\nBlack Caucus\nArab and Middle Eastern members\nAsian Pacific American "| __truncated__ NA "Physicians" ...
## ..$ : chr [1:49] NA "Gender and sexual identity" NA "Religion" ...
## ..$ : chr [1:49] NA "LGBT members\nLGBT Equality Caucus\nWomen\nSenate\nHouse\nIssues Caucus\nCurrent House" NA "Buddhist members\nHindu members\nJewish members\nMormon (LDS) members\nMuslim members\nQuaker members" ...
## ..$ : chr [1:49] NA "Occupation" NA "Related" ...
## ..$ : chr [1:49] NA "Physicians" NA "By length of service historically\nCurrent members by wealth\nFrom multiple states\nDied in office\n1790–1899\n"| __truncated__ ...
## ..$ : chr [1:49] NA "Religion" NA NA ...
## ..$ : chr [1:49] NA "Buddhist members\nHindu members\nJewish members\nMormon (LDS) members\nMuslim members\nQuaker members" NA NA ...
## ..$ : chr [1:49] NA "Related" NA NA ...
## ..$ : chr [1:49] NA "By length of service historically\nCurrent members by wealth\nFrom multiple states\nDied in office\n1790–1899\n"| __truncated__ NA NA ...
## $ : tibble [16 × 32] (S3: tbl_df/tbl/data.frame)
## ..$ Members and leaders: chr [1:16] "MembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting member"| __truncated__ "Membership" "Members" "Senate" ...
## ..$ Members and leaders: chr [1:16] "MembershipMembers\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting member"| __truncated__ "Members\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseate"| __truncated__ "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" ...
## ..$ : chr [1:16] "Membership" "Members" NA NA ...
## ..$ : chr [1:16] "Members\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseate"| __truncated__ "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" NA NA ...
## ..$ : chr [1:16] "Members" "Senate" NA NA ...
## ..$ : chr [1:16] "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" NA NA ...
## ..$ : chr [1:16] "Senate" "House" NA NA ...
## ..$ : chr [1:16] "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" "Members\nseniority\nDean\nFormer\nExpelled, censured, and reprimanded\nServed a single term\nSwitched parties\n"| __truncated__ NA NA ...
## ..$ : chr [1:16] "House" NA NA NA ...
## ..$ : chr [1:16] "Members\nseniority\nDean\nFormer\nExpelled, censured, and reprimanded\nServed a single term\nSwitched parties\n"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Leaders" NA NA NA ...
## ..$ : chr [1:16] "Senate\nPresident\nlist\nPresident pro tempore\nlist\nLeaders\nDemocratic Caucus\nChair\nSecretary\nPolicy Comm"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Senate" NA NA NA ...
## ..$ : chr [1:16] "President\nlist\nPresident pro tempore\nlist\nLeaders\nDemocratic Caucus\nChair\nSecretary\nPolicy Committee Ch"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "House" NA NA NA ...
## ..$ : chr [1:16] "Speaker\nlist\nLeaders\nBipartisan Legal Advisory Group\nDemocratic Caucus\nRepublican Conference" NA NA NA ...
## ..$ : chr [1:16] "Districts" NA NA NA ...
## ..$ : chr [1:16] "List\nApportionment\nGerrymandering" NA NA NA ...
## ..$ : chr [1:16] "Groups" NA NA NA ...
## ..$ : chr [1:16] "Congressional caucus\nCaucuses of the United States CongressEthnic and racial\nAfrican-American members\nSenate"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Congressional caucus" NA NA NA ...
## ..$ : chr [1:16] "Caucuses of the United States Congress" NA NA NA ...
## ..$ : chr [1:16] "Ethnic and racial" NA NA NA ...
## ..$ : chr [1:16] "African-American members\nSenate\nHouse\nBlack Caucus\nArab and Middle Eastern members\nAsian Pacific American "| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Gender and sexual identity" NA NA NA ...
## ..$ : chr [1:16] "LGBT members\nLGBT Equality Caucus\nWomen\nSenate\nHouse\nIssues Caucus\nCurrent House" NA NA NA ...
## ..$ : chr [1:16] "Occupation" NA NA NA ...
## ..$ : chr [1:16] "Physicians" NA NA NA ...
## ..$ : chr [1:16] "Religion" NA NA NA ...
## ..$ : chr [1:16] "Buddhist members\nHindu members\nJewish members\nMormon (LDS) members\nMuslim members\nQuaker members" NA NA NA ...
## ..$ : chr [1:16] "Related" NA NA NA ...
## ..$ : chr [1:16] "By length of service historically\nCurrent members by wealth\nFrom multiple states\nDied in office\n1790–1899\n"| __truncated__ NA NA NA ...
## $ : tibble [15 × 12] (S3: tbl_df/tbl/data.frame)
## ..$ X1 : chr [1:15] "Membership" "Members" "Senate" "House" ...
## ..$ X2 : chr [1:15] "Members\nBy length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseate"| __truncated__ "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" "Members\nseniority\nDean\nFormer\nExpelled, censured, and reprimanded\nServed a single term\nSwitched parties\n"| __truncated__ ...
## ..$ X3 : chr [1:15] "Members" NA NA NA ...
## ..$ X4 : chr [1:15] "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" NA NA NA ...
## ..$ X5 : chr [1:15] "Senate" NA NA NA ...
## ..$ X6 : chr [1:15] "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" NA NA NA ...
## ..$ X7 : chr [1:15] "House" NA NA NA ...
## ..$ X8 : chr [1:15] "Members\nseniority\nDean\nFormer\nExpelled, censured, and reprimanded\nServed a single term\nSwitched parties\n"| __truncated__ NA NA NA ...
## ..$ X9 : chr [1:15] NA NA NA NA ...
## ..$ X10: chr [1:15] NA NA NA NA ...
## ..$ X11: chr [1:15] NA NA NA NA ...
## ..$ X12: chr [1:15] NA NA NA NA ...
## $ : tibble [3 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:3] "Members" "Senate" "House"
## ..$ X2: chr [1:3] "By length of service\nBy shortness of service\nFreshmen\nYoungest members\nNon-voting members\nUnseated members" "Members\nseniority\nDean\nFormer\nExpelled or censured\nClasses\nBorn outside the U.S.\nResigned\nAppointed\nSwitched parties" "Members\nseniority\nDean\nFormer\nExpelled, censured, and reprimanded\nServed a single term\nSwitched parties\n"| __truncated__
## $ : tibble [2 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:2] "Senate" "House"
## ..$ X2: chr [1:2] "President\nlist\nPresident pro tempore\nlist\nLeaders\nDemocratic Caucus\nChair\nSecretary\nPolicy Committee Ch"| __truncated__ "Speaker\nlist\nLeaders\nBipartisan Legal Advisory Group\nDemocratic Caucus\nRepublican Conference"
## $ : tibble [5 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:5] "Congressional caucus" "Ethnic and racial" "Gender and sexual identity" "Occupation" ...
## ..$ X2: chr [1:5] "Caucuses of the United States Congress" "African-American members\nSenate\nHouse\nBlack Caucus\nArab and Middle Eastern members\nAsian Pacific American "| __truncated__ "LGBT members\nLGBT Equality Caucus\nWomen\nSenate\nHouse\nIssues Caucus\nCurrent House" "Physicians" ...
## $ : tibble [10 × 20] (S3: tbl_df/tbl/data.frame)
## ..$ Powers, privileges, procedure, committees, history, media: chr [1:10] "Powers\nArticle I\nCopyright\nCommerce (Dormant)\nContempt of Congress\nDeclaration of war\nImpeachment\nInquir"| __truncated__ "Powers" "Privileges" "Procedure" ...
## ..$ Powers, privileges, procedure, committees, history, media: chr [1:10] "Powers\nArticle I\nCopyright\nCommerce (Dormant)\nContempt of Congress\nDeclaration of war\nImpeachment\nInquir"| __truncated__ "Article I\nCopyright\nCommerce (Dormant)\nContempt of Congress\nDeclaration of war\nImpeachment\nInquiries\nTri"| __truncated__ "Salaries\nFranking\nImmunity" "Act of Congress\nlist\nAppropriation bill\nBill\nBlue slip\nBudget process\nCensure\nClosed sessions\nHouse\nSe"| __truncated__ ...
## ..$ : chr [1:10] "Powers" NA NA NA ...
## ..$ : chr [1:10] "Article I\nCopyright\nCommerce (Dormant)\nContempt of Congress\nDeclaration of war\nImpeachment\nInquiries\nTri"| __truncated__ NA NA NA ...
## ..$ : chr [1:10] "Privileges" NA NA NA ...
## ..$ : chr [1:10] "Salaries\nFranking\nImmunity" NA NA NA ...
## ..$ : chr [1:10] "Procedure" NA NA NA ...
## ..$ : chr [1:10] "Act of Congress\nlist\nAppropriation bill\nBill\nBlue slip\nBudget process\nCensure\nClosed sessions\nHouse\nSe"| __truncated__ NA NA NA ...
## ..$ : chr [1:10] "Senate-specific" NA NA NA ...
## ..$ : chr [1:10] "Advice and consent\nClasses\nExecutive communication\nExecutive session\nFilibuster\nJefferson's Manual\nSenate"| __truncated__ NA NA NA ...
## ..$ : chr [1:10] "Committees" NA NA NA ...
## ..$ : chr [1:10] "Chairman and ranking member\nOf the Whole\nConference\nDischarge petition\nHearings\nMarkup\nOversight\nList (J"| __truncated__ NA NA NA ...
## ..$ : chr [1:10] "Items" NA NA NA ...
## ..$ : chr [1:10] "Gavels\nMace of the House\nSeal of the Senate" NA NA NA ...
## ..$ : chr [1:10] "History" NA NA NA ...
## ..$ : chr [1:10] "House history\nMemoirs\nSpeaker elections\nSenate history\nElection disputes\nMemoirs\nContinental Congress\nFe"| __truncated__ NA NA NA ...
## ..$ : chr [1:10] "House history\nMemoirs\nSpeaker elections\nSenate history\nElection disputes\nMemoirs\nContinental Congress\nFe"| __truncated__ NA NA NA ...
## ..$ : chr [1:10] "House history\nMemoirs\nSpeaker elections\nSenate history\nElection disputes\nMemoirs\nContinental Congress\nFe"| __truncated__ NA NA NA ...
## ..$ : chr [1:10] "Media" NA NA NA ...
## ..$ : chr [1:10] "C-SPAN\nCongressional Quarterly\nThe Hill\nPolitico\nRoll Call" NA NA NA ...
## $ : tibble [9 × 4] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:9] "Powers" "Privileges" "Procedure" "Senate-specific" ...
## ..$ X2: chr [1:9] "Article I\nCopyright\nCommerce (Dormant)\nContempt of Congress\nDeclaration of war\nImpeachment\nInquiries\nTri"| __truncated__ "Salaries\nFranking\nImmunity" "Act of Congress\nlist\nAppropriation bill\nBill\nBlue slip\nBudget process\nCensure\nClosed sessions\nHouse\nSe"| __truncated__ "Advice and consent\nClasses\nExecutive communication\nExecutive session\nFilibuster\nJefferson's Manual\nSenate"| __truncated__ ...
## ..$ X3: chr [1:9] NA NA NA NA ...
## ..$ X4: chr [1:9] NA NA NA NA ...
## $ : tibble [1 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr "House history\nMemoirs\nSpeaker elections\nSenate history\nElection disputes\nMemoirs\nContinental Congress\nFe"| __truncated__
## ..$ X2: chr "House history\nMemoirs\nSpeaker elections\nSenate history\nElection disputes\nMemoirs\nContinental Congress\nFe"| __truncated__
## $ : tibble [16 × 32] (S3: tbl_df/tbl/data.frame)
## ..$ Capitol Complex (Capitol Hill): chr [1:16] "Legislativeoffices\nCongressional staff\nGov. Accountability Office (GAO)\nComptroller General\nArchitect of th"| __truncated__ "Legislativeoffices" "Offices" "Senate" ...
## ..$ Capitol Complex (Capitol Hill): chr [1:16] "Legislativeoffices\nCongressional staff\nGov. Accountability Office (GAO)\nComptroller General\nArchitect of th"| __truncated__ "Congressional staff\nGov. Accountability Office (GAO)\nComptroller General\nArchitect of the Capitol\nCap. Poli"| __truncated__ "Senate\nCurator\nHistorical\nLibraryHouse\nCongr. Ethics\nEmergency Planning, Preparedness, and Operations\nInt"| __truncated__ "Curator\nHistorical\nLibrary" ...
## ..$ : chr [1:16] "Legislativeoffices" NA "Senate" NA ...
## ..$ : chr [1:16] "Congressional staff\nGov. Accountability Office (GAO)\nComptroller General\nArchitect of the Capitol\nCap. Poli"| __truncated__ NA "Curator\nHistorical\nLibrary" NA ...
## ..$ : chr [1:16] "Offices" NA "House" NA ...
## ..$ : chr [1:16] "Senate\nCurator\nHistorical\nLibraryHouse\nCongr. Ethics\nEmergency Planning, Preparedness, and Operations\nInt"| __truncated__ NA "Congr. Ethics\nEmergency Planning, Preparedness, and Operations\nInterparliamentary Affairs\nLaw Revision Couns"| __truncated__ NA ...
## ..$ : chr [1:16] "Senate" NA NA NA ...
## ..$ : chr [1:16] "Curator\nHistorical\nLibrary" NA NA NA ...
## ..$ : chr [1:16] "House" NA NA NA ...
## ..$ : chr [1:16] "Congr. Ethics\nEmergency Planning, Preparedness, and Operations\nInterparliamentary Affairs\nLaw Revision Couns"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Employees" NA NA NA ...
## ..$ : chr [1:16] "Senate\nSecretary\nChaplain\nCurator\nHistorian\nLibrarian\nPages\nParliamentarian\nSergeant at Arms and Doorke"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Senate" NA NA NA ...
## ..$ : chr [1:16] "Secretary\nChaplain\nCurator\nHistorian\nLibrarian\nPages\nParliamentarian\nSergeant at Arms and Doorkeeper" NA NA NA ...
## ..$ : chr [1:16] "House" NA NA NA ...
## ..$ : chr [1:16] "Chaplain\nChief Administrative Officer\nClerk\nDoorkeeper\nFloor Operations\nFloor Services Chief\nHistorian\nP"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Library ofCongress" NA NA NA ...
## ..$ : chr [1:16] "Congressional Research Service\nReports\nCopyright Office\nRegister of Copyrights\nLaw Library\nPoet Laureate\n"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Gov.Publishing Office" NA NA NA ...
## ..$ : chr [1:16] "Public Printer\nCongressional Pictorial Directory\nCongressional Record\nOfficial Congressional Directory\nU.S."| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Capitol Building" NA NA NA ...
## ..$ : chr [1:16] "Brumidi Corridors\nCongressional Prayer Room\nCrypt\nDome\nStatue of Freedom\nRotunda\nHall of Columns\nStatuar"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Officebuildings" NA NA NA ...
## ..$ : chr [1:16] "Senate\nDirksen\nHart\nMountains and Clouds\nRussellHouse\nBuilding Commission\nCannon\nFord\nLongworth\nO'Neill\nRayburn" NA NA NA ...
## ..$ : chr [1:16] "Senate" NA NA NA ...
## ..$ : chr [1:16] "Dirksen\nHart\nMountains and Clouds\nRussell" NA NA NA ...
## ..$ : chr [1:16] "House" NA NA NA ...
## ..$ : chr [1:16] "Building Commission\nCannon\nFord\nLongworth\nO'Neill\nRayburn" NA NA NA ...
## ..$ : chr [1:16] "Otherfacilities" NA NA NA ...
## ..$ : chr [1:16] "Botanic Garden\nHealth and Fitness Facility\nHouse Recording Studio\nSenate chamber\nOld Senate Chamber\nOld Su"| __truncated__ NA NA NA ...
## ..$ : chr [1:16] "Related" NA NA NA ...
## ..$ : chr [1:16] "Capitol Hill\nUnited States Capitol cornerstone laying" NA NA NA ...
## $ : tibble [15 × 6] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:15] "Legislativeoffices" "Offices" "Senate" "House" ...
## ..$ X2: chr [1:15] "Congressional staff\nGov. Accountability Office (GAO)\nComptroller General\nArchitect of the Capitol\nCap. Poli"| __truncated__ "Senate\nCurator\nHistorical\nLibraryHouse\nCongr. Ethics\nEmergency Planning, Preparedness, and Operations\nInt"| __truncated__ "Curator\nHistorical\nLibrary" "Congr. Ethics\nEmergency Planning, Preparedness, and Operations\nInterparliamentary Affairs\nLaw Revision Couns"| __truncated__ ...
## ..$ X3: chr [1:15] NA "Senate" NA NA ...
## ..$ X4: chr [1:15] NA "Curator\nHistorical\nLibrary" NA NA ...
## ..$ X5: chr [1:15] NA "House" NA NA ...
## ..$ X6: chr [1:15] NA "Congr. Ethics\nEmergency Planning, Preparedness, and Operations\nInterparliamentary Affairs\nLaw Revision Couns"| __truncated__ NA NA ...
## $ : tibble [2 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:2] "Senate" "House"
## ..$ X2: chr [1:2] "Curator\nHistorical\nLibrary" "Congr. Ethics\nEmergency Planning, Preparedness, and Operations\nInterparliamentary Affairs\nLaw Revision Couns"| __truncated__
## $ : tibble [2 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:2] "Senate" "House"
## ..$ X2: chr [1:2] "Secretary\nChaplain\nCurator\nHistorian\nLibrarian\nPages\nParliamentarian\nSergeant at Arms and Doorkeeper" "Chaplain\nChief Administrative Officer\nClerk\nDoorkeeper\nFloor Operations\nFloor Services Chief\nHistorian\nP"| __truncated__
## $ : tibble [2 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ X1: chr [1:2] "Senate" "House"
## ..$ X2: chr [1:2] "Dirksen\nHart\nMountains and Clouds\nRussell" "Building Commission\nCannon\nFord\nLongworth\nO'Neill\nRayburn"
## $ : tibble [2 × 2] (S3: tbl_df/tbl/data.frame)
## ..$ vteOrder of precedence in the United States*: chr [1:2] "PresidentVice President\nGovernor (of the state in which the event is held)\nSpeaker of the House\nChief Justic"| __truncated__ "*not including acting officeholders, visiting dignitaries, auxiliary executive and military personnel and most diplomats"
## ..$ vteOrder of precedence in the United States*: chr [1:2] "PresidentVice President\nGovernor (of the state in which the event is held)\nSpeaker of the House\nChief Justic"| __truncated__ "*not including acting officeholders, visiting dignitaries, auxiliary executive and military personnel and most diplomats"Here, the table I want is the sixth one. We can grab it by either using double square brackets – [[6]] – or purrr’s pluck(6).
library(janitor)
senators <- tables %>%
pluck(6)
glimpse(senators)## Rows: 100
## Columns: 12
## $ State <chr> "Alabama", "Alabama", "Alaska", "Alaska",…
## $ Portrait <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ Senator <chr> "Tommy Tuberville", "Katie Britt", "Lisa …
## $ Party <lgl> NA, NA, NA, NA, NA, NA, NA, NA, NA, NA, N…
## $ Party <chr> "Republican", "Republican", "Republican",…
## $ Born <chr> "(1954-09-18) September 18, 1954 (age 68)…
## $ `Occupation(s)` <chr> "College football coachInvestment managem…
## $ `Previous electiveoffice(s)` <chr> "None", "None", "Alaska House of Represen…
## $ Education <chr> "Southern Arkansas University (BS)", "Uni…
## $ `Assumed office` <chr> "January 3, 2021", "January 3, 2023", "De…
## $ Class <chr> "2026Class 2", "2028Class 3", "2028Class …
## $ `Residence[1]` <chr> "Auburn[2]", "Montgomery", "Girdwood", "A…## alternative approach using css
senators <- us_senators %>%
html_elements(css = "#senators") %>%
html_table() %>%
pluck(1) %>%
clean_names()You can see that the tibble contains “dirty” names and that the party column appears twice – which will make it impossible to work with the tibble later on. Hence, I use clean_names() from the janitor package to fix that.
2.2.3.3.3 Extracting attributes
You can also extract attributes such as links using html_attrs(). An example would be to extract the headlines and their corresponding links from r-bloggers.com.
rbloggers <- read_html("https://www.r-bloggers.com")A quick check with the SelectorGadget told me that the element I am looking for is of class “.loop-title” and the child of it is “a”, standing for normal text. With html_attrs() I can extract the attributes. This gives me a list of named vectors containing the name of the attribute and the value:
r_blogger_postings <- rbloggers %>% html_elements(css = ".loop-title a")
r_blogger_postings %>% html_attr("href")## [1] "https://www.r-bloggers.com/2023/01/python-for-r-users-workshop/"
## [2] "https://www.r-bloggers.com/2023/01/how-to-become-a-data-scientist-in-2023/"
## [3] "https://www.r-bloggers.com/2023/01/the-falling-of-ark-innovation-etf-forecasting-with-boosted-arima-regression-model/"
## [4] "https://www.r-bloggers.com/2023/01/curl-5-0-0-massive-concurrent-downloads-and-http-2/"
## [5] "https://www.r-bloggers.com/2023/01/end-to-end-testing-with-shinytest2/"
## [6] "https://www.r-bloggers.com/2023/01/designing-accessible-research-with-r-shiny-ui-part-2/"
## [7] "https://www.r-bloggers.com/2023/01/check-whether-any-values-of-a-logical-vector-are-true/"
## [8] "https://www.r-bloggers.com/2023/01/mran-is-getting-shutdown-what-else-is-there-for-reproducibility-with-r-or-why-reproducibility-is-on-a-continuum/"
## [9] "https://www.r-bloggers.com/2023/01/expanding-our-community-through-multilingual-publishing/"
## [10] "https://www.r-bloggers.com/2023/01/git-and-github-in-r-for-the-casual-user/"
## [11] "https://www.r-bloggers.com/2023/01/satrdays-london-2023/"
## [12] "https://www.r-bloggers.com/2023/01/how-a-user-2014-experience-led-to-the-development-of-a-1700-member-r-community-in-budapest/"
## [13] "https://www.r-bloggers.com/2023/01/top-data-science-skills-step-by-step-guide/"
## [14] "https://www.r-bloggers.com/2023/01/progressr-0-13-0-cli-progressr-%e2%99%a5/"
## [15] "https://www.r-bloggers.com/2023/01/mran-time-machine-will-be-retired-on-july-1/"
## [16] "https://www.r-bloggers.com/2023/01/imputation-in-r-top-3-ways-for-imputing-missing-data/"
## [17] "https://www.r-bloggers.com/2023/01/rtutor-public-infrastructure-spending-and-voting-behaviour/"
## [18] "https://www.r-bloggers.com/2023/01/fast-track-your-next-move-with-r/"
## [19] "https://www.r-bloggers.com/2023/01/dplyr-style-without-dplyr/"
## [20] "https://www.r-bloggers.com/2023/01/escaping-the-malthusian-trap/"Links are stored as attribute “href” – hyperlink reference. html_attr() allows me to extract the attribute’s value. Hence, building a tibble with the article’s title and its corresponding hyperlink is straight-forward now:
tibble(
title = r_blogger_postings %>% html_text2(),
link = r_blogger_postings %>% html_attr(name = "href")
)## # A tibble: 20 × 2
## title link
## <chr> <chr>
## 1 Python for R users workshop http…
## 2 How to Become a Data Scientist in 2023 http…
## 3 The Falling of ARK Innovation ETF: Forecasting with Boosted ARIMA Regr… http…
## 4 curl 5.0.0: massive concurrent downloads and HTTP/2 http…
## 5 End-to-end testing with shinytest2 http…
## 6 Designing Accessible Research with R/Shiny UI – Part 2 http…
## 7 Check whether any values of a logical vector are TRUE http…
## 8 MRAN is getting shutdown – what else is there for reproducibility with… http…
## 9 Expanding our Community through Multilingual Publishing http…
## 10 git and GitHub in R for the casual user http…
## 11 SatRdays London 2023 http…
## 12 How a UseR! 2014 Experience Led to the Development of a 1,700-member R… http…
## 13 Top Data Science Skills- step by step guide http…
## 14 progressr 0.13.0: cli + progressr = ♥ http…
## 15 MRAN Time Machine will be retired on July 1 http…
## 16 Imputation in R: Top 3 Ways for Imputing Missing Data http…
## 17 RTutor: Public Infrastructure Spending and Voting Behaviour http…
## 18 Fast-Track Your Next Move with R http…
## 19 Dplyr-style without dplyr http…
## 20 Escaping the Malthusian Trap http…Another approach for this would be using the polite package and its function html_attrs_dfr() which binds together all the different attributes column-wise and the different elements row-wise.
library(polite)
rbloggers %>%
html_elements(css = ".loop-title a") %>%
html_attrs_dfr() %>%
select(title = 3,
link = 1) %>%
glimpse()## Rows: 20
## Columns: 2
## $ title <chr> "Python for R users workshop", "How to Become a Data Scientist i…
## $ link <chr> "https://www.r-bloggers.com/2023/01/python-for-r-users-workshop/…2.2.3.4 Automating scraping
Well, grabbing singular points of data from websites is nice. However, if you want to do things such as collecting large amounts of data or multiple pages, you will not be able to do this without some automation.
An example here would again be the R-bloggers page. It provides you with plenty of R-related content. If you were now eager to scrape all the articles, you would first need to acquire all the different links leading to the blog postings. Hence, you would need to navigate through the site’s pages first to acquire the links.
In general, there are two ways to go about this. The first is to manually create a list of URLs the scraper will visit and take the content you need, therefore not needing to identify where it needs to go next. The other one would be automatically acquiring its next destination from the page (i.e., identifying the “go on” button). Both strategies can also be nicely combined with some sort of session().
2.2.3.4.1 Looping over pages
For the first approach, we need to check the URLs first. How do they change as we navigate through the pages?
url_1 <- "https://www.r-bloggers.com/page/2/"
url_2 <- "https://www.r-bloggers.com/page/3/"
initial_dist <- adist(url_1, url_2, counts = TRUE) %>%
attr("trafos") %>%
diag() %>%
str_locate_all("[^M]")
str_sub(url_1, start = initial_dist[[1]][1]-5, end = initial_dist[[1]][1]+5) # makes sense for longer urls## [1] "page/2/"str_sub(url_2, start = initial_dist[[1]][1]-5, end = initial_dist[[1]][1]+5)## [1] "page/3/"There is some sort of underlying pattern and we can harness that. url_1 refers to the second page, url_2 to the third. Hence, if we just combine the basic URL and, say, the numbers from 1 to 10, we could then visit all the pages (exercise 3a) and extract the content we want.
urls <- str_c("https://www.r-bloggers.com/page/", 1:10, "/") # this is the stringr equivalent of paste()
urls## [1] "https://www.r-bloggers.com/page/1/" "https://www.r-bloggers.com/page/2/"
## [3] "https://www.r-bloggers.com/page/3/" "https://www.r-bloggers.com/page/4/"
## [5] "https://www.r-bloggers.com/page/5/" "https://www.r-bloggers.com/page/6/"
## [7] "https://www.r-bloggers.com/page/7/" "https://www.r-bloggers.com/page/8/"
## [9] "https://www.r-bloggers.com/page/9/" "https://www.r-bloggers.com/page/10/"You can run this in a for-loop. For the loop to run efficiently, space for every object should be pre-allocated (i.e., you create a list beforehand, and its length can be determined by an educated guess).
result_list <- vector(mode = "list", length = length(urls)) # pre-allocate space!!!
for (i in seq_along(urls)){
result_list[[i]] <- read_html(urls[[i]])
}2.2.4 Conclusion
To sum it up: when you have a good research idea that relies on Digital Trace Data that you need to collect, ask yourself the following questions:
- Is there an R package for the web service?
- If 1. == FALSE: Is there an API where I can get the data (if TRUE, use it)
- If 1. == FALSE & 2. == FALSE: Is screen scraping an option and any structure in the data that you can harness?
If you have to rely on screen scraping, also ask yourself the question how you can minimize the number of requests you make to the server. Going back and forth on web pages or navigating through them might not be the best option since it requires multiple requests. The most efficient way is usually to try to get a list of URLs of some sort which you can then just loop over.
2.3 Further links
- The
stringrcheatsheet. - A YouTube video on regexes by Johns Hopkins professor Roger Peng.
- And a chapter by Roger Peng.
- A website for practicing regexes.
- You can also consult the
introversepackage if you need help with the packages covered here –introverse::show_topics("stringr")will give you an overview of thestringrpackage’s functions, andget_help("name of function")will help you with the respective function. - More on HTML
- More on CSS selectors
- The
rvestvignette - A “laymen’s guide” on web scraping (blog post)
2.4 Exercises
2.4.1 Regexes
1.Write a regex for Swedish mobile numbers. Test it with str_detect("+46 71-738 25 33", "[insert your regex here]").
Solution. Click to expand!
library(tidyverse)
#1
str_detect("+46 71-738 25 33", "\\+46 [0-9]{2}\\-[0-9]{3} [0-9]{2} [0-9]{2}")- Remember the vector of heights?
- How can you extract the meters using the negative look behind?
- Bring it into numeric format (i.e., so that
your_solution == c(1.3, 2.01, 3.1)yieldsTRUE) using regexes andstringrcommands.
Solution. Click to expand!
heights <- c("1m30cm", "2m01cm", "3m10cm")
#a
meters <- str_extract(heights, "(?<!m)[0-9]")
#b
for_test <- str_replace(heights, "(?<=[0-9])m", "\\.") %>%
str_replace("cm", "") %>%
as.numeric()
for_test == c(1.3, 2.01, 3.1)- Find all Mercedes in the
mtcarsdata set.
Solution. Click to expand!
mtcars %>%
rownames_to_column("model") %>%
filter(str_detect(model, "Merc"))- Take the IMDb file(
read_csv("https://www.dropbox.com/s/81o3zzdkw737vt0/imdb2006-2016.csv?dl=1")) and split theGenrecolumn into different columns (hint: look at thetidyr::separate()function). How would you do it ifGenrewere a vector usingstr_split_fixed()?
Solution. Click to expand!
imdb <- read_csv("https://www.dropbox.com/s/81o3zzdkw737vt0/imdb2006-2016.csv?dl=1")
imdb %>%
separate(Genre, sep = ",", into = c("genre_1", "genre_2", "genre_3"))
imdb$Genre %>%
str_split_fixed(pattern = ",", 3)2.4.2 Structured Scraping
- Start a session with the tidyverse Wikipedia page. Adapt your user agent to some sort of different value. Proceed to Hadley Wickham’s page. Go back. Go forth. Check the
session_history()to see if it has worked.
Solution. Click to expand!
tidyverse_wiki <- "https://en.wikipedia.org/wiki/Tidyverse"
hadley_wiki <- "https://en.wikipedia.org/wiki/Hadley_Wickham"
user_agent <- httr::user_agent("Hi, I'm Felix and I try to steal your data.")wiki_session <- session(tidyverse_wiki, user_agent)
wiki_session %<>% session_jump_to(hadley_wiki) %>%
session_back() %>%
session_forward()
wiki_session %>% session_history()- Start a session on “https://www.scrapethissite.com/pages/advanced/?gotcha=login”, fill out, and submit the form. Any value for login and password will do. (Disclaimer: you have to add the URL as an “action” attribute after creating the form, see this tutorial. –
login_form$action <- url)
url <- "https://www.scrapethissite.com/pages/advanced/?gotcha=login"
#extract and set login form here
login_form$action <- url # add url as action attribute
# submit form
base_session <- session("https://www.scrapethissite.com/pages/advanced/?gotcha=login") %>%
session_submit(login_form) Solution. Click to expand!
url <- "https://www.scrapethissite.com/pages/advanced/?gotcha=login"
#extract and set login form here
login_form <- read_html(url) %>%
html_form() %>%
pluck(1) %>%
html_form_set(user = "123",
pass = "456")
login_form$action <- url # add url as action attribute
# submit form
base_session <- session("https://www.scrapethissite.com/pages/advanced/?gotcha=login") %>%
session_submit(login_form)
base_session %>% read_html() %>% html_text()base_session %>% read_html() %>% html_elements(".col-md-offset-4") %>% html_text2()- Scrape 10 profile timelines from the following list. Check the documentation for instructions.
Solution. Click to expand!
library(rtweet)
uk_mps <- lists_members(
list_id = "217199644"
)
# get 10 profiles
sample_uk_mps <- uk_mps %>%
slice_sample(n = 10) %>%
pull(screen_name)
# get timelines
## using purrr::map
timelines <- sample_uk_mps %>%
map(get_timeline)
## using for-loop
timelines <- vector(mode = "list", length = length(sample_uk_mps))
for (i in seq_along(sample_uk_mps)){
timelines[[i]] <- get_timeline(user = sample_uk_mps[[i]])
}2.4.3 Scraping unstructured data
- Download all movies from the (IMDb Top250 best movies of all time list)[https://www.imdb.com/chart/top/?ref_=nv_mv_250]. Put them in a tibble with the columns
rank– in numeric format,title,urlto IMDb entry,rating– in numeric format.
Solution. Click to expand!
imdb_top250 <- read_html("https://www.imdb.com/chart/top/?ref_=nv_mv_250")
tibble(
rank = imdb_top250 %>%
html_elements(".titleColumn") %>%
html_text2() %>%
str_extract("^[0-9]+(?=\\.)") %>%
parse_integer(),
title = imdb_top250 %>%
html_elements(".titleColumn a") %>%
html_text2(),
url = imdb_top250 %>%
html_elements(".titleColumn a") %>%
html_attr("href") %>%
str_c("https://www.imdb.com", .),
rating = imdb_top250 %>%
html_elements("strong") %>%
html_text() %>%
parse_double()
)
imdb_top250 %>%
html_elements("a") %>%
html_text2()- Scrape the (British MPs Wikipedia page)[https://en.wikipedia.org/wiki/List_of_MPs_elected_in_the_2019_United_Kingdom_general_election].
- Scrape the table containing data on all individual MPs. (Hint: you can select and remove single lines using
slice(), e.g.,tbl %>% slice(1)retains only the first line,tbl %>% slice(-1)removes only the first line.) - Extract the links linking to individual pages. Make them readable using
url_absolute()–url_absolute("/kdfnndfkok", base = "https://wikipedia.com")–>"https://wikipedia.com/kdfnndfkok". Add the links to the table from 1a. - Scrape the textual content of the first MP’s Wikipedia page.
- (ADVANCED!) Wrap 1c. in a function called
scrape_mp_wiki().
Solution. Click to expand!
# scrape uk mps
library(tidyverse)
library(rvest)
library(janitor)
#a
mp_table <- read_html("https://en.wikipedia.org/wiki/List_of_MPs_elected_in_the_2019_United_Kingdom_general_election") %>%
html_element("#elected-mps") %>%
html_table() %>%
select(-2, -6) %>%
slice(-651) %>%
clean_names()
#b
mp_links <- read_html("https://en.wikipedia.org/wiki/List_of_MPs_elected_in_the_2019_United_Kingdom_general_election") %>%
html_elements("#elected-mps b a") %>%
html_attr("href")
table_w_link <- mp_table %>%
mutate(link = mp_links %>%
paste0("https://en.wikipedia.org", .))
#c
link <- table_w_link %>%
slice(1) %>%
pull(link)
link %>%
read_html() %>%
html_elements("p") %>%
html_text2() %>%
str_c(collapse = " ") %>%
enframe(name = NULL, value = "wiki_article") %>%
mutate(link = link)
#d
scrape_page <- function(link){
Sys.sleep(0.5)
read_html(link) %>%
html_elements("p") %>%
html_text2() %>%
str_c(collapse = " ") %>%
enframe(name = NULL, value = "wiki_article") %>%
mutate(link = link)
}
mp_wiki_entries <- table_w_link$link[[1]] %>%
map(~{scrape_page(.x)})
mp_wiki_entries_tbl <- table_w_link %>%
left_join(mp_wiki_entries %>% bind_rows())- Scrape the 10 first pages of R-bloggers in an automated fashion. Make sure to take breaks between requests by including
Sys.sleep(2).
- Do so using the url vector we created above.
- Do so by getting the link for the next page and a loop.
- Do so by using
session()in a loop.
Solution. Click to expand!
# a: check running number in URL, create new URLs, map()
urls <- str_c("https://www.r-bloggers.com/page/", 1:10, "/")
scrape_r_bloggers <- function(url){
Sys.sleep(2)
read_html(url) %>%
html_elements(css = ".loop-title a") %>%
html_attrs_dfr()
}
map(urls, scrape_r_bloggers)
# b: extract the next page and move forward in the loop
rbloggers %>% html_elements(css = ".next") %>% html_attr("href")
url <- "https://www.r-bloggers.com/"
content <- vector(mode = "list", length = 10L)
for (i in 1:10){
page <- read_html(url)
content[[i]] <- page %>%
html_elements(css = ".loop-title a") %>%
html_attrs_dfr()
url <- page %>% html_elements(css = ".next") %>% html_attr("href")
Sys.sleep(2)
}
# c: session()
session(url) %>% session_follow_link(css = ".next")
content <- vector(mode = "list", length = 10L)
session_page <- session(url)
session_page$response$status_code
for (i in 1:10){
content[[i]] <- session_page %>%
html_elements(css = ".loop-title a") %>%
html_attrs_dfr()
session_page <- session_page %>% session_follow_link(css = ".next")
}