Chapter 1 DNA Methylation
A compilation of resources on understanding why the addition of that little methyl group matters.
What is DNA methylation?
From compgenomr:
10.1 What is DNA methylation?
Cytosine methylation (5-methylcytosine, 5mC) is one of the main covalent base modifications in eukaryotic genomes, generally observed on CpG dinucleotides. Methylation can also rarely occur in a non-CpG context, but this was mainly observed in human embryonic stem and neuronal cells (Lister, Pelizzola, Dowen, et al. 2009; Lister, Mukamel, Nery, et al. 2013). DNA methylation is a part of the epigenetic regulation mechanism of gene expression. It is cell-type-specific DNA modification. It is reversible but mostly remains stable through cell division. There are roughly 28 million CpGs in the human genome, 60–80% are generally methylated. Less than 10% of CpGs occur in CG-dense regions that are termed CpG islands in the human genome (Smith and Meissner 2013). It has been demonstrated that DNA methylation is also not uniformly distributed over the genome, but rather is associated with CpG density. In vertebrate genomes, cytosine bases are usually unmethylated in CpG-rich regions such as CpG islands and tend to be methylated in CpG-deficient regions. Vertebrate genomes are largely CpG deficient except at CpG islands. Conversely, invertebrates such as Drosophila melanogaster and Caenorhabditis elegans do not exhibit cytosine methylation and consequently do not have CpG rich and poor regions but rather a steady CpG frequency over their genomes (Deaton and Bird 2011).
How does DNA methylation work?
Diagram I made of methylation and de-methylation: found here.
From compgenomr:
10.1.1 How DNA methylation is set?
DNA methylation is established by DNA methyltransferases DNMT3A and DNMT3B in combination with DNMT3L and maintained through cell division by the methyltransferase DNMT1 and associated proteins. DNMT3a and DNMT3b are in charge of the de novo methylation during early development. Loss of 5mC can be achieved passively by dilution during replication or exclusion of DNMT1 from the nucleus. Recent discoveries of the ten-eleven translocation (TET) family of proteins and their ability to convert 5-methylcytosine (5mC) into 5-hydroxymethylcytosine (5hmC) in vertebrates provide a path for catalyzed active DNA demethylation (Tahiliani, Koh, Shen, et al. 2009). Iterative oxidations of 5hmC catalyzed by TET result in 5-formylcytosine (5fC) and 5-carboxylcytosine (5caC). 5caC mark is excised from DNA by G/T mismatch-specific thymine-DNA glycosylase (TDG), which as a result reverts cytosine residue to its unmodified state (He, Li, Li, et al. 2011). Apart from these, mainly bacteria, but possibly higher eukaryotes, contain base modifications on bases other than cytosine, such as methylated adenine or guanine (Clark, Spittle, Turner, et al. 2011).

Location of methylation
Invertebrates: location is non-random and mainly located in gene bodies (Li et al 2018). From Eirin-Lopez and Putnam 2019: Additionally, DNA methylation can commonly be found in gene bodies, where it hypothetically contributes to the reduction of transcriptional variation, reduction of spurious transcription, and facilitation of alternative splicing (reviewed in Roberts & Gavery 2012). Methylation is spread evenly across a gene.
Vertebrates: located in promoters regions and usually associated with turning “on and off” genes or gene silencing. From Eirin-Lopez and Putnam 2019: Repetitive CpG sequences (CpG islands) can be found located upstream in promoter regions, and here the presence of methylation can silence transcription by blocking the binding of transcription factors (but see Ford et al. 2017).
From Eirin-Lopez and Putnam 2019: Beyond the coding portions of the genome, DNA methylation can also be associated with intergenic regions and transposable elements, contributing to the silencing of transposable elements and viral elements that would otherwise result in genome disruption by sequence and DNA mutation (Rey et al. 2016).
Differentially expressed methylated regions are not necessarily the same as differentially expressed genes (Li et al 2018). DNA methylation changes don’t necessarily correlated with gene expression changes.
DNA Methylation location in invertebrates (Li et al 2018)

Consequences of methylation
Gene body methylation is established and maintained as a result of active transcription
Li et al 2018: as a result of active transcription by RNA polymerase II and recruitment of histone-modifying protein SetD2 that trimethylates histone H3 at lysine 36 (H3K36me3). This histone mark is bound via PWWP (Pro-Trp-Trp-Pro) domain present in DNA methyltransferase DNMT3b (de novo) which then methylates surrounding DNA.
Sarda et al 2012
DNA Methylation regulates transcriptional noise
DNA methylation regulates transcriptional homeostasis of algal symbiosis in the coral model Aiptasia (Li et al 2018):
- Methylated genes show significant reduction of spurius transcription and transcriptional noise.
- Spurious transcription can produce partial transcripts that when translated results in truncated proteins that would interfere with native function.
- Prefers CpG-poor regions
- Gene body methylation is (+) correlated with expression: DNA methylation either: 1.) increases expression or 2.) is increased as a consequence of transcription.
Higher DNA methylation equals lower transcriptional variability.
Methylation’s role in differing life stages
Larvae and adult life stages can differ in methylation patterning:
- Sea lamprey (Covelo-Soto et al 2015)
Sex determination linked to differential methylation patterns: Half-smooth tongue sole (Shao et al 2014), temperature-mediated sex determination in fish (Ellison et al 2015, Navarro-Martin et al 2011) and sea turtles (Venegas et al 2016).
Methylation occurs in a bimodal distribution
Over time, methylation causes hypermutability of cytosines to thymines. Which results in a reduction of CpG dinucleotides; therefore, if an organism has heavy historical methylation on a gene set then the abundance of CpG dinucleotides observed (O) would lower than expected (E). A higher CpG O/E value means there is low historical methylation (lower observed # creates a higher CpG O/E value). A lower CpG O/E value means there is high historical methylation (higher observed # creates a lower CpG O/E value).
CpG O/E calculation from Dimond and Roberts 2016:

CpG O/E calculation from Dixon et al 2014:
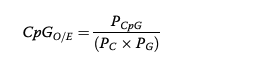
CpG O/E distribution from an eastern oyster in Gavery and Roberts 2010:
wk-hv
CpG O/E distributions in corals from Dimond and Roberts 2016:

cpgcorals
Function annotation of gene sets in weakly and heavily methylated categories from Dimond and Roberts 2016:

cpg corals
Functional annotation of CpG O/E genes from Dixon et al 2014. Genes with high CpG O/E values are more likely to be environmentally inducible.

fn
Relationship between measured methylation and CpG O/E values from Gavery and Roberts 2012:
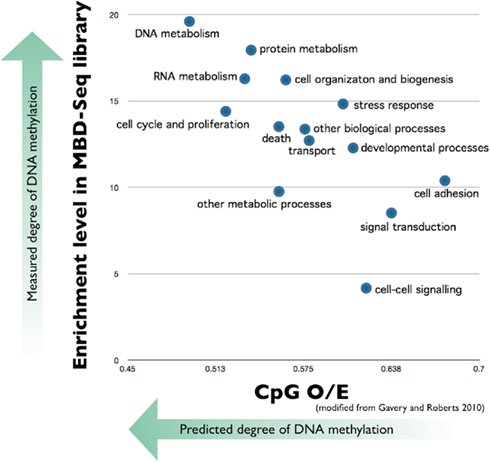
cpgoe
Methylation is modulator of alternative splicing
Come back to Lev Maor et al 2015
Coral methylation patterns are dynamic
Methylation profiles (RADSeq techniques) of Porites asteroides converged in a common garden setting (decreased in % methylated CpGs). Corals from several reefs in Belize were transplanted to a common garden for 1 year. Positive & significant relationship b/w genetic and epigenetic variation = evidence for methylation heritability. Differentially methylated loci = intracellular signaling, apoptosis, gene regulation, epigenetic crosstalk (Dimond and Roberts 2019).
- calcium-independent protein kinase C-like: involved in intracellular signaling (typically associated with hypomethylation)
- tax1-binding protein homolog: negative regulation of apoptotic processes via negative regulation of NF=kB transcription factor activity. Loss of symbionts is associated with elevated levels of NF-kB.
- U5 small nuclear ribonucleoprotein 200 kDa helicase: mRNA splicing via role in the spliceosome.
- putative FAM98A protein: positive regulation of cell proliferation, gene expression, and protein methylation.
The above functions seem to be housekeeping functions that were differentially methylated? This involved in heritability? Long term change in housekeeping methylation levels. Not short-term env. inducible?
In reduced pH conditions, Montipora capitata global methylation levels did not change, but Pocillopora acuta’s increased after 6 week exposure. Caveat: Mcap could have undergone increases and decreases that leveled out to an appearance of no response(Putnam et al 2016).
3 month reciprocal transplant of Acropora millepora from GBR; reefs were different in mean temperature and other abiotic factors. MBD-seq methods. Fitness higher at colder reef (possibly b/c higher concentration of nutrients). Methylation shift in genic regions. Transplant from hot to cold resulted in highly methylated genes becoming less methylated and less methylated genes becoming more methylated. This was mirrored but less pronounced from cold to hot reef. Transplated corals GBM shifted to reflect the location transplanted to: aka dynamic not fixed patterns. Dixon et al 2018; Dixon et al 2017 > cold reef was also higher in nutrients – more nutrients allowing for more methylation activity via more metabolites and energy?
Increase in genome-wide methylation in Stylophora pistillata when exposed to pH conditions for 2 years. Changes modified gene expression and altered pathways in cell cycle regulation. Liew et al 2018
Comparison of genetic and epigenetic variation in Porites spp. of different morphotypes and variety of habitats in Belize. Stronger evidence for genetic structuring rather than epigenetic. Weak positive correlation b/w pairwise genetic and epigenetic distance. 2 pairs of clone showed substantial epigenetic difference = strong env. effect. Methylation is likely strong mix of Environment x Genotype. RRBS methods used. Dimond et al 2017
Seasonal changes in methylation
Acropora cervicornis, over a 17-month period, displayed methylation changes that correlated to sea surface temperature change and this is consist across genets, source sites, and site-specific conditions. Methylation-Sensitive Amplified Polymorphism (MSAP) methods. (Rodriquez-Casariego et al 2020).
Ocean acidification induced methylation changes
(Venkataraman et al 2020) description below.
7 day exposure to extremely elevated pCO2 conditions in pteropod resulted in reduced global methylation after 1 day exposure and leveled off to control after 6 days. MethylFlash Quantification Kit methods (Bogan et al 2020).
Transgenerational methylation
Sea urchins were acclimated during gametogenesis in up-welling and non-upwelling conditions and progeny from these populations were raised in high and low pCO2 treatments. Sampled across 3 developmental stages. Differential progeny methylation correlated to differential paternal condition and progeny condition correlated very little to progeny methylation. Methods: Pico methyl-seq library prep kit, Illumina HiSeq4000 lanes (Strader et al 2019).
Transmission of CpG methylation from adults to sperm and larvae demonstrates genome-wide inheritance. (Liew et al 2020)
Rondon et al 2017
Ocean acidification conditions for 28 days resulted in 598 differentially methylated loci (DML), mostly in exons. Changes seen in reproductive tissue in eastern oyster means that these changes could be inherited. Measured by MBD-BS methods. DML involved in protein ubiquitination (Venkataraman et al 2020).