Chapter 8 Biochemical Adaptation: Somero et al
Chapter 1: Conceptual Foundation for Understanding Biochemical Adaptation
Stressor: env. factor that perturbs biochem structures/fxn
Stress: change induced in the system by stressor
biochemical systems: efficient: enzymatic enhancement of chemical reaction rates; accurate: copying, transcribing, translating genetically coded information in the cell; responsive: rapid and sophisticated signaling and subsequent effector responses that make changes internally or externally.
- This all depends on reversible changes in molecular shape during function
Marginal stability and fluidity:
- a middle ground between stability to ensure existence but not so rigid that a molecular cannot undergo changes when needed. a balancing act and spectrum of rigid conformation to marginally stable conformation to too flexible a conformation
- fluidity reflects a capacity for altering shape
ligand: substrate of enzymatic rxn
protein “softness”: ease at which a protein’s surface can alter geometry
Ultimate vs proximate causes:
- Ultimate: selective advantages that led to the origin and retention of a trait; “why?”
- Proximate: refer to the “mechanism”/the means by which the beneficial trait is achieved.
Intermediate level of protein structural stability (not too rigid or too flexible) is important to ensure optimal protein function. the proximate cause would be amino acid substitutions that modify the inherent structural stability of the protein OR modulating the intracellular fluid that bathes the protein. Either way the same result could occur of intermediate level of protein stability
Exaptations: subsequent exploitations of a trait (that was adapted and had beneifical function) for other functions
- BAT from previous mechanisms that reduced production of ROS
Sources of genetic and phenotypic variation that provide the “raw material” for adaptive change
Horizontal gene transfer (HGT):
Genes are exchanged either between different evolutionary lineages or between conspecifics
- organisms can acquire types of genes needed for local environments
- can favor rapid and extensive rebuilding of genetic “toolkit” (in microbiome)
Organisms can shuffle genes?? how does that even work?? come back to this..
Gene duplication
Duplication of genes followed by sequence changes that lead to adaptive variations on different protein themes – major mechanism for facilitating exaptation at the protein level.
ploidy: the entire genome is duplicated
tandem gene duplication: individual gene is copied but this can also include blocks of genes that support specific biochemical functions
Interaction with methylation? If a gene is methylated/modified epigenetically, is it physically possible to copy that? Does the methyl mark continue or does it have to expend the energy to get rid of it?
Increases in ploidy or tandem gene duplication creates paralogous genes that encode protein paralogs.
Allelic polymorphism
allozyme: when non-synonymous nucleotide changes in one allele for a protein-coding gene lead to alterations in amino acid sequence.
In a changing environment, it would be advantageous to keep allozyme variants that have different environmental optima
Lyon effect: one of two X chromosomes is silened by epigenetic mechanisms
Allel-specific expression (ASE): regulation involving specific genes rather than full chromosomes.
Local adaptation
Assembling the right types of genetic variation to enhance the population’s success under its particular set of local habitat conditions, which likely differs significantly form another location.
- selection for allelic variations of proteins that differ in thermal optima
- altered patterns of gene expression
RNA editing: splice variants
Post transcription, pre translation; during preparation to be translated
introns are removed and exons are joined
Intron-free RNA: has been seen to be stress-induced proteins; these have to be expressed without delay so it would be advantageous to skip the splicing step
Splice variants: one expressed gene can be spliced in different patterns and thus result in mature mRNA variants from the same original mRNA
RNA editing: changing base composition
mRNA can be chemically modified to alter amino acid sequence. This involves a change in codons and thus can give rise to multiple protein isoforms.
i.e. adenine to inosine by the enzyme adenosine deaminase
Small regulatory (noncoding) RNAs
gene that encode small RNA molecules that are transcibed but not translated.
- determine whether a specific mRNA gets translated into a protein
- controlling half-lives of mRNA
Stress-induced unmasking of cryptic genetic variation
Hsp90 example: in not stressful conditions, this protein is a part of signal transduction pathways as a molecular chaperone to montior the protein folding in that pathway. In stressful conditions, Hsp90 moves to be a chaperone of another class of proteins. Upon leaving the previous function, proteins could be folded incorrectly and lead to disrupted processes and potentially change the phenotype. This has the potential to unmask genetic variation, which has the potential to be adaptive.
Transgenerational inheritance of epigenetic changes (TGI)
epigenetic changes that modify gene transcription pass from one generation to the next = TGI
Do epigenetic changes to a chromosome “wash out” after a few generations or are they long-lasting?
Epigenetic modifications, if inherited, exist on chromosomes that segregate in the same manner that all nuclear genetic material and thus are subject to the same evolutionary genetic processes that ultimately determine whether they will persist or be lost from a population. Caused by proteins that are themselves genetically encoded and subject to all evolutionary forces like mutuations, genetic drift, and natural selection
Symbiosis with other organisms
Symbionts and microbiome.
Eukaryotes have a slower translation rate, which could enable more complex protein structures.
Steno vs. eury therm: stenos may 1.) not have the appropriate proteins any more b/c of evolving in a highly predictable environment 2.) lost the ability to upregulated or expression those genes if even they have them
Smaller changes that can have a big impact: micromolecules (low-molecular-mass constituents) can regulate function or stability of macromolecules, and adjustments to micromolecules themselves – proton activity (pH) organic osmolyte concentrations
Chapter 2: Oxygen and Metabolism
Metabolic depression (hypometabolism): reducing the requirement for ATP, often in response to reduced levels of oxygen.
Energy metabolism pathways
Cytochrome c oxidase system: supports enhanced yield of ATP per substrate molecule but also a mechanism for decreasing the amounts of toxic oxygen by-products produced during aerobic ATP generation.
- via electron transport system (ETS), serving as electron capacitor that stores 4 electrons until the system can be discharged to generate the nontoxic end product, water.
- tetravalent reduction of oxygen (4 electrons to dioxygen molecule to from 2 molecules of water)
- capacitor function reduces risk of univalent reduction of dioxygen, which generates ROS
- five components in oxidase enzyme: 2 heme irons, 2 copper centers with 2 and 1 coppers each
- cytochrome c enzyme + cytochome oxidase enzyme
therefore efficiently functioning cytochrome c oxidase systems is essential
Oxygen’s “good side” is generating biologically useful forms of energy (ATP) and “bad side” is ROS production.
Can a coral go into an oxygen depleted state if the symbionts are expelled? Lower biproduct consumption of oxygen from photosynthetic pathways of the symbiont?
Oxygen-dependent biosynthetic pathways includes ones that produce collagen, steroid compounds like cholesterol.
Dioxygen (O2) can accept pairs of e- (later used in ATP production) or single e- which leads to production of reactive oxygen species (ROS).
- highly reactive; damage DNA, lipids, proteins, integrity of genome and biomolecules
- increases as result of cellular stress
- env stress from high temperature, osmotic challenges, transition between high and low levels of oxygen
- to reduce (scavenge) ROS, requires NADPH as a co-factor (or reducing equivalent)
Oxygen demands per unit mass are reduced in larger species. > how does this differ with tissue thickness? How does oxygen enter the coral’s body?
Critical oxygen partial pressure (Pcrit): Oxygen concentration below a certain point where performance begins to decline. Higher Pcrit = greater sensitivity to oxygen limitation. This limit is driven by metabolic requirements of partiuclar developmental stages or modes of activity
Organismal properties that establish animals’ matabolic requirements
Energy metabolism: base function is to provide adequate supply of ATP
Standard metabolic rate: ectotherms (depend on external sources of heat) i.e. corals
Basal metabolic rate: endothermic homeotherm
ATP consuming pathways:
Specific dynamic action (SDA): postabsorptive increase in energy metabolism
- thought to be caused by production of proteins
Factors that establish metabolic rate:
1. physical activity (i.e. locomotory activity driven by ATP turnover in muscle)
2. digestion
3. reproduction
4. time of day
5. age
6. abiotic factors: temperature, oxygen concentration, cellular stress b/c of the need to maintain cellular homeostasis and repair tissue damage
2 types of biochemical responses: types of metabolic pathways AND alterations in the flux rates through those pathways. > Are all coral pathways the same?
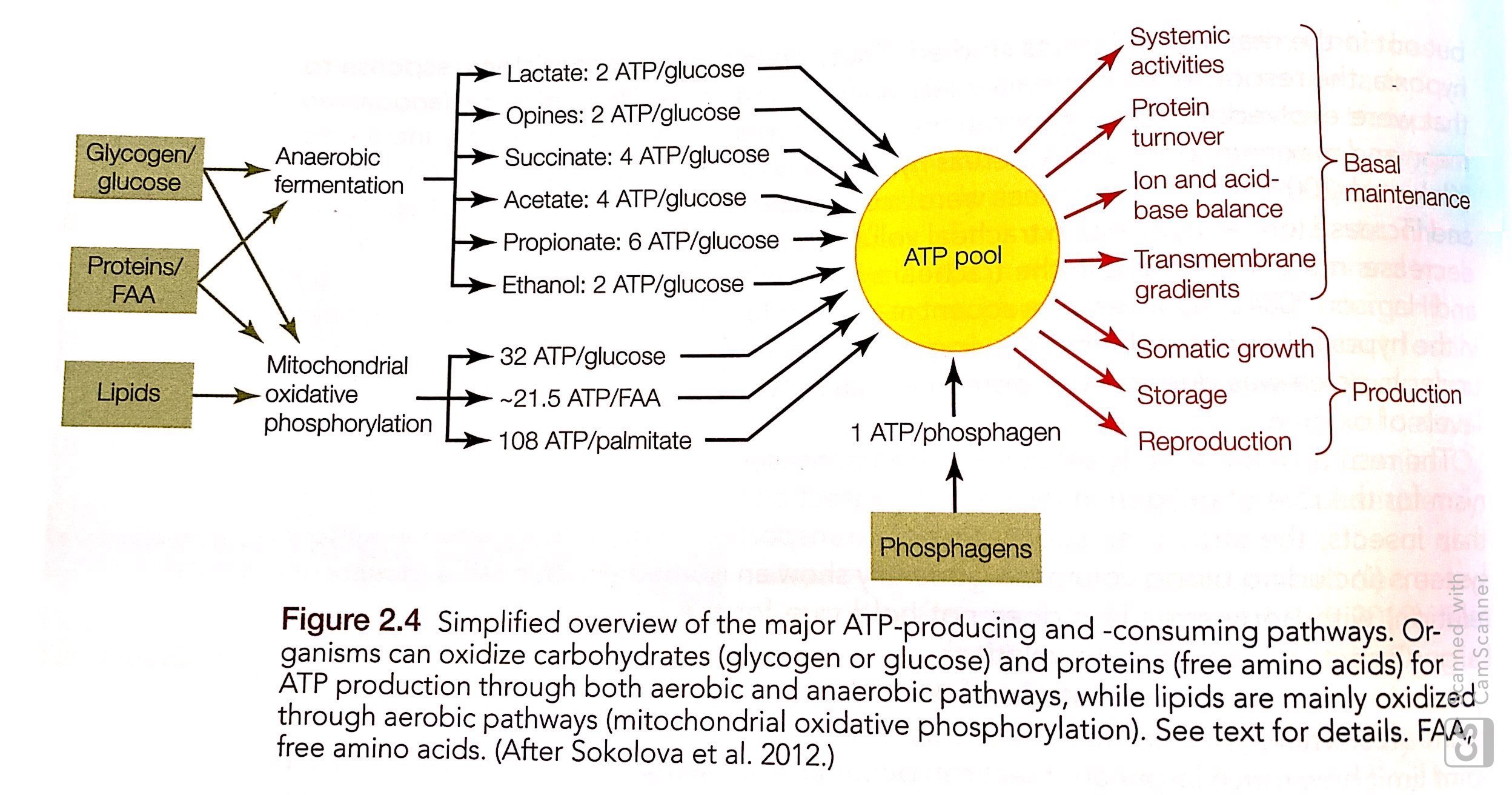
Main pathways of ATP production
see DNA methylation PDF figure for the below pathways
- Glycolysis
- Tricarboxylic acid (TCA) cycle or Krebs or Citric Acid Cycle
- Mitochondrial ATP synthase complex: uses proton gradient generated by the electron transport system (ETS) across the inner mitochondrial membrane to synthesize ATP.
2 mechanisms to add the terminal phosphate to ADP:
1. Substrate-level phosphorylation: transfer of “high-energy” phosphate group from metabolite (i.e. PEP) to ADP = ATP. Used in 2 rxns in glycolysis and 1 rxn in TCA cycle
2. Oxidative phosphorylation: phosphorylation of ADP by the ATP synthase complex of mitochondrial inner membrane. E- for this come from reducing equivalents in the form of NADH produced by glycolysis, TCA cycle, B-oxidation of fatty acids.
Short-term increase in ATP production / buffering ATP fluctuation: 1. phosphagens creatine and 2. arginine-phosphate kinases: catalyze exchange of terminal phosphate groups between ADP and creatine-phosphate or arginine-phosphate. These are reversible phosphorylation rxns
Production of reducing equivalents: NADH, NADPH, FADH2. Functions of these:
- deliver e- to ETS; therefore provide force to drive transport of protons across inner membrane and establishing proton gradient whose subsequent dissipation drives oxidative phosphorylation
NADH: essential at complex I in ETS to maintain flux rates through catabolic pathways that break down substrates like glucose. Rxns in TCA cycle are primary source of NADH
NAD+: essential cofactor in one of rxns in glycosis and rxns in TCA cycle
Oxidative phosphorylation: involving reduction of oxygen through the cytochrome c oxidase rxn: highest yield of ATP per substrate molecule catabolized BUT there needs to be oxygen to support this pathway
Reduced oxygen availability can arise via:
1. physiological (functional) hypoxia: transport of O2 from environment to respiring tissues cannot keep up with oxygen demand
2. oxygen levels below what is needed to support ATP production, environmental hypoxia occurs
In either type of hypoxic situation, molecules other than oxygen serve as electron acceptors for continued catabolic activity
Fermentation: reduction of an organic molecule to regenerate the oxidized cofactor
Lactate dehydrogenase (LDH): enzyme used in NADH-requiring fermentation rxn which reduces pyruvate to lactate
NADPH: used in many anabolic reduction rxns; pentose phosphate pathway (PPP) is major supplier of NADPH.
- used to reduce ROS so when ROS is high, demand for NADPH increases
Flavin-containing nucleotides (FAD): important in reversible oxidation-reduction rxns; ATP generation
- FADH2 used in TCA cycle and deliver e- to ETS in complex II
Voltage-dependent anion channel (VDAC): regulates transport of ADP and ATP across inner and outer membranes of the mitochondria

fig2
Bi products of metabolism
metabolite of glycolysis can be coverted to yield fatty acids, triacylglycerides for fat storage and phosphyoglyceride lipids for membranes
Glycosis products can be reversible thermodynamically and energy storage molecules can be converted to sustain glucose levels when needed.
TCA cycle = provides building blocks for fatty acid synthesis and carboxylic acid skeletons for amino acid biosynthesis
PPP pathway = building blocks for nucleotides of DNA and RNA
Temporal and spatial ATP buffering by phosphagens
High ATP turnover rates results in first line of defense = increase in rxn rates of creatine kinase (CK) and arginine kinase (AK). AK is the main source in invertebrates
- AK most effective at buffering ATP during prolonged conditions of lower ATP/ADP ratios and low pH.
inorganic phospate (Pi):
- assists in intracellular pH buffering mediated by activity of AK
- substrate for glycogen phosphorylase and thus promote glycogenolysis
Different isoforms of these kinases can be expressed within one cell; localized kinases are utilized for optimal fxn
Recycling of mitochondrial ADP by creatine kinases activites oxidative phosphorylation, this enzyme regulates rates of mitochondrial respiration and therefore reduces ROS production.
Reactive Nitrogen Species (RNS) is similar to ROS.
Posttranslational modifications and signaling by metabolic pathways
Posttranslational modifications (PTMs) change the activity, location, turnover of proteins in response to shifts in metabolism.
Acetyl-CoA: central metabolite of both carbohydrate and fatty acid catabolism and key building block for synthesis of lipids and cholesterol.
Acetylases: use ACoA as substrate to acetylate proteins, deacetylases remove the acetyl group from proteins
- affects almost all metabolic enzymes and nuclear regulatory proteins
- increase in levels of ACoA indicate a high energy charge, signaling to the cell the potential for enhancing activities
- high levels in nucleus trigger acetylation on histones which leads to loosening of chromatin structure and activation of gene expression
HDACs reverse this process
Sirtuins are are a type of HDAC and used NAD+ as a substrate to remove the acetyl group
Metabolic control theory
suggests that all enzyme-catalyzed steps in a pathway can contribute to the regulation of flux
Aerobic metabolism
Infoldings of inner membrane form cristase that reach into the matrix and serve to increase the surface area of the inner membrane (forms classic shape of mitochondria). Products of metabolic pathways need to be transported across several membranes.
Exchange of metabolites b/w cytosol and intermembrane space is controlled by a porin (voltage-dependent anion channel VDAC). VDAC has low selectivity.
Exchange across intermembrane into matrix is controlled by adenine nucleotide translactor (ANT), mainly controls exchange of ATP and ADP. NADH is transported into the matrix through malate-aspartate shuttle (below).

fig3
Pyruvate oxidized by PDH rxn, forming NADH and Acetyl-CoA. Acetyl-CoA feeds into TCA/citrate synthase reaction to form citrate (below)

fig4
Electron transport system (ETS) use reducing equivalents: NADH and FADH2 at complexes I and II. Complex IV = cytochrome c oxidase (see notes above). This builds a chemical and electrical gradient called proton motive force that is used in oxidative phosphorylation.

fig5
B-oxidation of fatty acids generates acetyl-CoA for the TCA cycle

fig6
Mitochondrial functions

fig7
Genetic aspects
Hybrid breakdown: breakdown in fitness in F2 hybrid generation in crosses between population.
Maternal inheritance of the mitochondrial genome is essentially clonal.
Intergenomic (nuclear and mitochondrial) coadaptation between gentically distinct populations of copepods caused hybrid breakdown (disrupted function) when populations were hybridized.
- divergence of mitochondrial sequences of COI of subunit of cytochrome c oxidase (complex IV) can be high in populations (especially in those locally adapted)
Oxidative Stress: Production and Scavenging of ROS
Cytochrome c oxidase (COX) is an electron capacitor, holding e- until the system can discharge a univalent reduction of molecular oxygen into water. Sometimes this reduction can produce ROS instead.
Reactive species: strong tendency to either provide (reduce) or extract (oxidize) e- to or from other molecules.
Radicals: unpaired e- on the outer shell and are ore strongly oxidizing
ROS:
1. Superoxide anion (O2 -): causes by leakage of e- from ETS
2. Hydrogen peroxide (H2O2): longer half life and less radical, allowing it to diffuse easily across inner mitochondrial membrane, ER, and peroxisomal membranes. This is facilitated by aquaporins. Therefore H2O2 can affect processes far from its origin.
- when reacts with ferrous iron (Fe 2+) its converted into OH (this is the Fenton rxn)
3. Hydroxyl radical (OH): most reactive ROS, most scavenging is targeted at H2O2 so that it doesn’t turn into OH
Generates from:
1. Complexes I and III of ETS
2. PDH (pyruvate dehydrogenase)
3. aKGH of the TCA cycle
In conditions of:
1. high CoQH2/CoQ ratios
2. high NADH/NAD+ ratios
3. large proton gradients
Manganese superoxide dismutase (Mn-SOD)
ROS scavenging systems:
- glutathione and thioredoxin-peroxiredoxin systems
- catalase
ROS causes oxidative damage which changes the structure and fxn of DNA. This is fixed by base excision repair and nucleotide excision repair mechanism – DNA damage recognition and repair enzymes.
ROS causes oxidation of fatty acids and cholesterol, causes leaking membranes and alteration of lipid-protein interactions with membranes.
Oxidation of proteins is a removal of hydrogen from alpha carbon atom of an amino acid residue. This triggers a downstream cycle that results in degradation of proteins and protein function.
Metabolic depression (reduction of mitochondrial activity) can reduce rates of ROS generation.
In mitochondria:

mito
Pyruvate and aKG function like antioxidants and facilitate scavenging of ROS. They protect neurons from H2O2
Example of response in mussels
Decreases in abundance, changes in subunit composition, and posttranslational modifications of metabolic enzymes are potential mechanisms of regulation. All towards down regulation of metabolism and possbily ROS production with a switch to production of NADPH from pro-oxidant NADH, which can serve as a reducing equivalent for ROS scavenging.

fig8
Uncoupling proteins
Supply of ADP to ATP synthase rxn is needed to allow reentry of protons that have been moved outward across inner membrane by ETS (see below). The supply of ADP depends on the use of ATP that drives various work in the cell. When ATP use rates are low, e- builds up and leakage of those e- will produce superoxide. “safety valve” = uncoupled proteins that allow transport of e- back into the inner membrane space for COX that isn’t dependent on ATP production and ADP use.

fi9
But potential is high to waste large amounts of E so it is very tightly regulated and only when danger of ROS generation arises.
ROS scavenging
First line of defense:
Manganese superoxide dismutase (MnSOD): converts superoxide into hydrogen peroxide in mitochondria
Copper-zinc SOD (Cu-Zn-SOD): does the above in the inner membrane space
Second line of defense:
Hydrogen peroxide dismutation by catalase in peroxisome, AND glutathione peroxidase (GPx) (glutathione system) and peroxiredoxin (Prx) in mitochondria.

fig10

fig11
Thioredoxin-peroxiredoxin system: H2O2 scavenging; Trx, Prx.
In polychaetes during acute thermal stress: heat tolerant species did not change SOD isoforms. GR = gutathione reductase. > This lines up with 1st chapter data. Mcap didn’t change TAC, but there is higher variation and higher values of TAC in Pacuta.

fig12
Low pH may cause increase in ROS production through release of heavy metals such as Fe2+ from intracellular stores, triggering the Fenton rxn, or through intxn of CO2 with peroxynritrite resulting in reactive O, N, C species.
Non-enzymatic antioxidant responses
Glutathione (main cellular non-enzymatic antioxidant), ascorbic acid (vitamin C), a-tocopherol (vitamin E), and carotenoids.
Ascorbate is a co-factor for several enzymes.
Antartic fishes might have higher levels of the above systems and a creater capacity to scavenge ROS.
Sources of reducing power (NADPH)
GR and TR require NADPH to maintain levels of reduced Glutathione and thioredoxin, therefore the pathways to produce NADPH are important in antioxidant response too.
Pathways to produce NADPH: cystolic pentose phosphate pathway (PPP), NADP-dependent dehyrdogenases, and transhydrogenase. NADP-IDH and ALDH.
Acute stress in Mytilus caused increase in the above and in the non-oxidative PPP process that resulted in many NADPH.
Oxidative stress in the ER
Secreted and membrane proteins fold and mature in the ER; newly translated proteins, formation and proof-reading and disulfide bond formation.
Maturation of proteins in the ER happens before glycosylation and then are exported can be a source of ROS.
ER is a compartment that is highly sensitive to environmental challenge. ER molecular chaperones: PDI, Grp78, Grp94. Studies in mussels have shown that these increase in response to acute salinity stress and decreased in recovery.
Rates of H2O2 production from disulfide bond formation would follow the same pattern of increase/decrease of use of molecular chaperone. Then, this effect would be mirrored in changes in antioxidant protein expression.
Metabolic Rates and Oxygen
States where metabolic rates drop well below the BMR/SMR is referred to metabolic depression / hypometabolism. Hypermetabolism = Maximum metabolic rates (MMR) achieved during exhaustive physical activity. Field metabolic rate = organisms going about their lives sustain varying metabolic rates for various activities.
This is measured by calorimetry: production of heat is most direct way to quantify metabolic rate = direct calorimetry
Measurements of oxygen consumption = indirect calorimetry

fig13
Metabolic rates of single cells decrease with increasing body mass in birds, animals, and non-avian reptiles. > I don’t think corals would follow the same trend? How is energy dissipated across the whole colony? I don’t think a polyp on one side is providing a major amount of resources to other side in the same way other animals’ bodies do.
Potential differences in metabolic function:
1. more mitochondria per unit of mass of tissue
2. more enzymes per unit of mass of mitochondria
3. enzymes with different specific activities
Differential enzyme function:
1. intrinsic catalytic power of an enzyme: rate at which an active site can convert substrate to product
2. posttranslational modifications that modify the Kcat value of the proteins
3. change the milieu in which enzymes work to alter the specific activity of the enzyme. In respiration, the milieu is the lipid composition of the inner mitochondrial membrane. The lipid environment surrounding the proteins involved in respiration modulates their activity = viscoptropic effect.
highly viscous lipid environment = more resistance for proteins. Decreasing viscuosity = increase rate of protein function
Metabolic depression and hypometabolism
Adverse conditions challenge organisms’ ability to maintain ATP turnover rates. One common response is to down-regulate metabolism or overall ATP turnover rate. Characterized by a reduction in mitochondrial respiration and ATP synthesis, down-regulating ATP-consuming processes, reduction in ion regulation (activity of NA+ - K+ - ATPase), protein synthesis, biosynthesis of other macromolecules and cellular assemblages. Increase in protein stabilization to prolong lifespan.
In brine shrimp during metabolic depression, they downregulated processes that generated Acetyl-CoA that goes into the TCA cycle: PDH. Which also generates ROS.
Drops in pH: response likely depends on function of vacuolar proton pump / V-ATPase. Its purpose is to acidify cellular components such as lysosomal vesicles.
Energetic cost of transcription is a lot smaller than protein synthesis. MicroRNAs regulate the expression of 60% of all protein-coding genes.
Nuclear factor erythroid 2-related factor 2 (Nrf2) = txn factor that increases expression of several antioxidant proteins. > Check this in invertebrates?
Energy Homeostasis and Environmental Stress: A Conceptual Framework
Dynamic Energy Budget (DEB) model: balances energy acquisition and allocation among competing phys processes within an individual.
Oxygen- and capacity- limited thermal tolerance (OCLTT): framework for how oxygen supply sets limits to stress tolerance.
The available energy flux through any organism will have to prioritize the maintenance of BMR/SMR. Sometimes to the detriment of growth, storage, reproduction.
Surplus metabolic capacity: difference between BMR and MMR = aerobic scope. An organism’s aerobic scope plays a role in setting an organism’s capacity to cope with stress.
Bioenergetic framework:

fig14
All of that to say that these metabolic pathways are critical to keep regulated, and oxygen levels are critical for the function of metabolism.