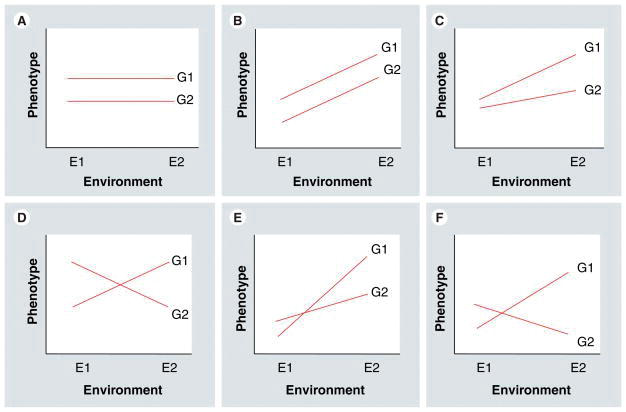
Chapter 5 Genotype x Environment Interactions
Panel A and B = no GxE interaction; A = Genotype interaction, B = environment interaction.
Panel C = Non-crossover G x E interaction; GxE but ranks remain unchanged.
Panels D-F = Crossover G x E interaction
Osei et al 2017
Baye et al 2011
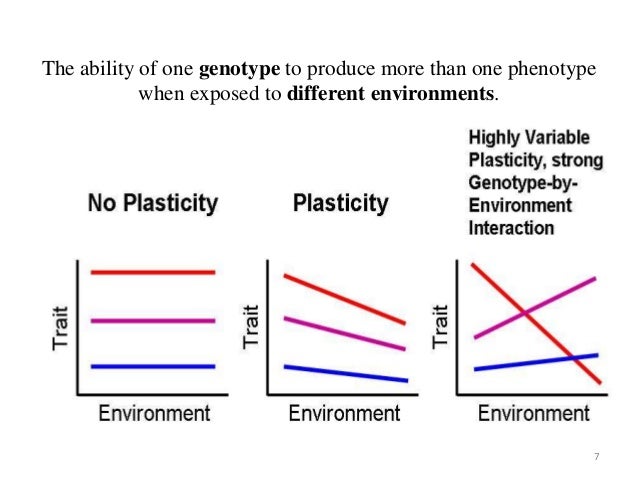
Plasticity and reaction norms (Nicotra et al 2010):

rxn
Adaptation: Content from Environmental Physiology of Animals: Willmer et al. lab copy here. Only accessible to Putnam Lab members.
The relationships of genotype x environment x physiology:

gxe
Adaptation
1. characters or traits observed in an organism that are a result of selection
2. process in which natural selection adjusts the frequency of genes that code for traits affecting fitness.
Evidence of adaptation: only if a trait has changed through time in ways that it will make it more effective at its task, therefore enhance fitness. The below are three types of correlative evidence (which is never concrete):
1. Correlation b/w character and environment/use.
2. Comparisons of individual differences w/in a species.
3. Observation of the effects of altering a character.
Selection for plasticity itself is an adaptation.
Traits become fixed b/c of natural selection, either presently selected on or previously selected on but now present for reasons that don’t have much to do with the original reason for selection. The latter = phylogenetic inertia
Optimality is a relative concept and a particular design need only to be sufficient to do better than the existing alternatives (there is such thing as good enough). Natural selection doesn’t necessarily provide maximal or even optimal solutions.
Genetic drift: chance shifts in gene frequencies
Genetic bottleneck: the above can be particularly true in small pop sizes
Phylogeny matters:

phylo
The environment can act on many developmental stages:

dev
Developmental switch: i.e. temperature dependent sex determination (TSD)
Understanding phenotypic plasticity in reaction norms:

rxn-norm
Reality of conforming and regulating to/in an environment:

conformer-norm
Genomic control
Amount of RNA made from a particular region of DNA must therefore be very tightly controlled, and is partly determined by the goodness of match b/w promoter region sequence and the RNA polymerase.
Six points where gene expression can be controlled:
1. Txn control: gene regulatory proteins (bind to specific sites on the DNA close to the gene coding sequence). Either transcriptional activators or transcriptional repressors.
Zinc-finger motif: where areas of alpha and beta helices are linked by a zinc molecule.

geneexp

txn
Protein degradation control:
- Ubiquitin links to destruction-bound proteins and serves as a degradation signal
Transposable elements:
- “jumping genes” that can move around the genome as DNA or an RNA intermediate
- some carry information for transposition and may accelerate mutation at the sites where they insert and exit. Others carry both instructions for transposition and some other gene
- i.e. antibiotic resistance in bacteria
Four types of acclimation
Solid line = initial rate-temperature curve
Dashed line = curve after thermal acclimation (to higher temp).
C = cold-acclimatized
W = warm-acclimatized

four