Chapter 2 Epigenetic Modifications
Page still in progress..
Epigenetic factors are involved in alternative mRNA splicing = major source of protein diversity and therefore, phenotypic variation (Luco et al 2011).
Types of Modifications
- DNA Methylation: See DNA-methylation.md for breakdown of DNA methylation.
- Histone variants and posttranslational modifications
- Non-coding RNAs
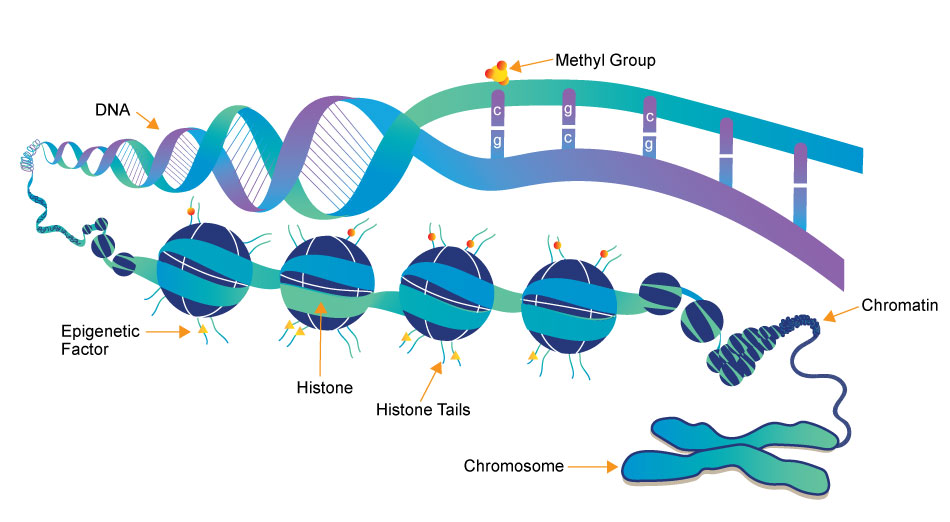
types
Chromatin Organization
Histone variants and posttranslational modifications
From Eirin-Lopez and Putnam 2019: Some of these variants have been identified in marine invertebrates, including H2A.X, H2A.Z, and macroH2A, which are involved in critical roles in the epigenetic regulation of DNA structure and metabolism (Gonzalez-Romero et al. 2012, Moosmann et al. 2011).
- histone H2A.Z is the most studied histone variant
- histone H2A.X is also well studied, partially because of its involvement in maintenance of genome integrity
- histone macroH2A is involved in gene repression at heterochromatic regions (Rivera-Casas et al 2016)
From Eirin-Lopez and Putnam 2019: These modifications:
- not only participate in the regulation of chromatin structure but also help recruit proteins and chromatin-remodeling complexes that influence transcription as well as many other DNA processes, such as repair, replication, and recombination (Bannister & Kouzarides 2011).
- chromatin metabolism and epigenetic memory (Feil & Fraga 2012).
Acetylation:
Methylation:
Phosphorylation:
Germinal chromatin
Highly specialized epigenetic reprogramming that differs in male and female germ cells.
From Eirin-Lopez and Putnam 2019: In order to be vertically transmitted to the offspring, epigenetic modifications need to be present in the germline and endure the extreme reorganization of chromatin during gametogenesis, especially in the case of males. Since nucleosomes are disassembled and histones are widely replaced by protamines in mature spermatozoa (Eirin-Lopez & Ausio 2009), the epigenetic contributions of sperm chromatin to embryo development have been considered highly limited.
[insert hollie’s sperm methylation data set]
Explain the below figure with no caption for comps

fig3
Noncoding RNAs
Environmental Epigenetics
The idea that epigenetic modifications and changes can lead to individual phenotypic changes (intragenerational plasticity) and phenotypic changes across progeny (transgenerational plasticity).
Marine Environmental Epigenetics (Eirin-Lopez and Putnam 2019):
- Challenge 1: Elucidating cause–effects relationships among environmental signals, epigenetic modifications, and the dynamics of wash-in and wash-out of epigenetic effects.
- Challenge 2: Generating a detailed mechanistic understanding of epigenetic mechanisms and their interplay.
- Challenge 3: Demonstrating the capacity for and mechanisms of transgenerational epigenetic inheritance.
- Challenge 4: Clarifying the interplay of genetic and epigenetic features and evolutionary consequences
For comps, be able to explain both of the below figures without the caption.

fig1

fig2
Epigenetic conditioning or hardening
Epigenetic Crosstalk H3K36me3 histone modification and gene body methylation is conserved in cnidarians
Epigenetic reprogramming