Chapter 6 Analysis of Data
6.1 Interactions plot
We plotted the reported interactions with the hierarchical edge bundling technique. Interactions that are oftenly reported are highlighted in purple.
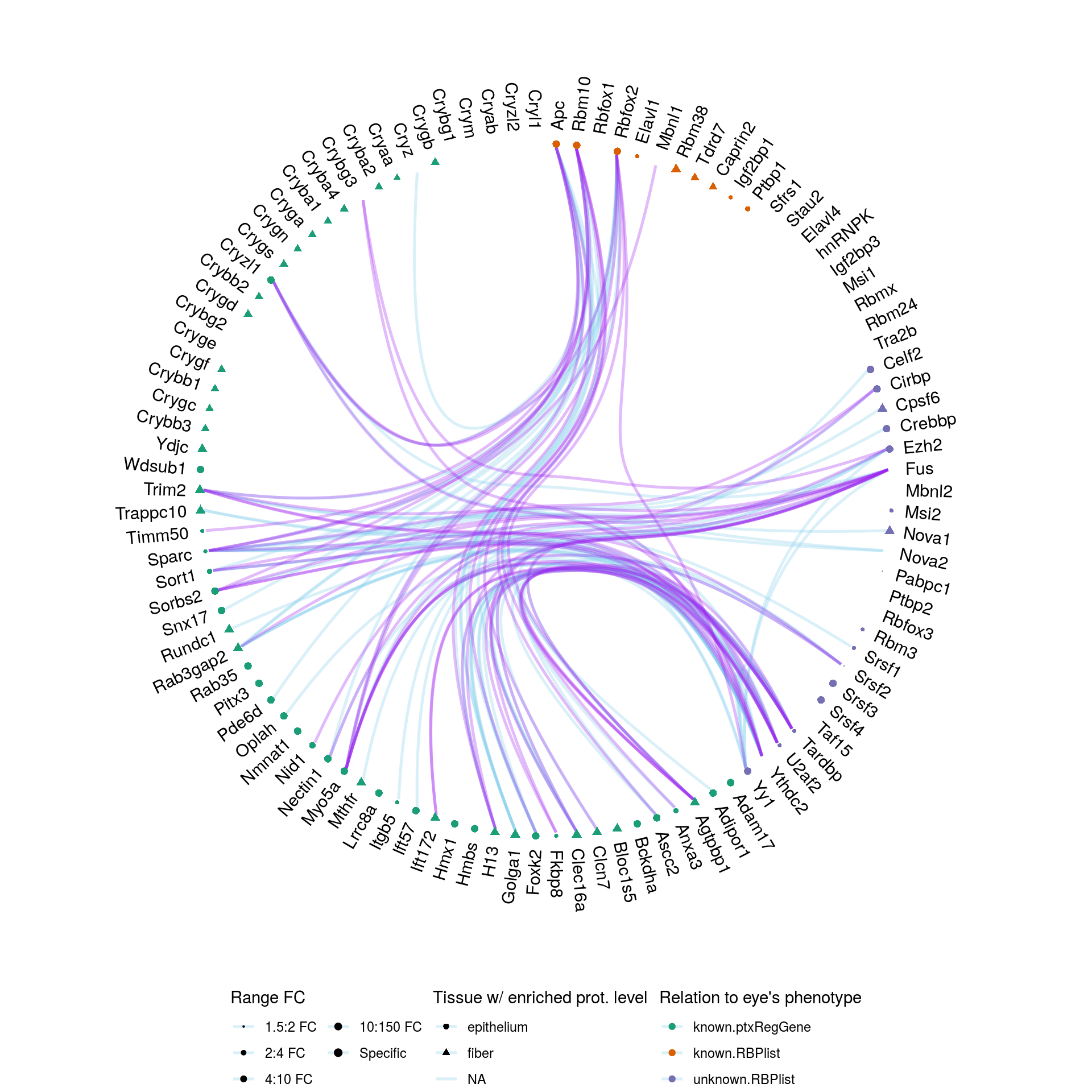
Figure 6.1: Reported interactions between genes potentially ptx-regulated and RNA binding proteins. Only interactions where at least one actor is known to be associated with eye’s phenotype are shown.
We note the absence of reported interactions of crystallin transcripts with RNA binding proteins. This is due to the tissue examined in datasets of POSTAR.
6.2 Ideas box
Using this approach we highlighted potential post-transcriptional gene regulation of our candidate genes via RNA binding proteins.
Next steps will be to investigate this list of actors, check for known regulations by RBP, eventually in human. Eventually, we could enrich the list of candidate genes with genes that are known to be ptx-regulated. For example, crystallin transcripts. In parallel, the same pipeline could be applied to POSTAR human dataset, which is 10 times larger than mouse dataset.
So as to do that I will have to find a way to convert my list of candidate genes, find the human homolog. Focusing on redundant conserved regulations between mice and human might be a good idea for a starter.
It might be informative to plot reported interactions on genes transcripts so as to assess the region that is regulated by the RBP.
- YY1 : As a regulator of cell proliferation, YY1 has been shown to act either through transcriptional activation of cell‐cycle progression genes, such as c‐myc (Riggs et al. 1993), or through direct interaction with – and resulting inactivation of – the tumor suppressor gene product p53 (Sui et al. 2004).
During Xenopus development, YY1 was found to be predominantly expressed in the anterior neural tube, developing brain, and optic vesicles (Kwon and Chung 2003). Inhibition of YY1 synthesis by injection of antisense oligonucleotides in Xenopus embryos caused a slight developmental delay and a marked reduction in the size of head structures and eyes (Kwon and Chung 2003). Additional evidence involving YY1 in eye development came from a study showing that the abnormal anterior eye formation observed in Ring1‐deficient mice was exacerbated in compound YY1+/− Ring1−/− mice (Lorente et al. 2006). A similar phenotype was recently reported in Rybp (Ring1 and YY1 binding protein) heterozygous null mouse embryos. Pirity et al. (2007)
A result suggesting that Rybp, Ring1, and YY1 may interact to orchestrate normal vertebrate eye development. In addition, in Xenopus and mouse embryos YY1 has been shown to activate the expression of Otx2 (Kwon and Chung 2003; Takasaki et al. 2007), a homeobox transcription factor that plays a central role in retinal cell differentiation (Nishida et al. 2003; Viczian et al. 2003).
NB: the expression of RYBP in melanoma cells was negatively regulated by miR‐9, which was suppressed by the RYBP binding protein, YY1. Thus, this study suggested that the YY1‐miR‐9‐RYBP axis plays a vital role in melanoma tumorigenesis. Zhan, Wang, and Ge (2018)
References
Pirity, Melinda K, Wei Lin Wang, Louise V Wolf, Ernst R Tamm, Nicole Schreiber-Agus, and Ales Cvekl. 2007. “Rybp, a polycomb complex-associated protein, is required for mouse eye development.” BMC Developmental Biology 7: 1–14. https://doi.org/10.1186/1471-213X-7-39.
Zhan, Shaohua, Tianxiao Wang, and Wei Ge. 2018. “Multiple roles of Ring 1 and YY1 binding protein in physiology and disease The roles of RYBP in development The roles of RYBP in the regulation of gene expression through the PcG complex and binding with” 22 (4): 2046–54. https://doi.org/10.1111/jcmm.13503.