Vignette 5 Evaluation by simulation
Gates Dupont & Chris Sutherland
5.1 Simulating SCR data
We can evaluate a design by simulating data and recovering parameter estimates.
We illustrate this using simulator(), which executes the following:
- Randomly assigns N activity centers to the state-space
- Calculates \(p\) for each \((x, s_i)\) pair based on distance
- Creates a 3D encounter history array, \([i,j,k]\), using a binomial model
- Fits a basic SCR model using
oSCR.fit() - Returns parameter estimates and summary statistics
Here’s the simulation function that we use. It’s fairly straightforward, but has a few extra elements to help with troubleshooting.
require(oSCR)
require(dplyr)
simulator<- function(traps, ss, N, p0, sigma, K, nsim, plot = TRUE) {
# Initialize data-collection matrix
simout1 <- matrix(NA, nrow=0, ncol=9) # Create empty matrix for output
colnames(simout1)<- c("p0","sig","d0", # Estimates
"nind", # Number of individuals (length of first dimmension of y)
"avg.caps","avg.spatial","mmdm", # Summary stats from oSCR (possibly different definitions)
"failures","EN") # Other components
# Initialize while loop starting values
sim = 1 # Starting value for nsim
sim_try = 0 # Feature to aid in tracking failures
total_its = 0 # Feature to insure different random seeds
# Get nsim "good" simulations (no failures)
while(sim < (nsim + 1)){
# Update loop for seed
total_its = total_its + 1
# Update status
print(paste("Simulation Number", sim, sep = " ")) # keep track
cat("size of state-space: ", nrow(ss), " pixels", fill=TRUE)
cat(paste0("\n Try ", sim_try + 1, "\n"))
# Re-assign name
statespace = ss
# Set seed for simulation
seed = total_its
set.seed(seed)
# Sampling activity centers
s <- statespace[base::sample(x = nrow(statespace), size = N),]
# Make the state space data frame (oSCR object)
myss <- as.data.frame(statespace)
myss$Tr <- 1
myss <- list(myss)
class(myss) <- "ssDF"
# Individual-trap distance matrix
D <- e2dist(s,traps)
# Compute detection probabilities:
pmat <- p0*exp(-D*D/(2*sigma*sigma)) # p for all inds-traps p_ij
ntraps <- nrow(traps)
# Setup encounter histories data frames
y <- array(0, dim=c(N, ntraps, K)) # empty 3D array (individuals by traps by sampling occasion)
# Simulate encounter histories
for(i in 1:N){ # loop through each individual/activity center
for(j in 1:ntraps){ # loop through each trap
y[i,j,1:K]<- rbinom(K, 1, pmat[i,j]) # y ~ binomial(p_ijk)
}
}
# Reduce encounter histories to only captured individuals
ncap <- apply(y,c(1), sum) # sum of captures for each individual
y.all = y # for summary stats
y <- y[ncap>0,,] # reduce the y array to include only captured individuals
# Some summary information, which is actually printed for you later with "print(scrFrame)"
caps.per.ind.trap <- apply(y,c(1,2),sum) # shows # capts for each indvidual across all traps
# Check for captures
check.y = length(dim(y)) %>%
if(. > 2){return(TRUE)} else {return(FALSE)}
# Check for spatial recaps
check.sp_recaps = as.matrix((caps.per.ind.trap > 0) + 0) %>%
rowSums() %>%
c(.,-1) %>% # This is just to avoid warning messages due to empty lists
max %>%
if(. > 1){return(TRUE)} else {return(FALSE)}
# Check should be 2 if the design obtained at least a capture AND a spatial recapture
check = 0 # Clear this value from previous iteration
check = check.y + check.sp_recaps
# Note: Checking for sp.recaps implies getting caps,
# but for troubleshooting good to keep both checks
if(check == 2){
# Make the SCRframe object
colnames(traps) <- c("X","Y")
sf <- make.scrFrame(caphist=list(y), traps=list(traps))
# Plotting
if(plot == TRUE){
# Make plot
plot(statespace, asp = 1, pch = 15, col = "gray85", cex = 0.9)
points(s, pch = 20, col = "blue", cex = 0.5)
spiderplot(sf, add=TRUE) # This plots captures and spatial recaptures between traps
}
# Fit a basic model SCR0 (null model, ~1 ~1 ~1)
out1 <- oSCR.fit(model=list(D~1,p0~1,sig~1), scrFrame = sf, ssDF=myss, trimS = 4*sigma)
# Obtain estimates from the model
stats <- print(sf)[[1]] # pulls avg caps, avg spatial caps, and mmdm
est <- out1$outStats$mle # pulls p0, sigma, and d0 estimates from the model
en = get.real(out1, type="dens", newdata=data.frame(session=factor(1)),
d.factor=nrow(out1$ssDF[[1]]))[1,1] # Total abundance
# Append to data-collection matrix
sim_vals = c(plogis(est[1]), exp(est[2]), exp(est[3]), dim(y)[1], stats, sim_try, en)
simout1 = rbind(simout1, sim_vals)
# Just add two blank lines to live status updates
cat("\n\n")
}
# Updating while() loop
if(check != 2){
sim_try <- sim_try + 1
} else {
sim <- sim + 1
sim_try = 0
}
}
# Return the resultis from the simulation
result = data.frame(simout1)
return(result)
}5.2 Run simulations
Running the simulations is straightforward with simulator().
5.3 Evaluate the results
One of the most immediately interpretable evaluation metrics is percent relative bias (“%RB”), shown below, calculated for each parameter.
\[ \%RB = 100 \sum{\frac{\hat{y} - y}{y}} \]
Other useful metrics include (though not limited to):
- coefficient of variation (precision)
- scaled root mean squared error (accuracy)
- coverage
5.4 Plot the evaluations
This design performs well on average for bias. Precision would improve with more traps.
ggplot(data = df, aes(x = parameter, y = value)) +
geom_boxplot(fill = "lightblue1", color = "skyblue4", width = 0.1) +
geom_hline(yintercept = c(-5, 5), linetype = "dashed", color = "gray70") +
geom_hline(yintercept = 0, lwd = 1, color = "gray70") +
xlab(NULL) + ylab("%RB") + ylim(-100, 100) +
theme_pubr() + theme(legend.position = "none")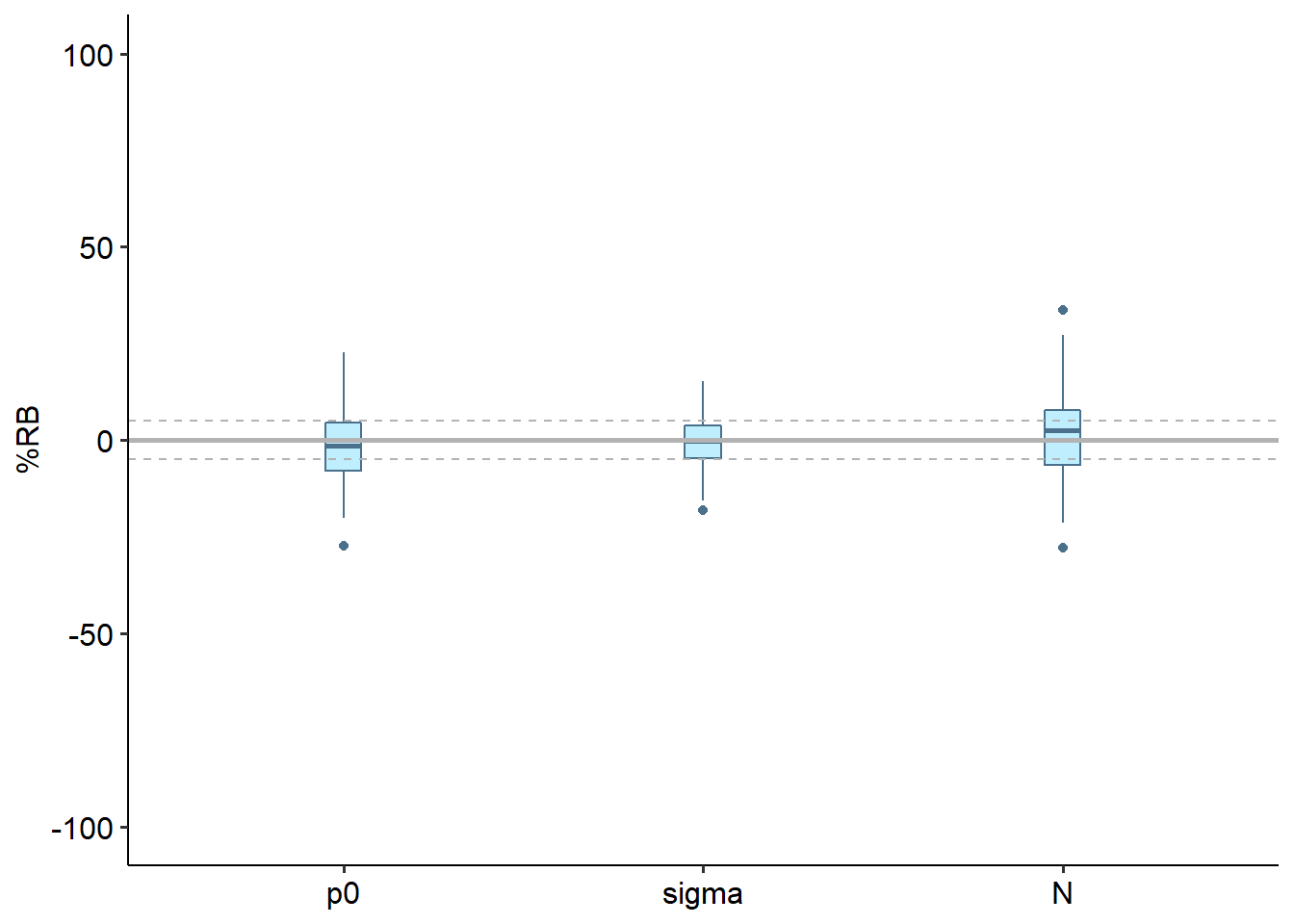
5.5 Take-home points
The oSCR framework:
- Conceptual and analytical framework for generating optimized designs.
- Three intuitive and statistically-grounded design criteria.
- Produce a greater amount of expected information, leading to more accurate estimates.
- Flexible for application to any species in any study area using SCR.
Johnson, Devin S, Dana L Thomas, Jay M Ver Hoef, and Aaron Christ. 2008. “A General Framework for the Analysis of Animal Resource Selection from Telemetry Data.” Biometrics 64 (3): 968–76.
Linden, Daniel W, Alexej PK Sirén, and Peter J Pekins. 2018. “Integrating Telemetry Data into Spatial Capture–Recapture Modifies Inferences on Multi-Scale Resource Selection.” Ecosphere 9 (4): e02203.
Royle, J Andrew, Richard B Chandler, Catherine C Sun, and Angela K Fuller. 2013. “Integrating Resource Selection Information with Spatial Capture–Recapture.” Methods in Ecology and Evolution 4 (6): 520–30.
Sun, Catherine. 2014. “Estimating Black Bear Population Density in the Southern Black Bear Range of New York with a Non-Invasive, Genetic, Spatial Capture-Recapture Study.” Master’s Thesis. Ithaca, New York, USA: Cornell University.