Chapter 3 May 31–Jun 5: Synchronous seminar
In this week of HE930, we will meet synchronously on Zoom to examine the predictive analytics process, discuss the ethics of using predictive analytics in HPEd, and apply predictive analytics together to a dataset.
Note that there is no assignment to complete and turn in this week. The only required work this week is to attend the synchronous seminar sessions on Zoom.
MOST IMPORTANTLY, please complete the following preparation before you attend the synchronous seminar sessions.
3.1 Preparation for synchronous seminar
Please watch the following two videos, which we will discuss in our synchronous seminar:
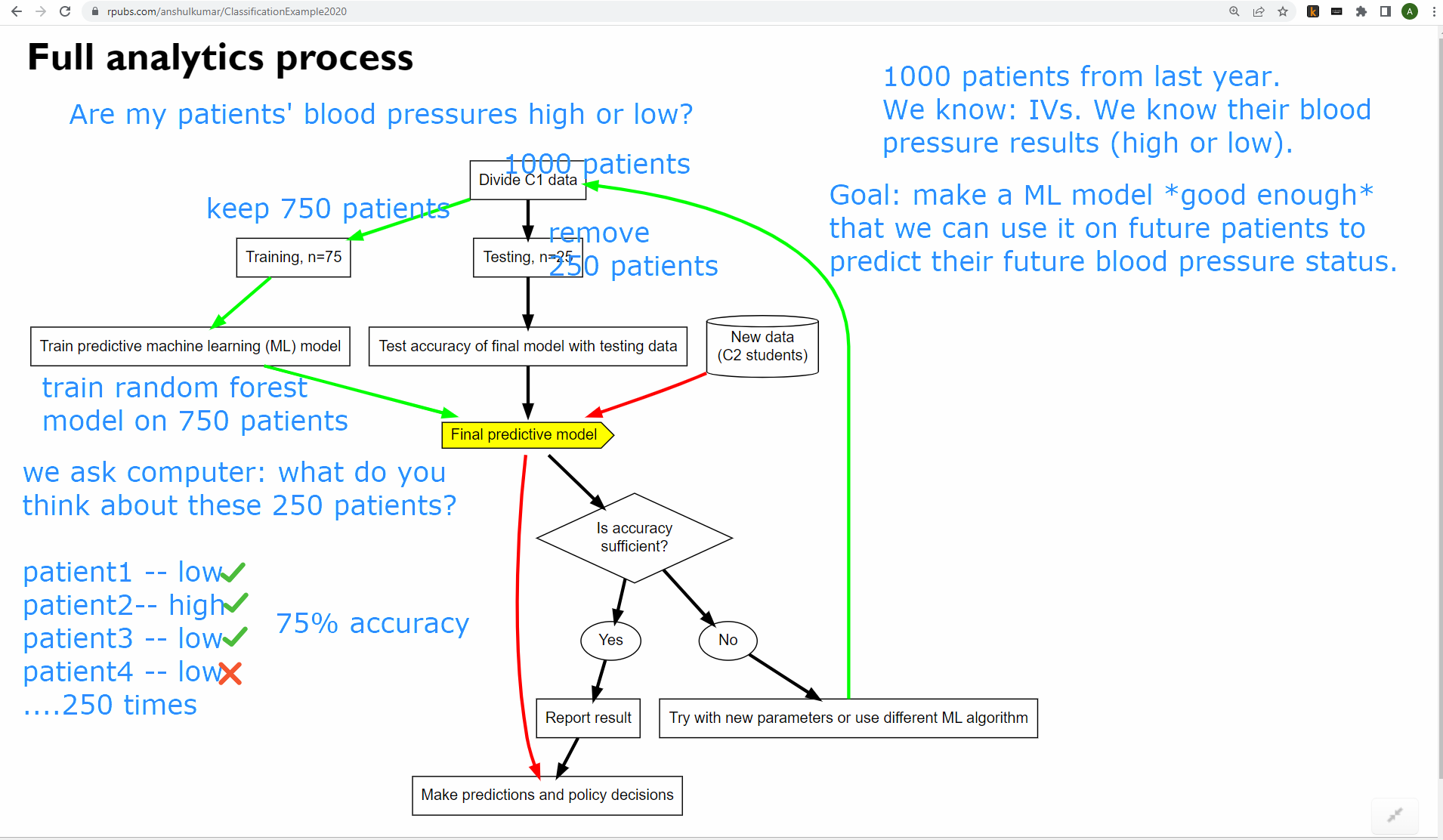
- “Predictive Analytics in Education: An introductory example”. https://youtu.be/wGE7C5w6hb4. Available on YouTube and also embedded below.
The slides for the video above are available at https://rpubs.com/anshulkumar/ClassificationExample2020.
- The Social Dilemma on Netflix. This link might take you there: https://www.netflix.com/title/81254224. If you do not have access to this on your own, inform the course instructors and we will arrange for you to watch.
Finally, in addition to watching the two videos above, please review all Week 1 readings, if you have not done so already.
3.2 Seminar details
Schedule as of May 24 2022:
The Analytics Process, Saturday May 28 2022, 4 p.m. Boston time. To prepare for this session, please review all Week 1 readings and watch this 26-minute video: https://youtu.be/wGE7C5w6hb4.
Machine Learning in R, Sunday May 29 2022, 4 p.m. Boston time. During this session, we will do a machine learning tutorial in R and RStudio on our computers. Please be prepared to run RStudio on your computer and share your screen. No other preparation for this session is needed.
The Social Dilemma and Ethical Concerns of ML, Monday May 30 2022, 12:30 p.m. Boston time. To prepare for this session, please watch The Social Dilemma on Netflix, which you can access at https://www.netflix.com/title/81254224.
You should have received calendar invitations to your email address for the sessions above. For now—as of May 24 2022—we will see who all is able to attend the sessions above. Then, for those who are not able to attend, we will schedule additional sessions in which the topics above will be repeated.
3.3 Session 1: The Analytics Process
This session is scheduled for:
- Saturday May 28 2022, 4 p.m. Boston time
- Additional repetitions of this session can be scheduled upon request (email the instructors).
3.3.1 Goal 1: Discuss the PA process and its applications
Reminder:
- PA = predictive analytics
- ML = machine learning
- AI = artificial intelligence
This discussion will primarily be driven by questions and topics raised by students during the sessions itself. The items listed below are meant to supplement our discussion.
3.3.1.1 Questions from week 1 discussion posts
What is PA/ML and how do we conduct it?
Item 1: Is predictive analytics part of regressions with all its types?- If yes, then what Is the difference between regressions and machine learning? (Ra A)
Item 2: The main questions are related to criteria of choosing one predictive analysis methodology over another and the ethical challenges in large data predictive analysis. (HL)
Item 3: There were a lot of terms and methods that I had to look up as I wasn’t familiar when working through the assignment. I look forward to learning more about them in the coming weeks and how I can apply use in my educational practice setting. (NF)
Item 4: I wish we started the course (this week) from conceptual understanding of Predictive analysis. Such what is Predictive analysis? Methods that can be utilized in predictive analysis? How to navigate through Predictive analysis? Why and when do we need predictive analysis? (SG)
Item 5: Will you walk us through these concepts that are very new to us? I tried to understand the content but certainly will benefit from a hands on approach to comprehension (RP)
Item 6: I would like to be exposed to the different predictive methods that are available. (LR)
Item 7: There were many new tests and algorithms referenced in the articles. Are there particular tests that we should have a better understanding more than others? (RS)
Using PA/ML in HPEd
Item 8: I want to learn more about the predictive methods and its connection to our own practice and setting. The articles provided some of the ideas but not clear yet how can I apply them to my clinical setting. (JF)
Item 9: How can you accurately design a re-usable analytics engine when the data being captured and the activities to be monitored can be vastly different from one learning management system to another and from one learning domain to another? It seems like developing an autonomous system is nearly impossible because someone has to manipulate the data for each scenario into a custom shape that fits the analysis method being used. (PS)
Ethics of PA/ML
Item 10: Will we hit on any ethical considerations? MIT brought back the SATs, which I assume had much to do with predictive analytics. How does one balance data and society? It makes sense that a test of current performance would predict future performance, but that does not account for an individual’s potential. There doesn’t seem to be a great way to pull potential out of a field of candidates, and most of the variables are surrogates which can hide the underlying structural causes of the issues. I recognize there is no answer here. And it is not fair to the studies we looked at to complain—they identify the at-risk people, not the reason they are at risk which can be discussed in the intervention. (MC)
Item 11: As we begin learning about predictive analytics, what are some approaches to minimize bias? (HD)
Item 12: I look forward to learning more about the predictive methods covered in the articles for week one. The article by Ekowo and Palmer (2016) emphasized the importance of institutions tackling evolving data privacy concerns. I do not have specific questions about this yet, but look forward to learning more about governance structures within institutions and best practices to support ethical and appropriate use of data. (CZ)
Disseminating/communicating results
Item 13: The articles are really good read, but as most of them are > 10 pages long with other readings, new concepts and many new terminologies (at least for me), the yield/outcome I gained may be minimized due to scattered focus (Su A)
3.3.1.2 Discussion questions from PA video/slides
How can predictive/learning analytics help you in your own work as an educator or at your organization/institution? What predictions would be useful for you to make?
How can you leverage data that your institution already collects (or that it is well-positioned to collect) using predictive analytic methods?
What would be the benefits and detriments of incorporating predictive analytics into your institution’s practices and processes?
Could predictive analysis complement any already-ongoing initiatives at your institution?
What would the ethical implications be of using predictive analytics at your institution? Would it cause unfair discrimination against particular learners? Would it help level the playing field for all learners?
3.4 Session 2: Machine Learning in R
This session is scheduled for:
- Sunday May 29 2022, 4 p.m. Boston time.
- Additional repetitions of this session can be scheduled upon request (email the instructors).
During this session, we will work together to complete and discuss the machine learning tutorial below.
3.4.1 Goals
Practice using decision tree and logistic regression models to predict which students are going to pass or fail, using the
student-por.csvdata.Fine-tune predictive models.
Compare the results of different predictive models and choose the best one.
Brainstorm about how the predictions would be used in an educational setting.
3.4.2 Important reminders
Anywhere you see the word
MODIFYis one place where you might consider making changes to the code.If you are not certain about any interpretations of results—especially confusion matrices, accuracy, sensitivity, and specificity—stop and ask an instructor for assistance.
3.4.3 Load relevant packages
Step 1: Load packages
if (!require(PerformanceAnalytics)) install.packages('PerformanceAnalytics')
if (!require(rpart)) install.packages('rpart')
if (!require(rpart.plot)) install.packages('rpart.plot')
if (!require(car)) install.packages('car')
if (!require(rattle)) install.packages('rattle')
library(PerformanceAnalytics)
library(rpart)
library(rpart.plot)
library(car)
library(rattle)3.4.4 Import and describe data
We will use the student-por.csv data.
Step 2: Import data
d <- read.csv("student-por.csv")The best place to download this data is from D2L.
Data source and details:
P. Cortez and A. Silva. Using Data Mining to Predict Secondary School Student Performance. In A. Brito and J. Teixeira Eds., Proceedings of 5th FUture BUsiness TEChnology Conference (FUBUTEC 2008) pp. 5-12, Porto, Portugal, April, 2008, EUROSIS, ISBN 978-9077381-39-7. Available at https://archive.ics.uci.edu/ml/datasets/Student+Performance.
Alternate source: https://www.kaggle.com/larsen0966/student-performance-data-set
If the code above does not work to load the data, you can try this code instead:
d <- read.csv(file = "student-por.csv", sep = ";")Step 3: List variables
names(d)## [1] "school" "sex" "age"
## [4] "address" "famsize" "Pstatus"
## [7] "Medu" "Fedu" "Mjob"
## [10] "Fjob" "reason" "guardian"
## [13] "traveltime" "studytime" "failures"
## [16] "schoolsup" "famsup" "paid"
## [19] "activities" "nursery" "higher"
## [22] "internet" "romantic" "famrel"
## [25] "freetime" "goout" "Dalc"
## [28] "Walc" "health" "absences"
## [31] "G1" "G2" "G3"Description of dataset and all variables: https://archive.ics.uci.edu/ml/datasets/Student+Performance.
All variables in formula (for easy copying and pasting):
(b <- paste(names(d), collapse="+"))## [1] "school+sex+age+address+famsize+Pstatus+Medu+Fedu+Mjob+Fjob+reason+guardian+traveltime+studytime+failures+schoolsup+famsup+paid+activities+nursery+higher+internet+romantic+famrel+freetime+goout+Dalc+Walc+health+absences+G1+G2+G3"Step 4: Calculate number of observations
nrow(d)## [1] 649Step 5: Generate binary version of dependent variable, G3 (final grade, 0 to 20).
d$passed <- ifelse(d$G3 > 9.99, 1, 0)We’re assuming that a score of over 9.99 is a passing score, and below that is failing.
Step 6: Descriptive statistics for G3 continuous numeric variable.
summary(d$G3)## Min. 1st Qu. Median Mean 3rd Qu.
## 0.00 10.00 12.00 11.91 14.00
## Max.
## 19.00sd(d$G3)## [1] 3.230656Step 7: Who all passed and failed, binary qualitative categorical variable.
with(d, table(passed, useNA = "always"))## passed
## 0 1 <NA>
## 100 549 0Step 8: Histogram
hist(as.numeric(d$G3))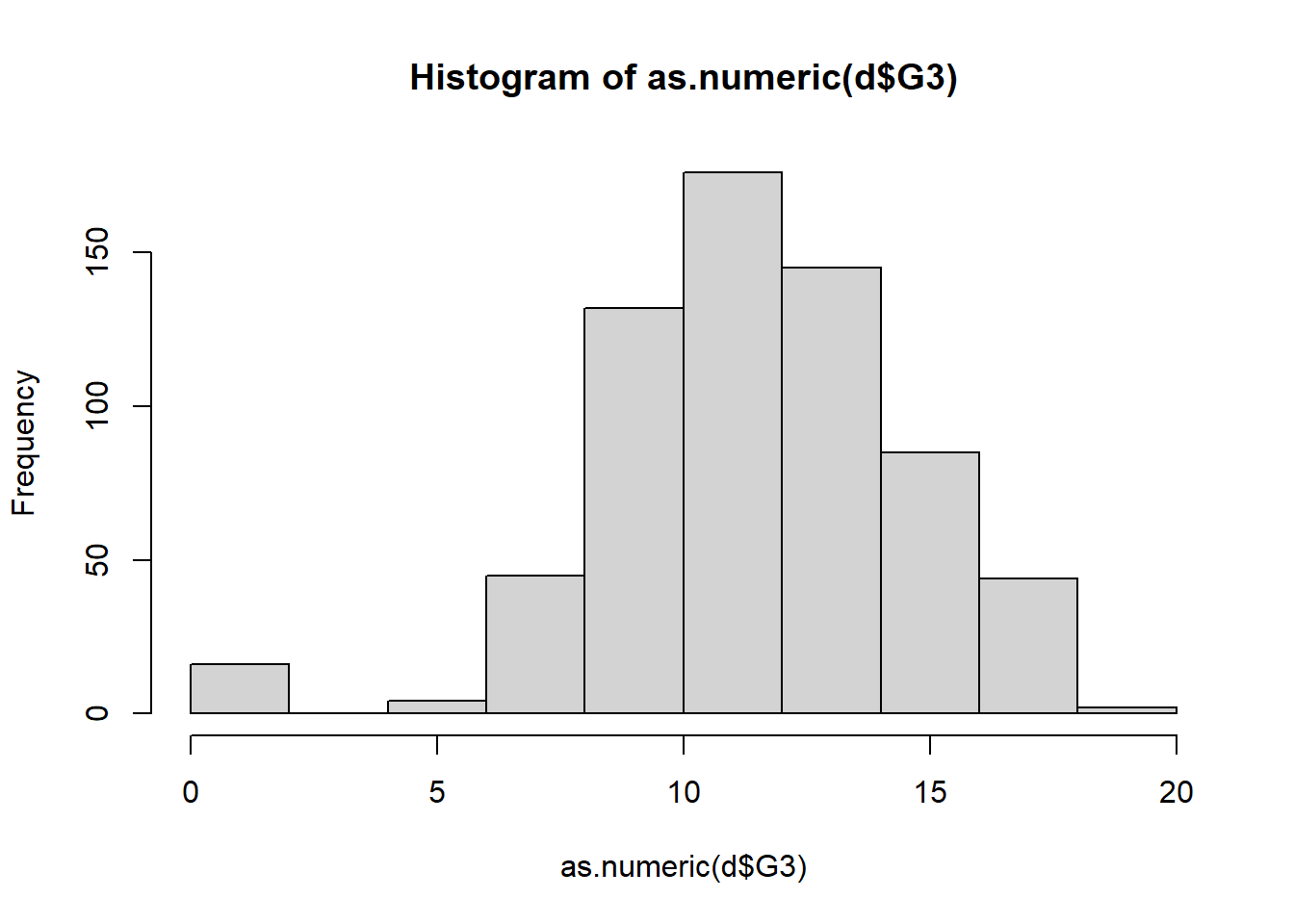
Step 9: Scatterplots
plot(d$G1 , d$G3) # MODIFY which variables you plot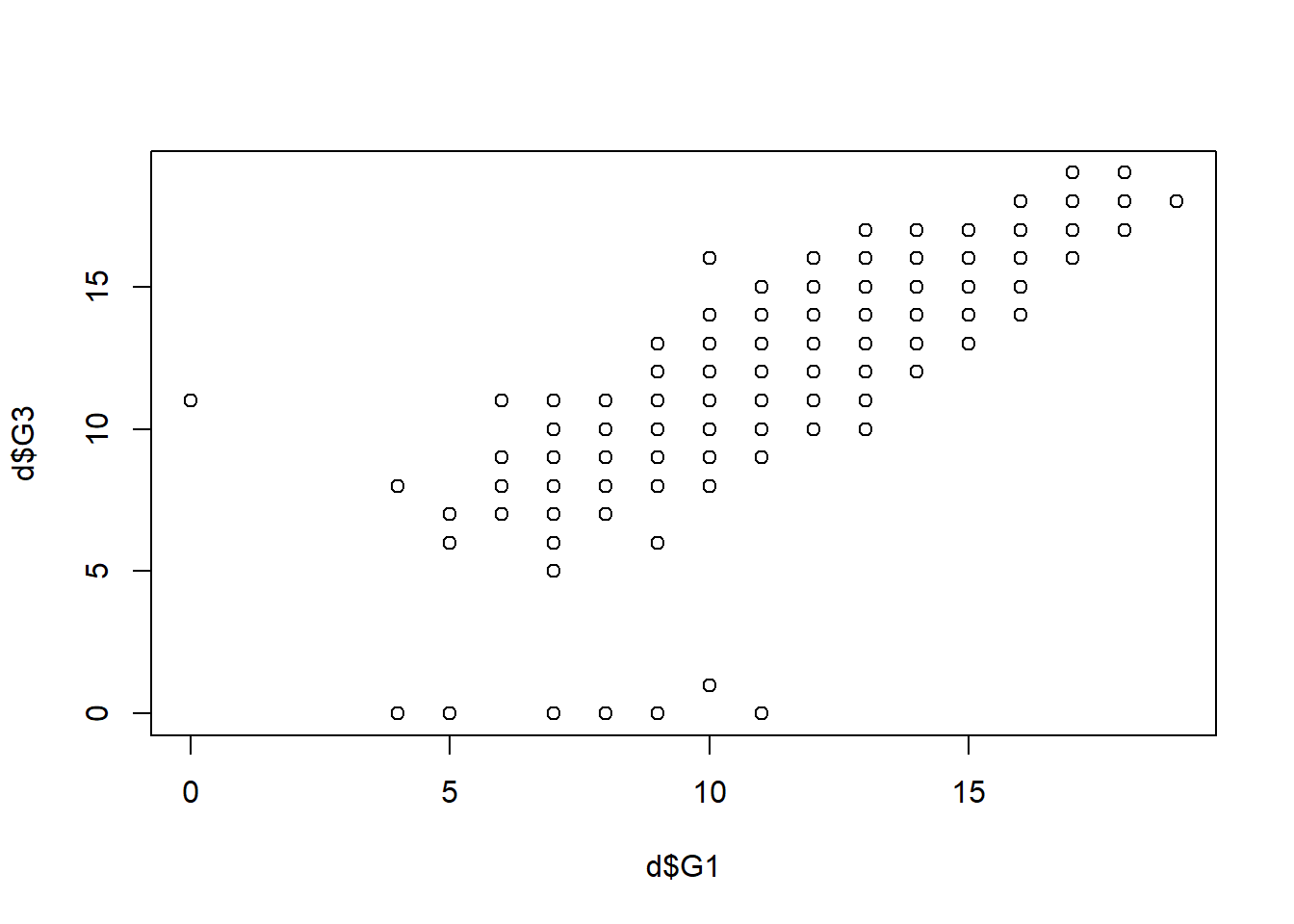
plot(d$health, d$G3)
Step 10: Selected correlations (optional)
chart.Correlation(d[c("G3","G1","Medu","failures")], histogram=TRUE, pch=19) ## Warning in par(usr): argument 1 does not name
## a graphical parameter
## Warning in par(usr): argument 1 does not name
## a graphical parameter
## Warning in par(usr): argument 1 does not name
## a graphical parameter
## Warning in par(usr): argument 1 does not name
## a graphical parameter
## Warning in par(usr): argument 1 does not name
## a graphical parameter
## Warning in par(usr): argument 1 does not name
## a graphical parameter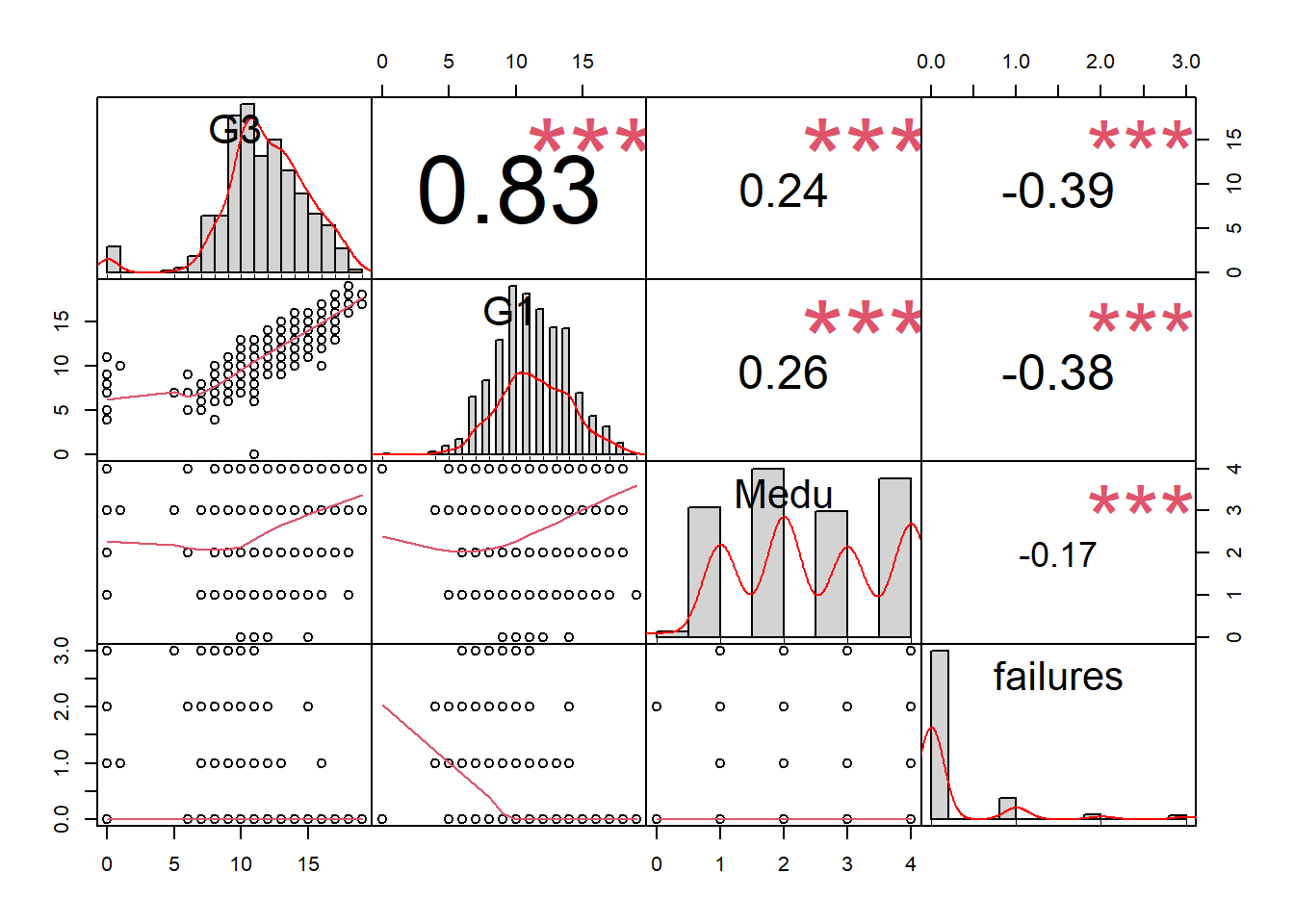
3.4.5 Divide training and testing data
Step 11: Divide data
trainingRowIndex <- sample(1:nrow(d), 0.75*nrow(d)) # row indices for training data
dtrain <- d[trainingRowIndex, ] # model training data
dtest <- d[-trainingRowIndex, ] # test data3.4.6 Decision tree model – regression tree
Activity summary:
- Goal: predict continuous outcome
G3, using a regression tree. - Start by using all variables to make decision tree. Check predictive capability.
- Remove variables to make predictions with less information.
- Modify cutoff thresholds and see how confusion matrix changes.
- Anywhere you see the word
MODIFYis one place where you might consider making changes to the code. - Figure out which students to remediate.
3.4.6.1 Train and inspect model
Step 14: Train a decision tree model
tree1 <- rpart(G3 ~ school+sex+age+address+famsize+Pstatus+Medu+Fedu+Mjob+Fjob+reason+guardian+traveltime+studytime+failures+schoolsup+famsup+paid+activities+nursery+higher+internet+romantic+famrel+freetime+goout+Dalc+Walc+health+absences+G1+G2, data=dtrain, method = 'anova')
# MODIFY. Complete the next step (visualize the tree) and
# then come back to this and run the model without G1 and
# G2. Then try other combinations.
summary(tree1)## Call:
## rpart(formula = G3 ~ school + sex + age + address + famsize +
## Pstatus + Medu + Fedu + Mjob + Fjob + reason + guardian +
## traveltime + studytime + failures + schoolsup + famsup +
## paid + activities + nursery + higher + internet + romantic +
## famrel + freetime + goout + Dalc + Walc + health + absences +
## G1 + G2, data = dtrain, method = "anova")
## n= 486
##
## CP nsplit rel error xerror
## 1 0.53749755 0 1.0000000 1.0096255
## 2 0.12066838 1 0.4625025 0.4691445
## 3 0.10749737 2 0.3418341 0.4436254
## 4 0.04493124 3 0.2343367 0.2603816
## 5 0.02353253 4 0.1894055 0.2157724
## 6 0.01000000 5 0.1658729 0.1917166
## xstd
## 1 0.09641264
## 2 0.06201578
## 3 0.05731408
## 4 0.04072088
## 5 0.03638119
## 6 0.03623172
##
## Variable importance
## G2 G1 failures Medu Fedu
## 44 28 7 7 6
## school age Walc
## 6 1 1
##
## Node number 1: 486 observations, complexity param=0.5374975
## mean=11.97119, MSE=9.669952
## left son=2 (249 obs) right son=3 (237 obs)
## Primary splits:
## G2 < 11.5 to the left, improve=0.53749750, (0 missing)
## G1 < 11.5 to the left, improve=0.46837770, (0 missing)
## failures < 0.5 to the right, improve=0.20487240, (0 missing)
## higher splits as LR, improve=0.15021990, (0 missing)
## school splits as RL, improve=0.06505104, (0 missing)
## Surrogate splits:
## G1 < 11.5 to the left, agree=0.889, adj=0.772, (0 split)
## failures < 0.5 to the right, agree=0.634, adj=0.249, (0 split)
## Medu < 3.5 to the left, agree=0.619, adj=0.219, (0 split)
## school splits as RL, agree=0.613, adj=0.207, (0 split)
## Fedu < 2.5 to the left, agree=0.603, adj=0.186, (0 split)
##
## Node number 2: 249 observations, complexity param=0.1206684
## mean=9.746988, MSE=5.450041
## left son=4 (11 obs) right son=5 (238 obs)
## Primary splits:
## G2 < 6.5 to the left, improve=0.41788320, (0 missing)
## G1 < 8.5 to the left, improve=0.28820220, (0 missing)
## failures < 0.5 to the right, improve=0.12327660, (0 missing)
## higher splits as LR, improve=0.08446519, (0 missing)
## school splits as RL, improve=0.05936389, (0 missing)
## Surrogate splits:
## G1 < 5.5 to the left, agree=0.964, adj=0.182, (0 split)
##
## Node number 3: 237 observations, complexity param=0.1074974
## mean=14.30802, MSE=3.44521
## left son=6 (123 obs) right son=7 (114 obs)
## Primary splits:
## G2 < 13.5 to the left, improve=0.61872030, (0 missing)
## G1 < 14.5 to the left, improve=0.45848660, (0 missing)
## age < 16.5 to the left, improve=0.08724415, (0 missing)
## schoolsup splits as RL, improve=0.05812433, (0 missing)
## Medu < 2.5 to the left, improve=0.03747041, (0 missing)
## Surrogate splits:
## G1 < 13.5 to the left, agree=0.848, adj=0.684, (0 split)
## age < 16.5 to the left, agree=0.599, adj=0.167, (0 split)
## Walc < 2.5 to the right, agree=0.586, adj=0.140, (0 split)
## Medu < 2.5 to the left, agree=0.578, adj=0.123, (0 split)
## Fedu < 3.5 to the left, agree=0.574, adj=0.114, (0 split)
##
## Node number 4: 11 observations
## mean=2.727273, MSE=13.10744
##
## Node number 5: 238 observations, complexity param=0.04493124
## mean=10.07143, MSE=2.713385
## left son=10 (95 obs) right son=11 (143 obs)
## Primary splits:
## G2 < 9.5 to the left, improve=0.32697950, (0 missing)
## G1 < 9.5 to the left, improve=0.24146310, (0 missing)
## failures < 0.5 to the right, improve=0.11576380, (0 missing)
## higher splits as LR, improve=0.08771469, (0 missing)
## goout < 4.5 to the right, improve=0.02768155, (0 missing)
## Surrogate splits:
## G1 < 8.5 to the left, agree=0.739, adj=0.347, (0 split)
## failures < 0.5 to the right, agree=0.676, adj=0.189, (0 split)
## higher splits as LR, agree=0.647, adj=0.116, (0 split)
## famrel < 1.5 to the left, agree=0.613, adj=0.032, (0 split)
## absences < 23 to the right, agree=0.609, adj=0.021, (0 split)
##
## Node number 6: 123 observations
## mean=12.90244, MSE=0.9173111
##
## Node number 7: 114 observations, complexity param=0.02353253
## mean=15.82456, MSE=1.741151
## left son=14 (87 obs) right son=15 (27 obs)
## Primary splits:
## G2 < 16.5 to the left, improve=0.55717020, (0 missing)
## G1 < 15.5 to the left, improve=0.43875750, (0 missing)
## age < 16.5 to the left, improve=0.07292958, (0 missing)
## studytime < 3.5 to the left, improve=0.04473009, (0 missing)
## reason splits as LLLR, improve=0.03190737, (0 missing)
## Surrogate splits:
## G1 < 16.5 to the left, agree=0.904, adj=0.593, (0 split)
## studytime < 3.5 to the left, agree=0.772, adj=0.037, (0 split)
##
## Node number 10: 95 observations
## mean=8.915789, MSE=3.108698
##
## Node number 11: 143 observations
## mean=10.83916, MSE=0.9741308
##
## Node number 14: 87 observations
## mean=15.27586, MSE=0.8894174
##
## Node number 15: 27 observations
## mean=17.59259, MSE=0.38957483.4.6.2 Tree visualization
Step 15: Visualize decision tree model in two ways.
prp(tree1)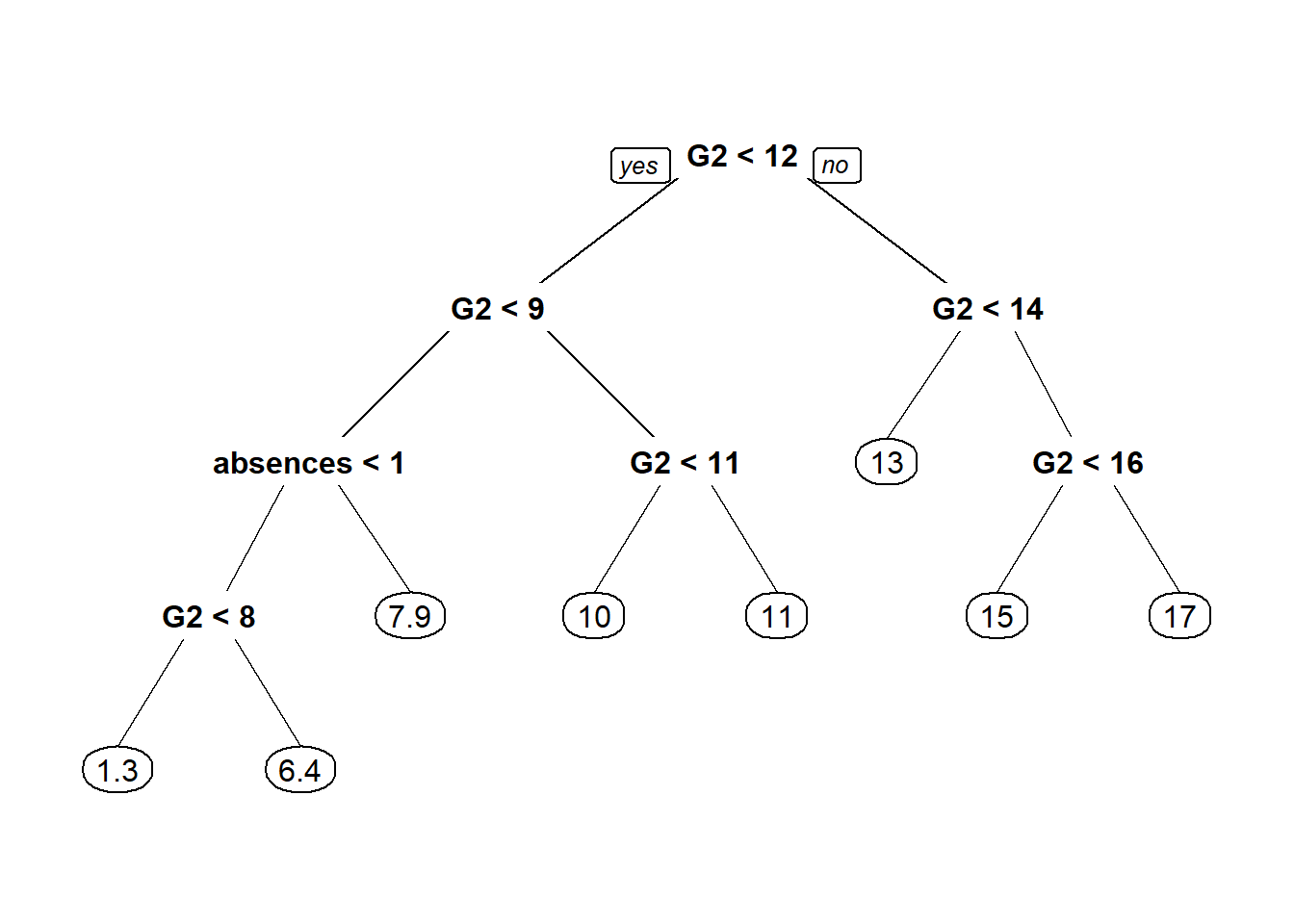
fancyRpartPlot(tree1, caption = "Regression Tree")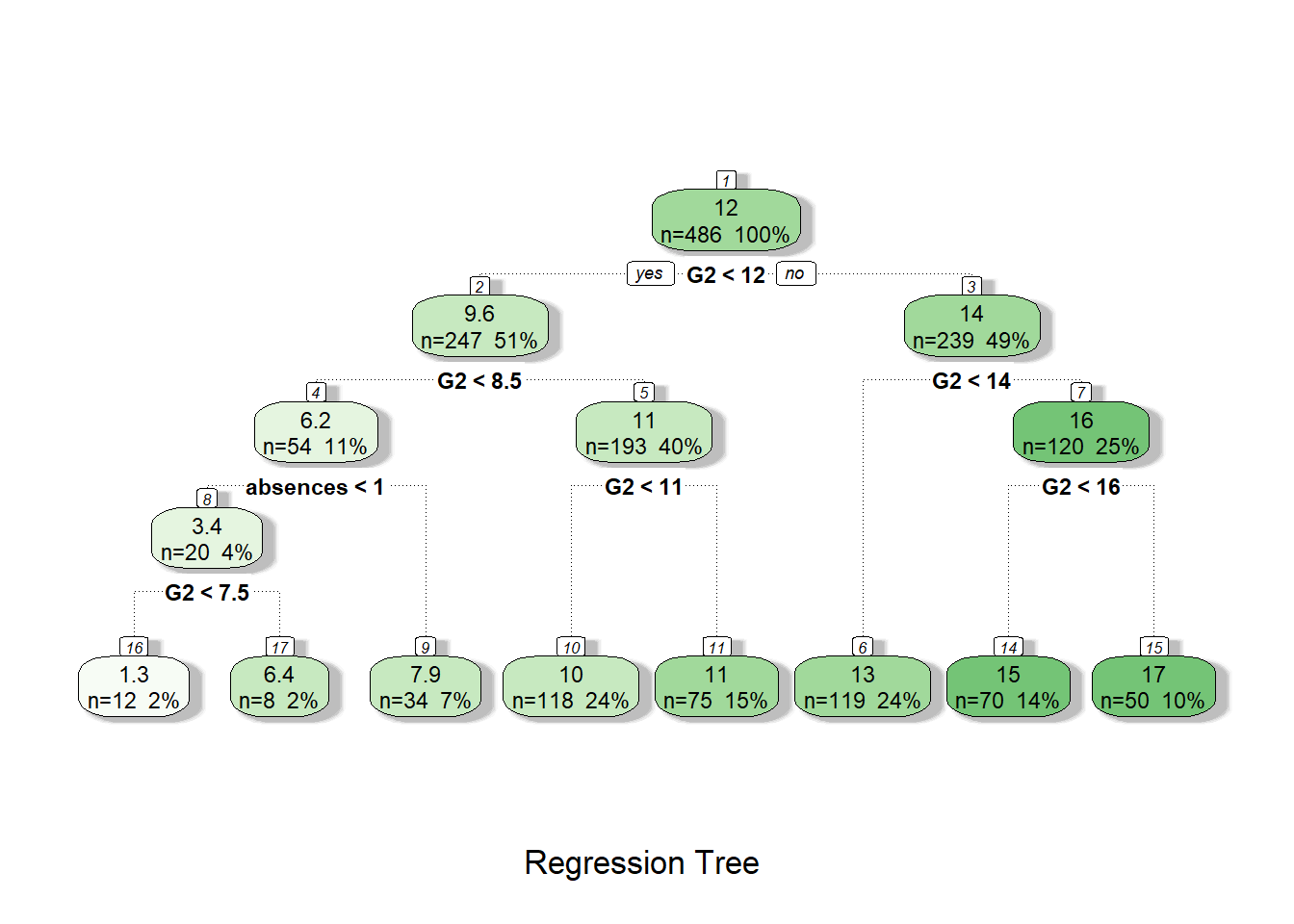
3.4.6.3 Test model
Step 16: Make predictions on testing data, using trained model
dtest$tree1.pred <- predict(tree1, newdata = dtest)Step 17: Visualize predictions
with(dtest, plot(G3,tree1.pred, main="Actual vs Predicted, testing data",xlab = "Actual G3",ylab = "Predicted G3"))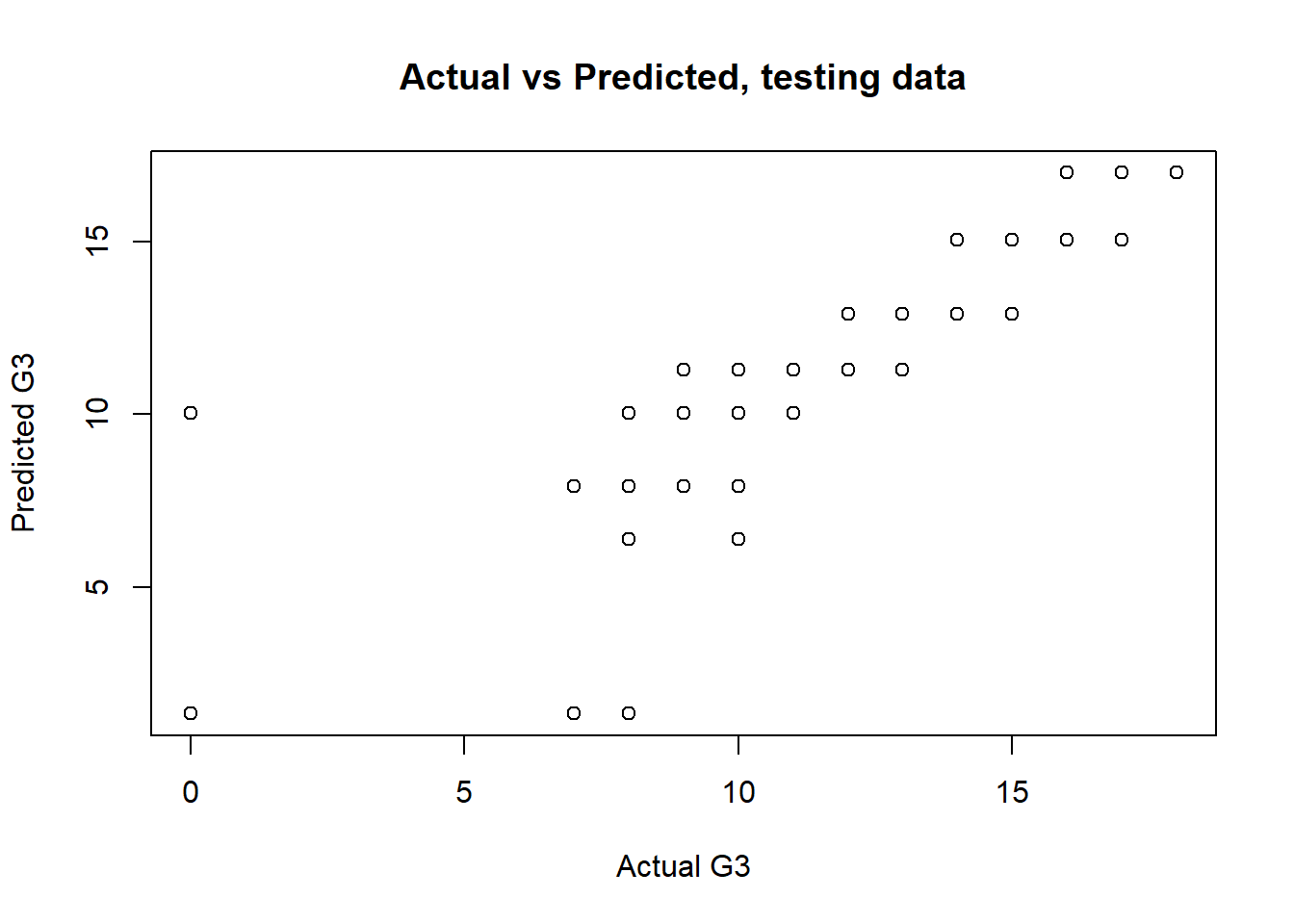
Step 18: Make confusion matrix.
PredictionCutoff <- 9.99 # MODIFY. Compare values in 9-11 range.
dtest$tree1.pred.passed <- ifelse(dtest$tree1.pred > PredictionCutoff, 1, 0)
(cm1 <- with(dtest,table(tree1.pred.passed,passed)))## passed
## tree1.pred.passed 0 1
## 0 26 13
## 1 2 122Step 19: Calculate accuracy
CorrectPredictions1 <- cm1[1,1] + cm1[2,2]
TotalStudents1 <- nrow(dtest)
(Accuracy1 <- CorrectPredictions1/TotalStudents1)## [1] 0.9079755Step 20: Sensitivity (proportion of people who actually failed that were correctly predicted to fail).
(Sensitivity1 <- cm1[1,1]/(cm1[1,1]+cm1[2,1]))## [1] 0.9285714Step 21: Specificity (proportion of people who actually passed that were correctly predicted to pass).
(Specificity1 <- cm1[2,2]/(cm1[1,2]+cm1[2,2]))## [1] 0.9037037BE SURE TO DOUBLE-CHECK THE CALCULATIONS ABOVE MANUALLY!
Step 22: It is very important for you, the data analyst, to modify the 9.99 cutoff assigned as PredictionCutoff above to see how you can change the predictions made by the model. Write down what you observe as you change this value and re-run the confusion matrix, accuracy, sensitivity, and specificity code above. What are the implications of your manual modification of this cutoff? Remind your instructors to discuss this, in case they forget!
3.4.7 Decision tree model – classification tree
Activity summary:
- Goal: predict binary outcome
passed, using a classification tree. - Start by using all variables to make decision tree. Check predictive capability.
- Remove variables to make predictions with less information.
- Modify cutoff thresholds and see how confusion matrix changes.
- Anywhere you see the word
MODIFYis one place where you might consider making changes to the code. - Figure out which students to remediate.
3.4.7.1 Train and inspect model
Step 23: Train a decision tree model
tree2 <- rpart(passed ~ school+sex+age+address+famsize+Pstatus+Medu+Fedu+Mjob+Fjob+reason+guardian+traveltime+studytime+failures+schoolsup+famsup+paid+activities+nursery+higher+internet+romantic+famrel+freetime+goout+Dalc+Walc+health+absences+G1+G2, data=dtrain, method = "class")
# MODIFY. Try without G1 and G2. Then try other combinations.
summary(tree2)## Call:
## rpart(formula = passed ~ school + sex + age + address + famsize +
## Pstatus + Medu + Fedu + Mjob + Fjob + reason + guardian +
## traveltime + studytime + failures + schoolsup + famsup +
## paid + activities + nursery + higher + internet + romantic +
## famrel + freetime + goout + Dalc + Walc + health + absences +
## G1 + G2, data = dtrain, method = "class")
## n= 486
##
## CP nsplit rel error xerror
## 1 0.54166667 0 1.0000000 1.0000000
## 2 0.02777778 1 0.4583333 0.4583333
## 3 0.01000000 5 0.3472222 0.6111111
## xstd
## 1 0.10877167
## 2 0.07702921
## 3 0.08785912
##
## Variable importance
## G2 G1 age Mjob Medu
## 57 24 3 3 3
## absences Fedu failures famsup higher
## 2 2 1 1 1
## Walc health
## 1 1
##
## Node number 1: 486 observations, complexity param=0.5416667
## predicted class=1 expected loss=0.1481481 P(node) =1
## class counts: 72 414
## probabilities: 0.148 0.852
## left son=2 (53 obs) right son=3 (433 obs)
## Primary splits:
## G2 < 8.5 to the left, improve=61.638130, (0 missing)
## G1 < 8.5 to the left, improve=54.059570, (0 missing)
## failures < 0.5 to the right, improve=18.666410, (0 missing)
## higher splits as LR, improve=14.310850, (0 missing)
## school splits as RL, improve= 9.271178, (0 missing)
## Surrogate splits:
## G1 < 7.5 to the left, agree=0.918, adj=0.245, (0 split)
## absences < 23 to the right, agree=0.893, adj=0.019, (0 split)
##
## Node number 2: 53 observations
## predicted class=0 expected loss=0.1320755 P(node) =0.1090535
## class counts: 46 7
## probabilities: 0.868 0.132
##
## Node number 3: 433 observations, complexity param=0.02777778
## predicted class=1 expected loss=0.06004619 P(node) =0.8909465
## class counts: 26 407
## probabilities: 0.060 0.940
## left son=6 (64 obs) right son=7 (369 obs)
## Primary splits:
## G1 < 9.5 to the left, improve=9.572720, (0 missing)
## G2 < 9.5 to the left, improve=8.209574, (0 missing)
## failures < 2.5 to the right, improve=3.155245, (0 missing)
## school splits as RL, improve=2.007149, (0 missing)
## higher splits as LR, improve=1.886526, (0 missing)
## Surrogate splits:
## failures < 0.5 to the right, agree=0.871, adj=0.125, (0 split)
## higher splits as LR, agree=0.866, adj=0.094, (0 split)
## G2 < 9.5 to the left, agree=0.859, adj=0.047, (0 split)
## age < 19.5 to the right, agree=0.855, adj=0.016, (0 split)
##
## Node number 6: 64 observations, complexity param=0.02777778
## predicted class=1 expected loss=0.3125 P(node) =0.1316872
## class counts: 20 44
## probabilities: 0.312 0.688
## left son=12 (8 obs) right son=13 (56 obs)
## Primary splits:
## age < 15.5 to the left, improve=3.500000, (0 missing)
## Mjob splits as RLRLL, improve=2.786449, (0 missing)
## G2 < 9.5 to the left, improve=2.293651, (0 missing)
## Medu < 2.5 to the right, improve=1.674972, (0 missing)
## failures < 0.5 to the left, improve=1.265182, (0 missing)
##
## Node number 7: 369 observations
## predicted class=1 expected loss=0.01626016 P(node) =0.7592593
## class counts: 6 363
## probabilities: 0.016 0.984
##
## Node number 12: 8 observations
## predicted class=0 expected loss=0.25 P(node) =0.01646091
## class counts: 6 2
## probabilities: 0.750 0.250
##
## Node number 13: 56 observations, complexity param=0.02777778
## predicted class=1 expected loss=0.25 P(node) =0.1152263
## class counts: 14 42
## probabilities: 0.250 0.750
## left son=26 (29 obs) right son=27 (27 obs)
## Primary splits:
## Mjob splits as LLRLL, improve=2.011494, (0 missing)
## Medu < 2.5 to the right, improve=1.682788, (0 missing)
## G2 < 9.5 to the left, improve=1.558630, (0 missing)
## G1 < 8.5 to the left, improve=1.285714, (0 missing)
## school splits as RL, improve=1.204836, (0 missing)
## Surrogate splits:
## Walc < 3.5 to the left, agree=0.696, adj=0.370, (0 split)
## health < 3.5 to the left, agree=0.661, adj=0.296, (0 split)
## Fjob splits as RRRLL, agree=0.643, adj=0.259, (0 split)
## Fedu < 2.5 to the right, agree=0.625, adj=0.222, (0 split)
## reason splits as LRLL, agree=0.625, adj=0.222, (0 split)
##
## Node number 26: 29 observations, complexity param=0.02777778
## predicted class=1 expected loss=0.3793103 P(node) =0.05967078
## class counts: 11 18
## probabilities: 0.379 0.621
## left son=52 (12 obs) right son=53 (17 obs)
## Primary splits:
## Medu < 2.5 to the right, improve=3.380663, (0 missing)
## G2 < 9.5 to the left, improve=3.139383, (0 missing)
## Walc < 2.5 to the right, improve=2.342041, (0 missing)
## school splits as RL, improve=1.877395, (0 missing)
## age < 17.5 to the left, improve=1.039788, (0 missing)
## Surrogate splits:
## Fedu < 2.5 to the right, agree=0.793, adj=0.500, (0 split)
## Mjob splits as RL-LL, agree=0.759, adj=0.417, (0 split)
## famsup splits as RL, agree=0.724, adj=0.333, (0 split)
## absences < 3.5 to the right, agree=0.724, adj=0.333, (0 split)
## G1 < 7.5 to the left, agree=0.724, adj=0.333, (0 split)
##
## Node number 27: 27 observations
## predicted class=1 expected loss=0.1111111 P(node) =0.05555556
## class counts: 3 24
## probabilities: 0.111 0.889
##
## Node number 52: 12 observations
## predicted class=0 expected loss=0.3333333 P(node) =0.02469136
## class counts: 8 4
## probabilities: 0.667 0.333
##
## Node number 53: 17 observations
## predicted class=1 expected loss=0.1764706 P(node) =0.03497942
## class counts: 3 14
## probabilities: 0.176 0.8243.4.7.2 Tree Visualization
Step 24: Visualize decision tree model in two ways
prp(tree2)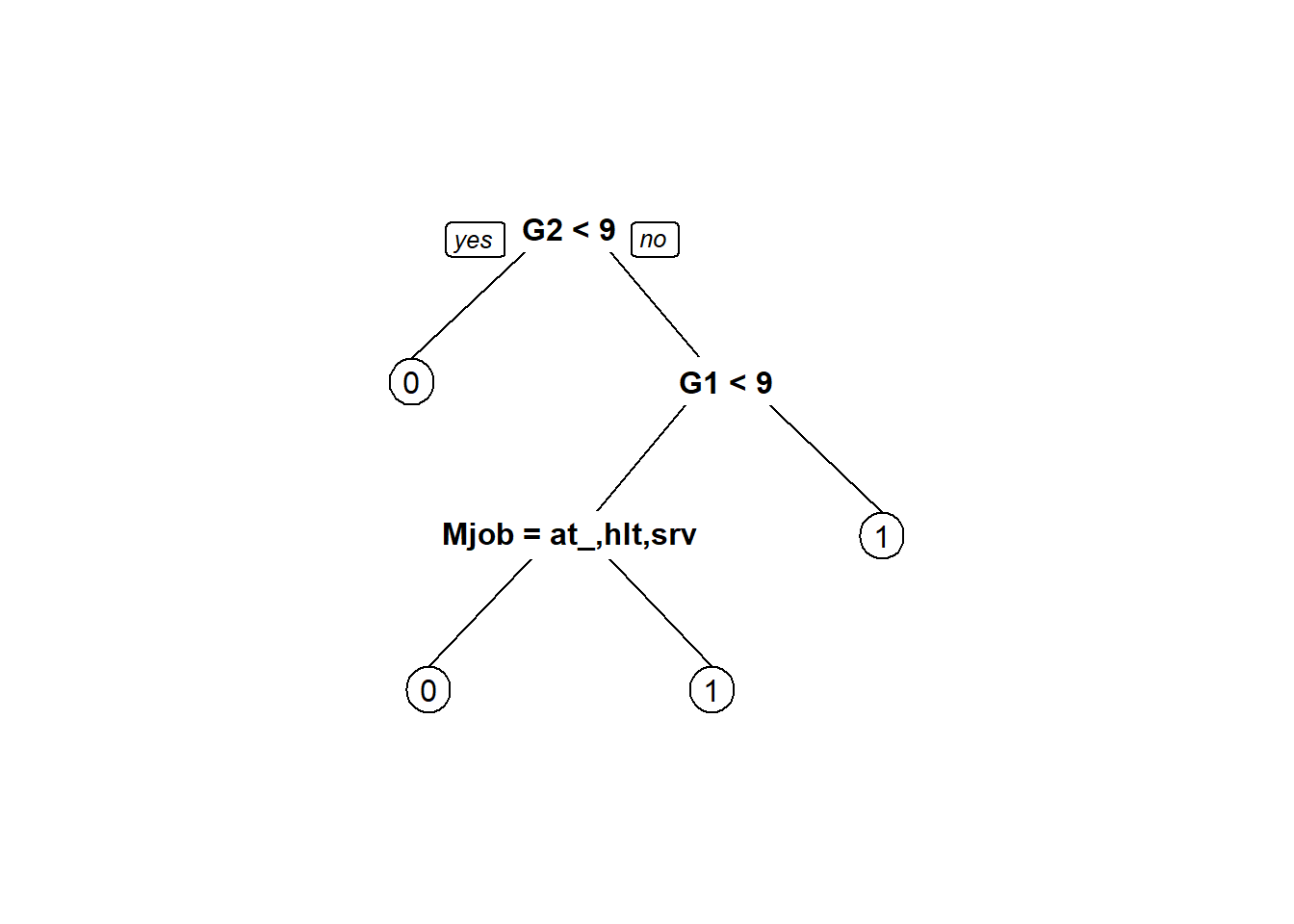
fancyRpartPlot(tree2, caption = "Classification Tree")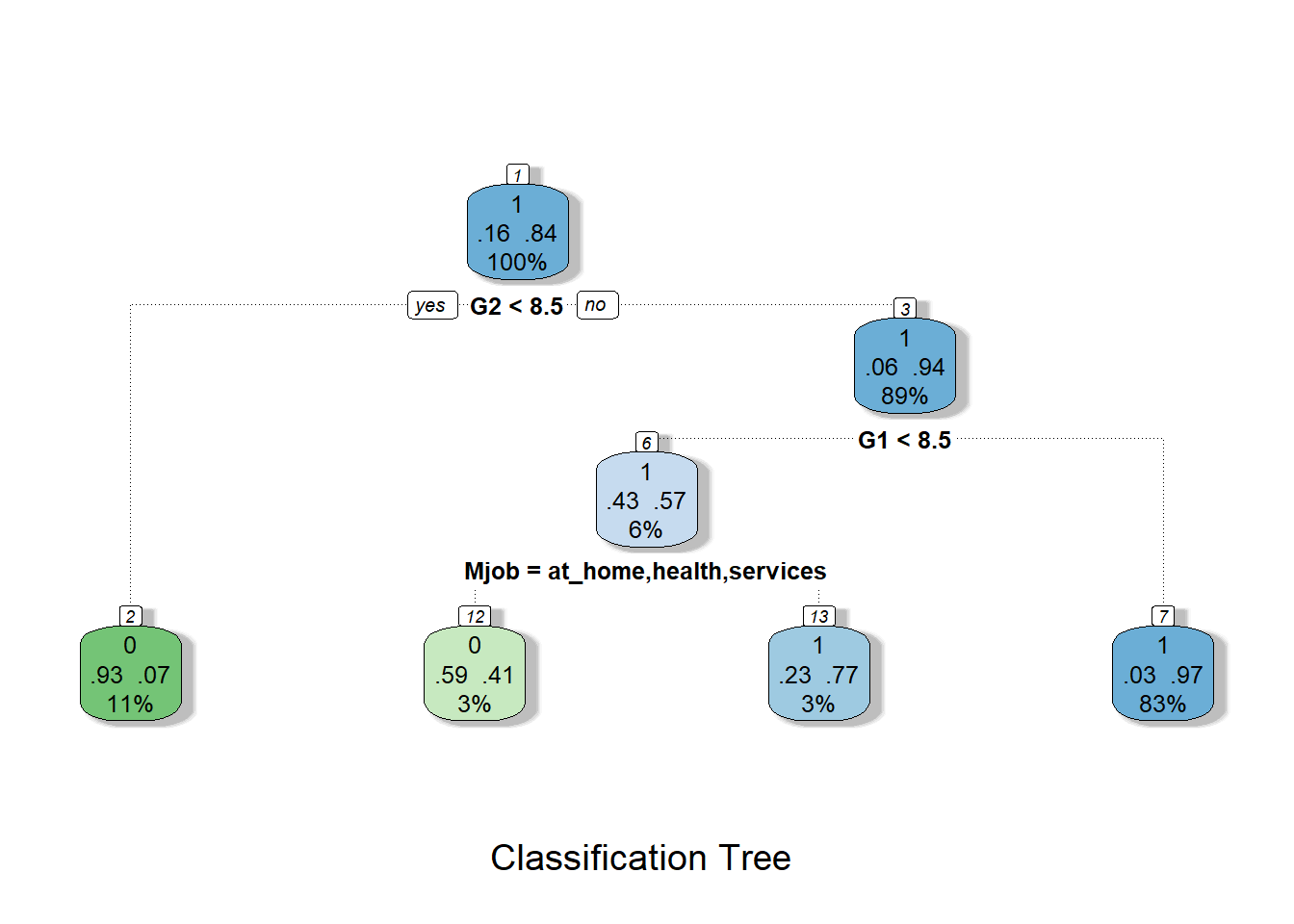
3.4.7.3 Test model
Step 25: Make predictions and confusion matrix on testing data classes, using trained model.
dtest$tree2.pred <- predict(tree2, newdata = dtest, type = 'class')
# MODIFY. change 'class' to 'prob'
(cm2 <- with(dtest,table(tree2.pred,passed)))## passed
## tree2.pred 0 1
## 0 22 7
## 1 6 128Step 26: Make predictions and confusion matrix on testing data using probability cutoffs. Optional; results not shown.
dtest$tree2.pred <- predict(tree2, newdata = dtest, type = 'prob')
ProbabilityCutoff <- 0.5 # MODIFY. Compare different probability values.
dtest$tree2.pred.probs <- 1-dtest$tree2.pred[,1]
dtest$tree2.pred.passed <- ifelse(dtest$tree2.pred.probs > ProbabilityCutoff, 1, 0)
(cm2b <- with(dtest,table(tree2.pred.passed,passed)))Step 27: Calculate accuracy
CorrectPredictions2 <- cm2[1,1] + cm2[2,2]
TotalStudents2 <- nrow(dtest)
(Accuracy2 <- CorrectPredictions2/TotalStudents2)## [1] 0.9202454Step 28: Sensitivity (proportion of people who actually failed that were correctly predicted to fail)
(Sensitivity2 <- cm2[1,1]/(cm2[1,1]+cm2[2,1]))## [1] 0.7857143Step 29: Specificity (proportion of people who actually passed that were correctly predicted to pass):
(Specificity2 <- cm2[2,2]/(cm2[1,2]+cm2[2,2]))## [1] 0.9481481ALSO DOUBLE-CHECK THE CALCULATIONS ABOVE MANUALLY!
3.4.8 Logistic regression model – classification
Activity summary:
- Goal: predict binary outcome
passed, using logistic regression. - Start by using all variables to make a logistic regression model. Check predictive capability.
- Remove variables to make predictions with less information.
- Modify cutoff thresholds and see how confusion matrix changes.
- Anywhere you see the word
MODIFYis one place where you might consider making changes to the code. - Figure out which students to remediate.
3.4.8.1 Train and inspect model
Step 30: Train a logistic regression model
blr1 <- glm(passed ~ school+sex+age+address+famsize+Pstatus+Medu+Fedu+guardian+traveltime+studytime+failures+schoolsup+famsup+paid+activities+nursery+higher+internet+romantic+famrel+freetime+goout+Dalc+Walc+health+absences+Mjob+reason+Fjob+G1+G2, data=dtrain, family = "binomial")
# MODIFY. Try without G1 and G2. Then try other combinations.
# also remove variables causing multicollinearity and see if it makes a difference!
summary(blr1)##
## Call:
## glm(formula = passed ~ school + sex + age + address + famsize +
## Pstatus + Medu + Fedu + guardian + traveltime + studytime +
## failures + schoolsup + famsup + paid + activities + nursery +
## higher + internet + romantic + famrel + freetime + goout +
## Dalc + Walc + health + absences + Mjob + reason + Fjob +
## G1 + G2, family = "binomial", data = dtrain)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -3.9535 0.0001 0.0060 0.0791 2.0724
##
## Coefficients:
## Estimate Std. Error
## (Intercept) -37.745302 9.282824
## schoolMS -2.026206 0.791563
## sexM -0.522535 0.859136
## age 0.861493 0.328548
## addressU -0.473080 0.732624
## famsizeLE3 0.413172 0.698261
## PstatusT -2.148203 1.427264
## Medu -0.102359 0.369692
## Fedu 0.194436 0.345439
## guardianmother -0.204879 0.823533
## guardianother 0.080149 1.557311
## traveltime 0.493957 0.402293
## studytime -0.039712 0.449457
## failures -0.045128 0.354732
## schoolsupyes 0.202339 0.983844
## famsupyes 1.018387 0.733199
## paidyes -0.863547 1.314252
## activitiesyes -0.464616 0.664420
## nurseryyes -0.430523 0.704578
## higheryes 1.745758 0.808090
## internetyes 0.240334 0.783576
## romanticyes -1.224801 0.714963
## famrel 0.088627 0.308760
## freetime 0.281816 0.294944
## goout 0.133326 0.315688
## Dalc 0.068650 0.396618
## Walc -0.263117 0.382453
## health 0.125104 0.247936
## absences -0.009120 0.074823
## Mjobhealth -0.145525 1.287657
## Mjobother 1.116503 0.855249
## Mjobservices -1.167946 1.094570
## Mjobteacher 0.008652 1.709678
## reasonhome -0.578204 0.946013
## reasonother 0.334645 0.996484
## reasonreputation 0.325221 1.031172
## Fjobhealth -4.476375 1.991217
## Fjobother -1.553272 1.244335
## Fjobservices -0.525646 1.310576
## Fjobteacher -2.679792 2.182808
## G1 0.808814 0.250675
## G2 1.976156 0.396360
## z value Pr(>|z|)
## (Intercept) -4.066 4.78e-05 ***
## schoolMS -2.560 0.01047 *
## sexM -0.608 0.54305
## age 2.622 0.00874 **
## addressU -0.646 0.51845
## famsizeLE3 0.592 0.55404
## PstatusT -1.505 0.13229
## Medu -0.277 0.78188
## Fedu 0.563 0.57353
## guardianmother -0.249 0.80353
## guardianother 0.051 0.95895
## traveltime 1.228 0.21950
## studytime -0.088 0.92959
## failures -0.127 0.89877
## schoolsupyes 0.206 0.83706
## famsupyes 1.389 0.16484
## paidyes -0.657 0.51114
## activitiesyes -0.699 0.48438
## nurseryyes -0.611 0.54117
## higheryes 2.160 0.03075 *
## internetyes 0.307 0.75906
## romanticyes -1.713 0.08669 .
## famrel 0.287 0.77408
## freetime 0.955 0.33933
## goout 0.422 0.67278
## Dalc 0.173 0.86258
## Walc -0.688 0.49147
## health 0.505 0.61385
## absences -0.122 0.90298
## Mjobhealth -0.113 0.91002
## Mjobother 1.305 0.19173
## Mjobservices -1.067 0.28596
## Mjobteacher 0.005 0.99596
## reasonhome -0.611 0.54107
## reasonother 0.336 0.73700
## reasonreputation 0.315 0.75247
## Fjobhealth -2.248 0.02457 *
## Fjobother -1.248 0.21193
## Fjobservices -0.401 0.68836
## Fjobteacher -1.228 0.21957
## G1 3.227 0.00125 **
## G2 4.986 6.17e-07 ***
## ---
## Signif. codes:
## 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 407.74 on 485 degrees of freedom
## Residual deviance: 107.12 on 444 degrees of freedom
## AIC: 191.12
##
## Number of Fisher Scoring iterations: 9car::vif(blr1)## GVIF Df GVIF^(1/(2*Df))
## school 2.535976 1 1.592475
## sex 3.052960 1 1.747272
## age 2.888823 1 1.699654
## address 2.000034 1 1.414226
## famsize 1.726103 1 1.313812
## Pstatus 2.228071 1 1.492672
## Medu 2.580093 1 1.606267
## Fedu 2.005726 1 1.416237
## guardian 3.150133 2 1.332239
## traveltime 1.876912 1 1.370004
## studytime 2.025777 1 1.423298
## failures 1.917451 1 1.384720
## schoolsup 1.977969 1 1.406403
## famsup 2.207537 1 1.485778
## paid 1.304981 1 1.142358
## activities 1.770642 1 1.330655
## nursery 1.471985 1 1.213254
## higher 1.969425 1 1.403362
## internet 2.379760 1 1.542647
## romantic 2.046356 1 1.430509
## famrel 1.997711 1 1.413404
## freetime 1.848877 1 1.359734
## goout 2.680688 1 1.637281
## Dalc 2.736602 1 1.654268
## Walc 4.392557 1 2.095843
## health 1.945098 1 1.394668
## absences 2.065073 1 1.437036
## Mjob 16.072036 4 1.415008
## reason 7.325690 3 1.393611
## Fjob 13.801042 4 1.388318
## G1 2.482536 1 1.575606
## G2 2.272489 1 1.5074783.4.8.2 Test model
Step 31: Make predictions on testing data, using trained model.
Predicting probabilities…
dtest$blr1.pred <- predict(blr1, newdata = dtest, type = 'response')
# MODIFY. change 'class' to 'prob'
ProbabilityCutoff <- 0.5 # MODIFY. Compare different probability values.
dtest$blr1.pred.probs <- 1-dtest$blr1.pred
dtest$blr1.pred.passed <- ifelse(dtest$blr1.pred > ProbabilityCutoff, 1, 0)
(cm3 <- with(dtest,table(blr1.pred.passed,passed)))## passed
## blr1.pred.passed 0 1
## 0 21 4
## 1 7 131Step 32: Make confusion matrix
(cm3 <- with(dtest,table(blr1.pred.passed,passed)))## passed
## blr1.pred.passed 0 1
## 0 21 4
## 1 7 131Step 33: Calculate accuracy
CorrectPredictions3 <- cm3[1,1] + cm3[2,2]
TotalStudents3 <- nrow(dtest)
(Accuracy3 <- CorrectPredictions3/TotalStudents3)## [1] 0.9325153Step 34: Sensitivity (proportion of people who actually failed that were correctly predicted to fail)
(Sensitivity3 <- cm3[1,1]/(cm3[1,1]+cm3[2,1]))## [1] 0.75Step 35: Specificity (proportion of people who actually passed that were correctly predicted to pass)
(Specificity3 <- cm3[2,2]/(cm3[1,2]+cm3[2,2]))## [1] 0.9703704ALSO DOUBLE-CHECK THE CALCULATIONS ABOVE MANUALLY!
Step 36: It is very important for you, the data analyst, to modify the 0.5 cutoff assigned as ProbabilityCutoff above to see how you can change the predictions made by the model. Write down what you observe as you change this value and re-run the confusion matrix, accuracy, sensitivity, and specificity code above. What are the implications of your manual modification of this cutoff? Remind your instructors to discuss this, in case they forget!