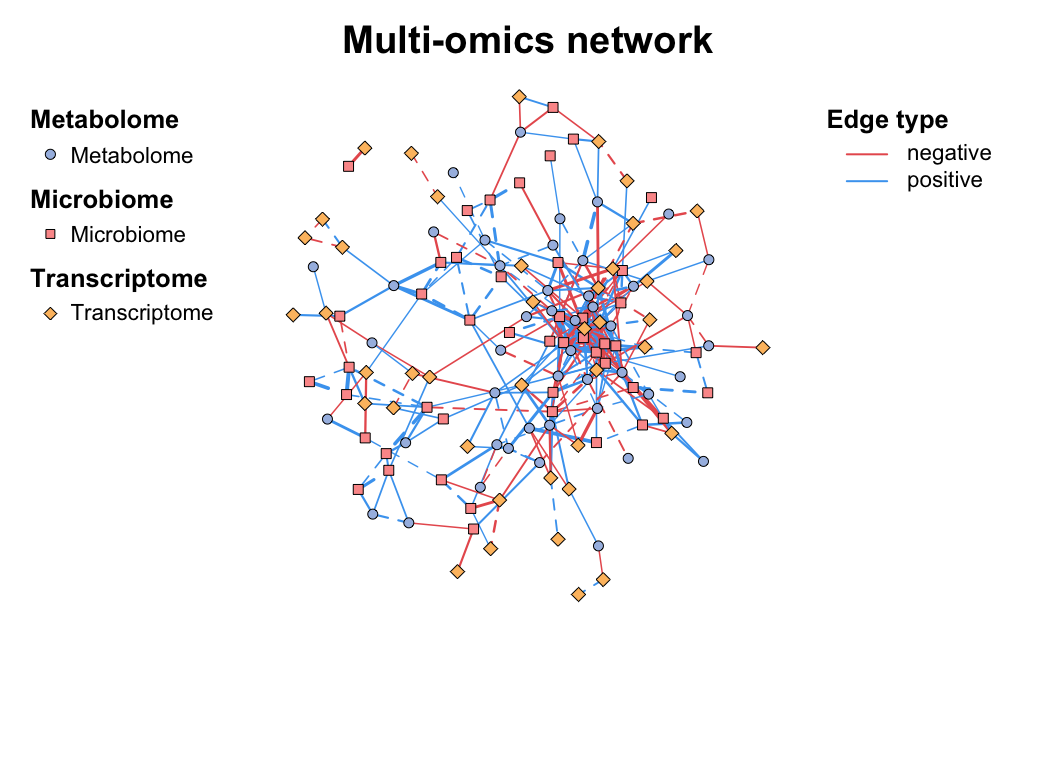
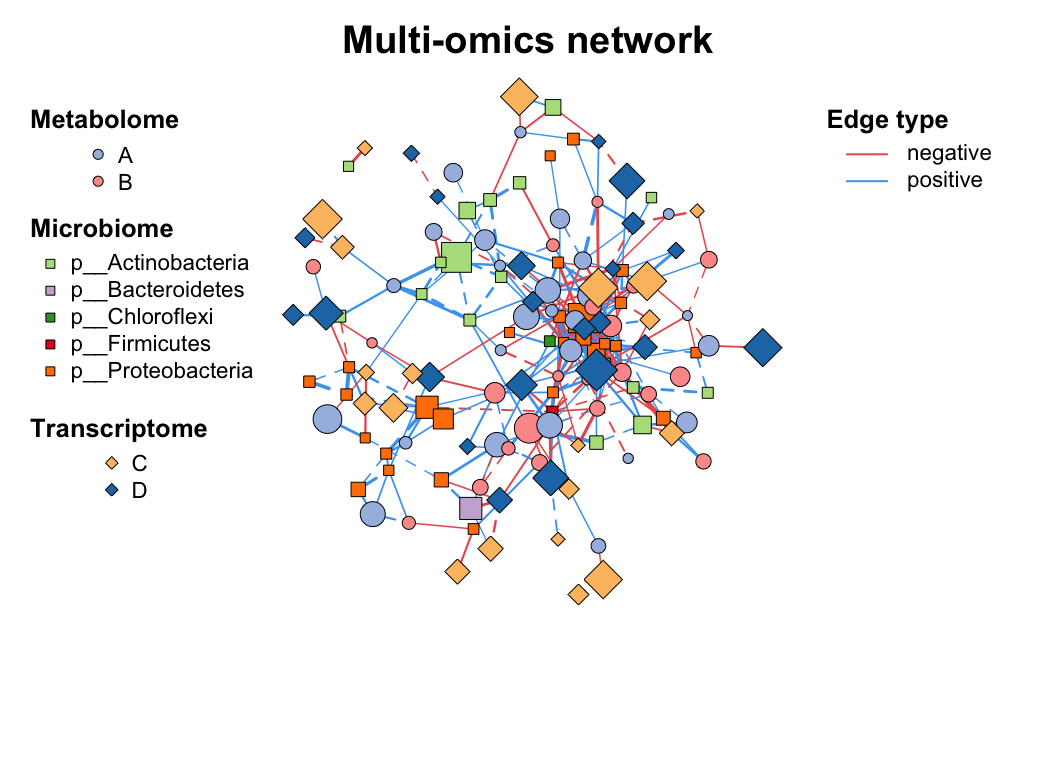
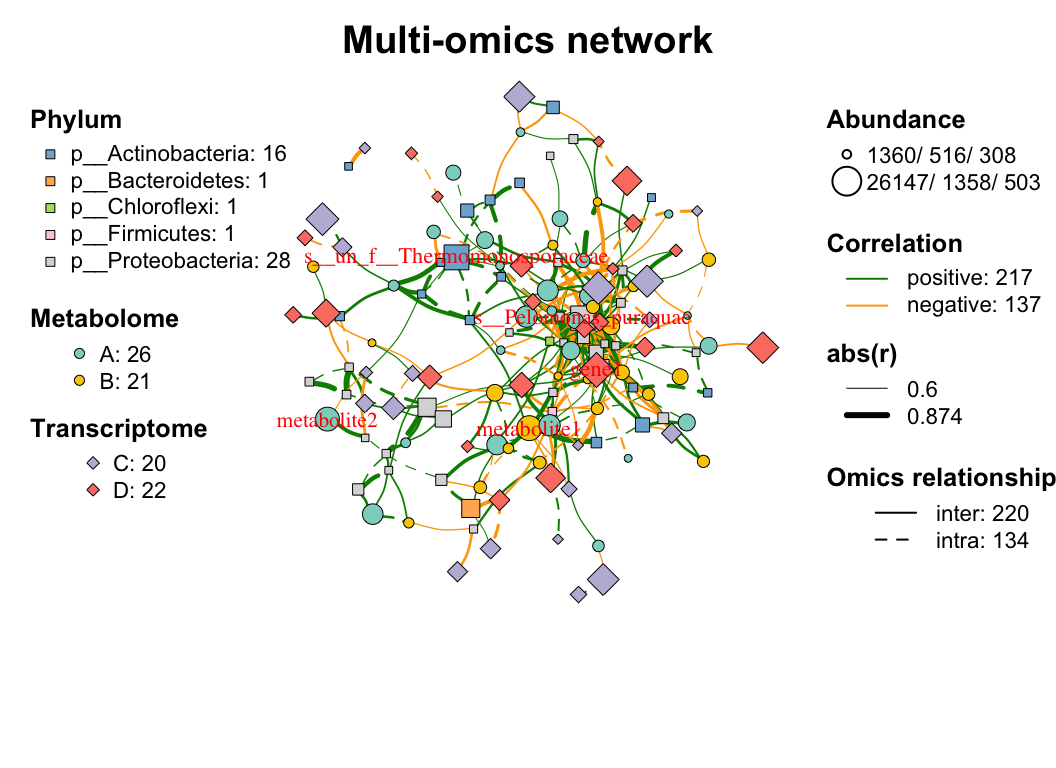
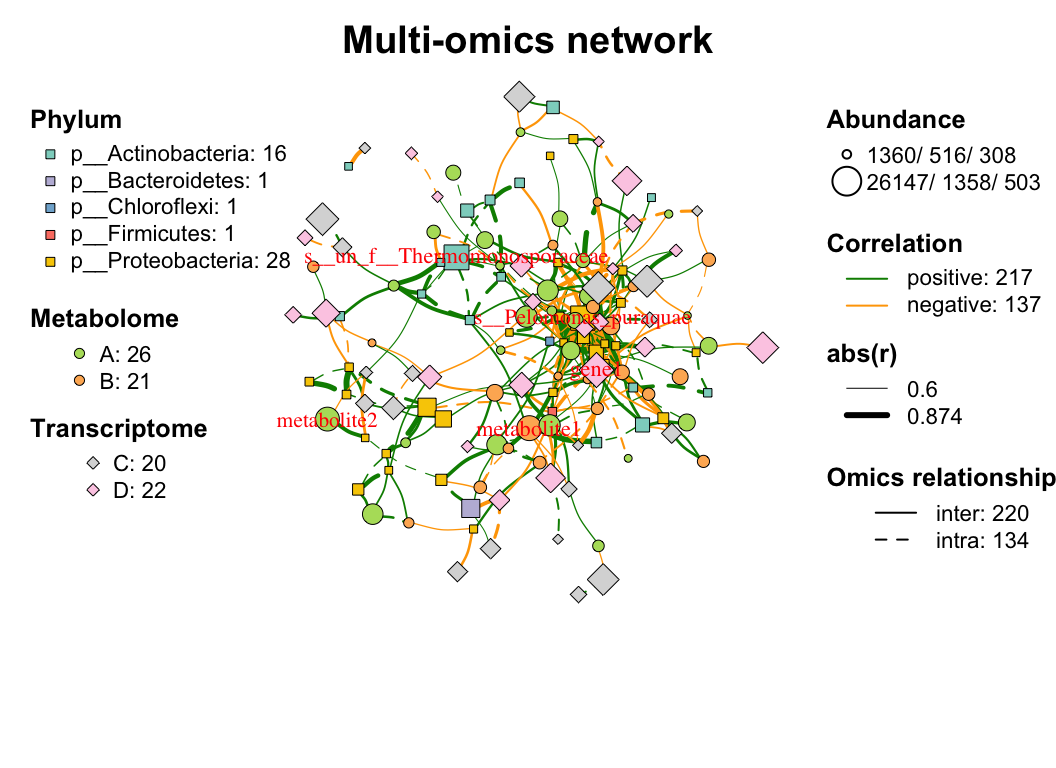
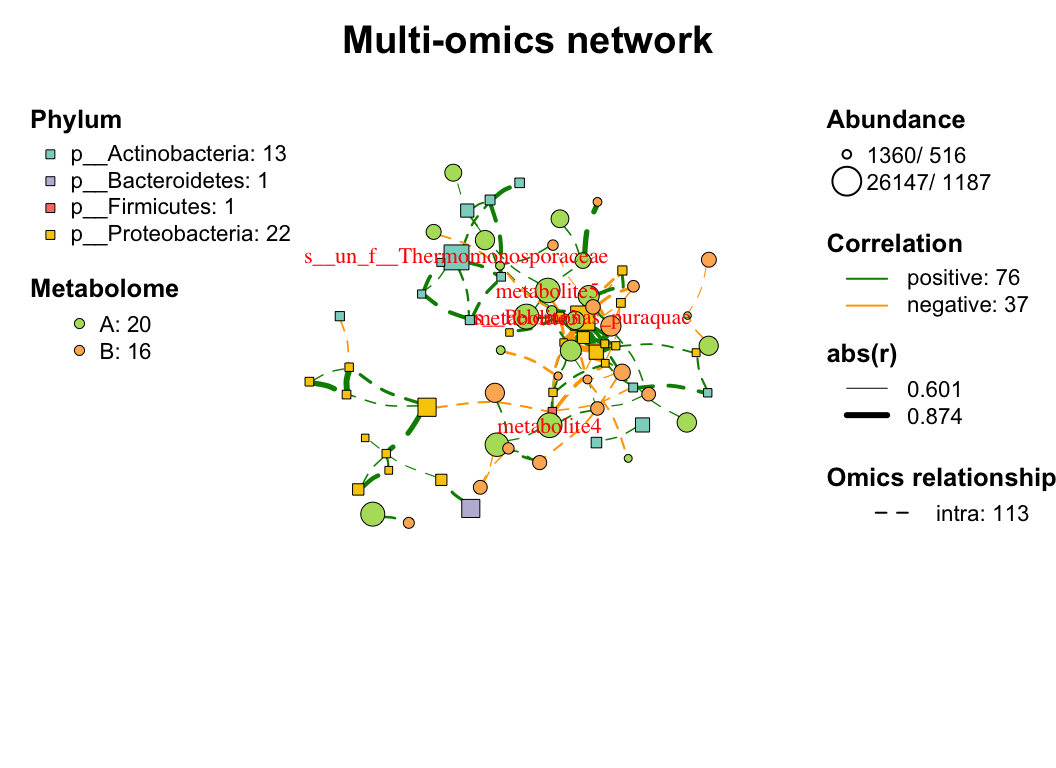
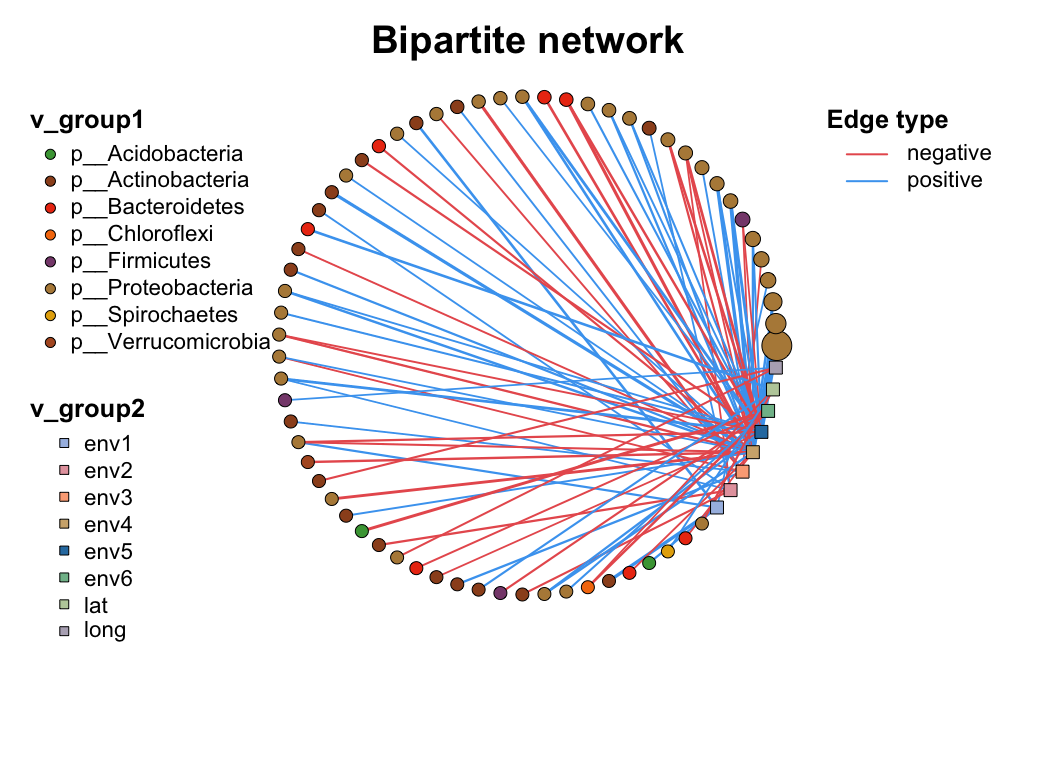
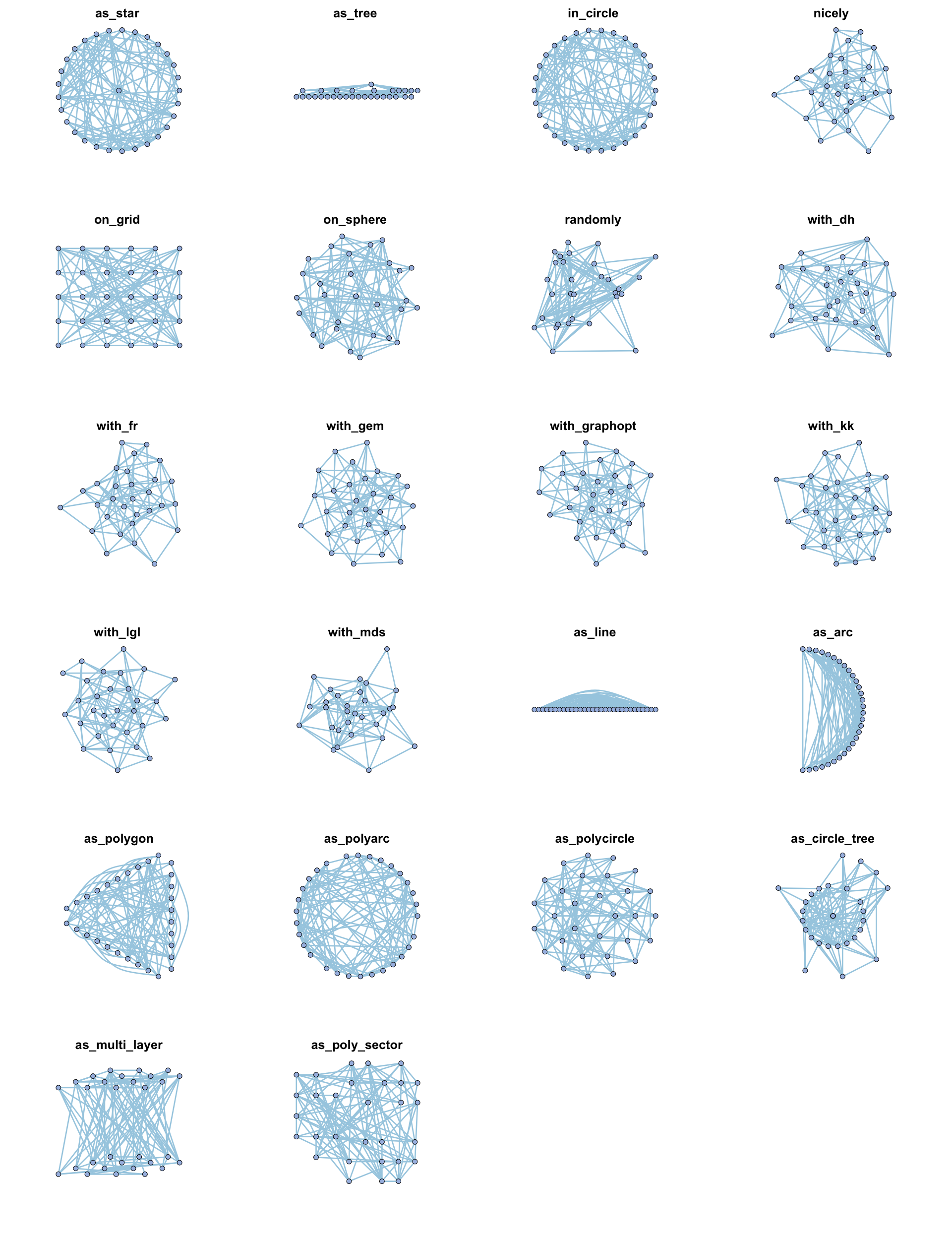
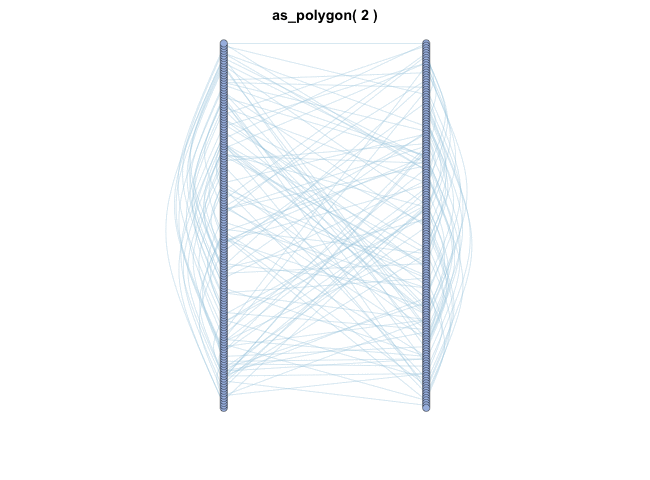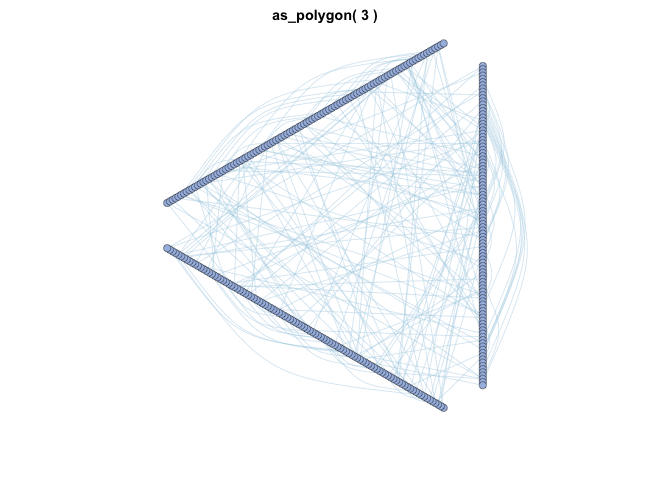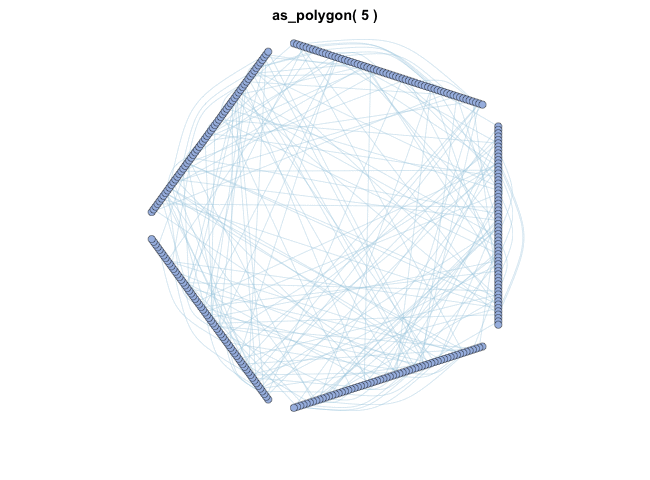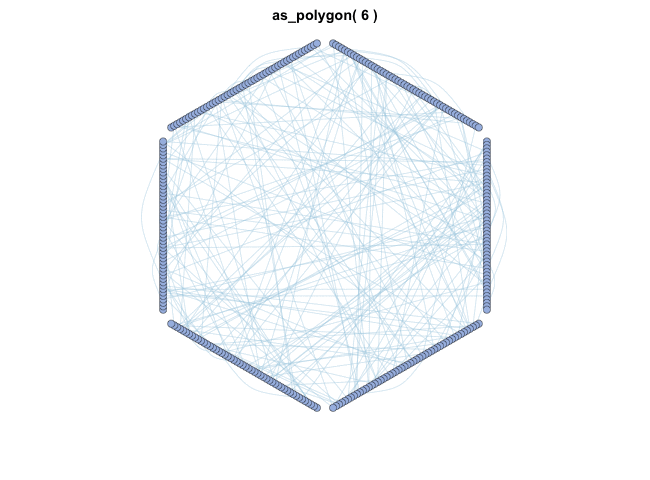
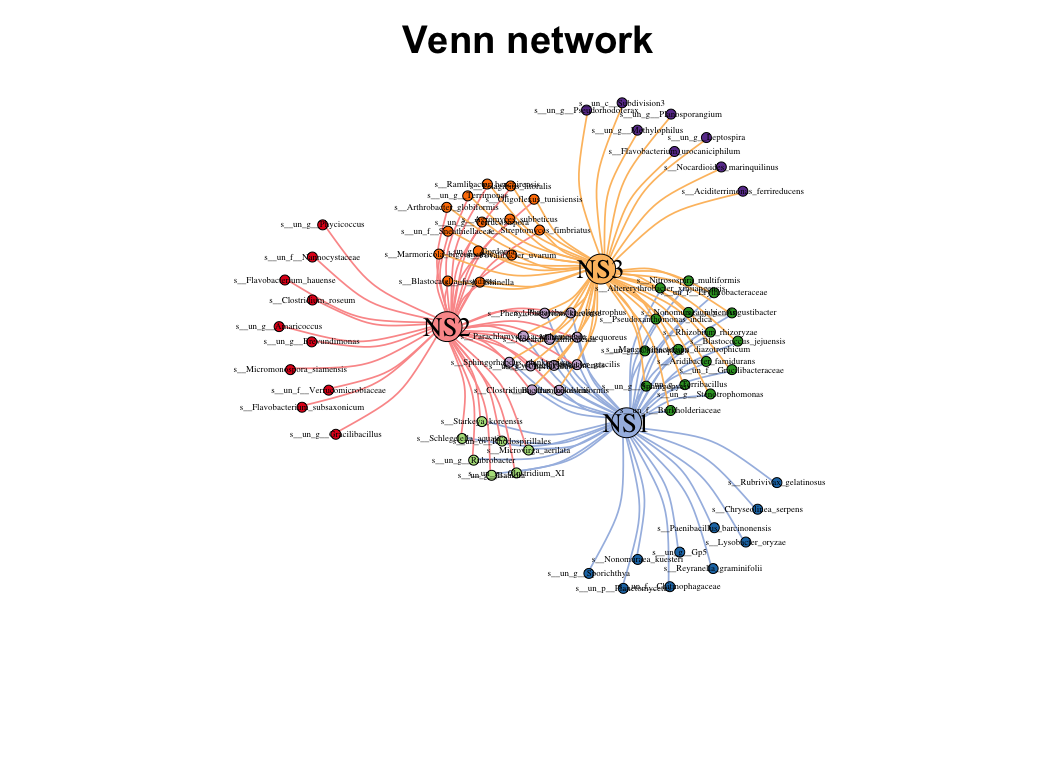
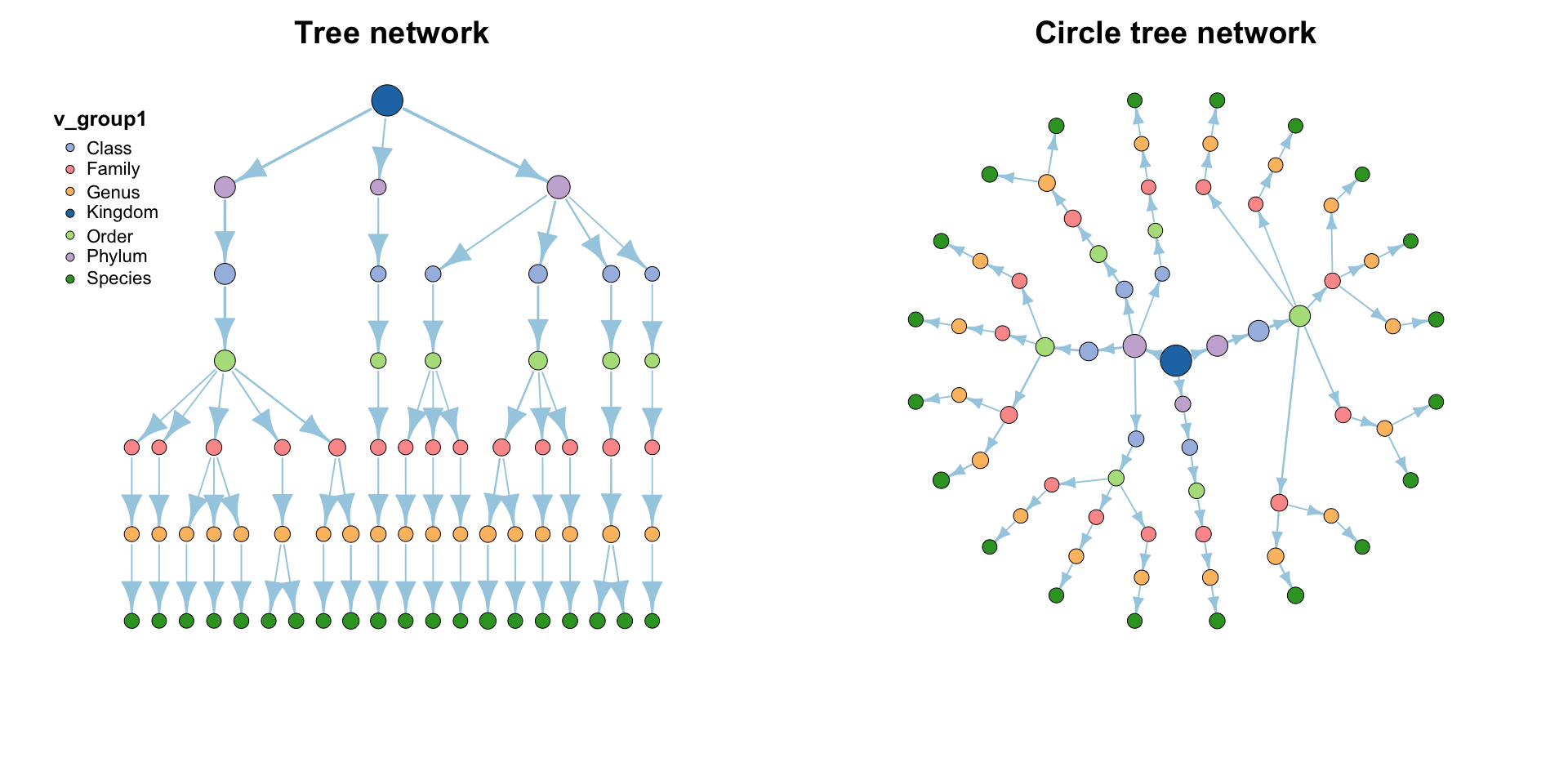
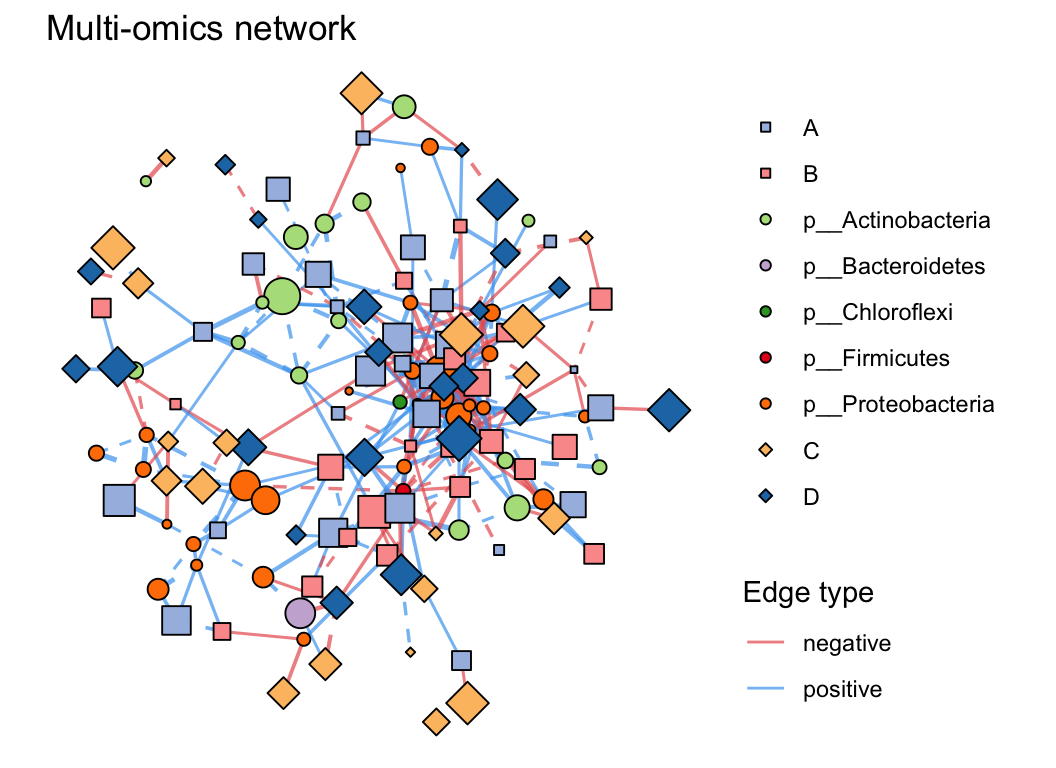
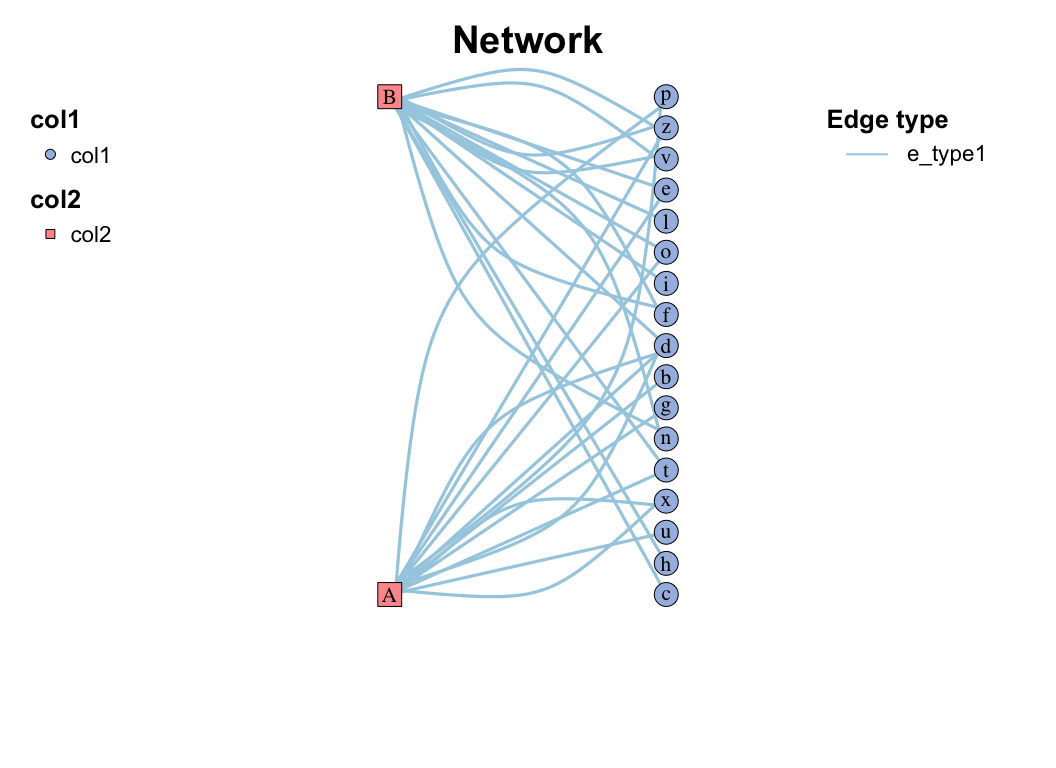
| coors |
1. cooridnates dataframe, 2. layout function (e.g. as_star(), as_tree(), in_circle(), nicely()... see help(c_net_lay)) |
| vertex.color |
color of nodes, receive vector (1. length same to number of nodes; 2. length same to numbers of v_class; 3. named verctor like c(A="red",B="blue")) |
| vertex. shape |
shape of nodes, receive vector (1. length same to number of nodes; 2. length same to numbers of v_group; 3. named verctor like c(v_group1="circle",B="square"))shape list: none, circle, square, csquare, rectangle, crectangle, vrectangle, pie, raster, or sphere |
| vertex.size |
size of nodes, receive numerical vector (length same to number of nodes)please use mmscale to control vertex.size, don't be too large. |
| vertex_size_range |
the vertex size range, e.g. c(1,10) |
| labels_num |
show how many labels,>1 indicates number, |
| vertex.label |
the label of nodes, NA indicates no label |
| vertex.label.family |
label font family |
| vertex.label.font |
label font, 1 plain, 2 bold, 3 italic, 4 bold italic, 5 symbol |
| vertex.label.cex |
label size |
| vertex.label.dist |
distance from label to nodes |
| vertex.label.color |
The color of the labels, The default value is black. |
| vertex.label.degree |
0:right, pi: left, pi/2:below, -pi/2:above |
| vertex.frame.color |
color of nodes frame |
| plot_module |
use module as the v_class |
| mark_module |
logical, mark the modules? |
| mark_color |
mark color |
| mark_alpha |
mark fill alpha, default 0.3 |
| module_label |
show module label? |
| module_label_cex |
module label cex |
| module_label_color |
module label color |
| module_label_just |
module label just, default c(0.5,0.5) |
| edge.color |
color of edges, receive vector (1. length same to number of edges; 2. length same to numbers of e_type; 3. named verctor like c(A="red",B="blue")) |
| edge.width |
width of edge, receive numerical vector (length same to number of edges)please use mmscale to control vertex.size, don't be too large. |
| edge_width_range |
the edge width range, e.g. c(1,10) |
| edge.lty |
linetype of edge, receive vector (1. length same to number of edges; 2. length same to numbers of e_class; 3. named verctor like c(A="red",B="blue")) |
| edge.arrow.size |
arrow size for directed network |
| edge.arrow.width |
arrow width for directed network |
| arrow.mode |
arrow mode, 0 no arrow, 1 back, 2 forward, 3 both |
| edge.label |
the label of edges, NA indicates no label |
| edge.label.family |
label font family |
| edge.label.font |
label font, 1 plain, 2 bold, 3 italic, 4 bold italic, 5 symbol |
| edge.label.cex |
label size |
| edge.label.x |
label x-axis |
| edge.label.y |
label y-axis |
| edge.label.color |
label color |
| edge.curved |
The curvature of the body, on a scale of 0-1, FALSE means 0, TRUE means 0.5 |
| legend |
show any legend? FALSE means close all legends |
| legend_cex |
character expansion factor relative to current par('cex'), default: 1 |
| legend_position |
legend_position, default: c(left_leg_x=-1.9,left_leg_y=1,right_leg_x=1.2,right_leg_y=1) |
| legend_number |
add numbers in legend? (v_class number, e_type number...) |
| lty_legend, lty_legend_title, lty_legend_order |
show lty_legend? and the title, the oreder of legend receives a vector |
| size_legend, size_legend_tiltle |
show size_legend? and the title |
| edge_legend, edge_legend_title, edge_legend_order |
show edge_legend? and the title, the order of legend receives a vector |
| width_legend, width_legend_title |
show width_legend? and the title |
| color_legend, color_legend_order |
show col_legend? and the title, the order of legend receives a vector |
| group_legend_title, group_legend_order |
the title of group, the order of legend receives a vector |
| margin |
margin, a vector whose length =4 |
| rescale |
scale the coors to [-1,1], default F |
| asp |
y/x ratio |
| frame |
if T, add frame |
| main |
the main title of graph |
| sub |
subtitle |
| xlab |
x-axis label |
| ylab |
y-axis label |
| seed |
random seed, default:1234, make sure each plot is the same |
| params_list |
a list of parameters, e.g. list(edge_legend = TRUE, lty_legend = FALSE), when the parameter is duplicated, the format argument will be used rather than the argument in params_list |