install.packages("pcutils")
install.packages("MetaNet")MetaNet: Network Analysis for Omics Data
Welcome
Install
MetaNet is a comprehensive network analysis package, especially in various biological omics.
Some functions of MetaNet are dependent with pcutils, so you also need to install pcutils.
The stable version can be installed from CRAN:
The latest development version can be found in https://github.com/Asa12138/MetaNet:
remotes::install_github("Asa12138/pcutils")
remotes::install_github("Asa12138/MetaNet", dependencies = T)For data manipulation, we recommend to use dplyr.
MetaNet
MetaNet is a comprehensive network analysis R package for omics data:
Support for integrated analysis for multi-omics data.
Calculate correlation network quickly, accelerate lots of analysis by parallel computing.
Handle bigger data, more than 10,000 nodes in each omics.
Offer various layout algorithms for multi-omics network and interfaces to other software (Gephi, Cytoscape, ggplot2), easy to visualize.
Provide comprehensive topology indexes calculation,including ecological network stability.
Citation

Please cite:
Chen Peng (2023). MetaNet: Network Analysis for Omics data. R package, https://github.com/Asa12138/MetaNet.
Quick Start
This is the overall development framework of MetaNet, including data processing, network construction, network analysis, network visualization and other functions:
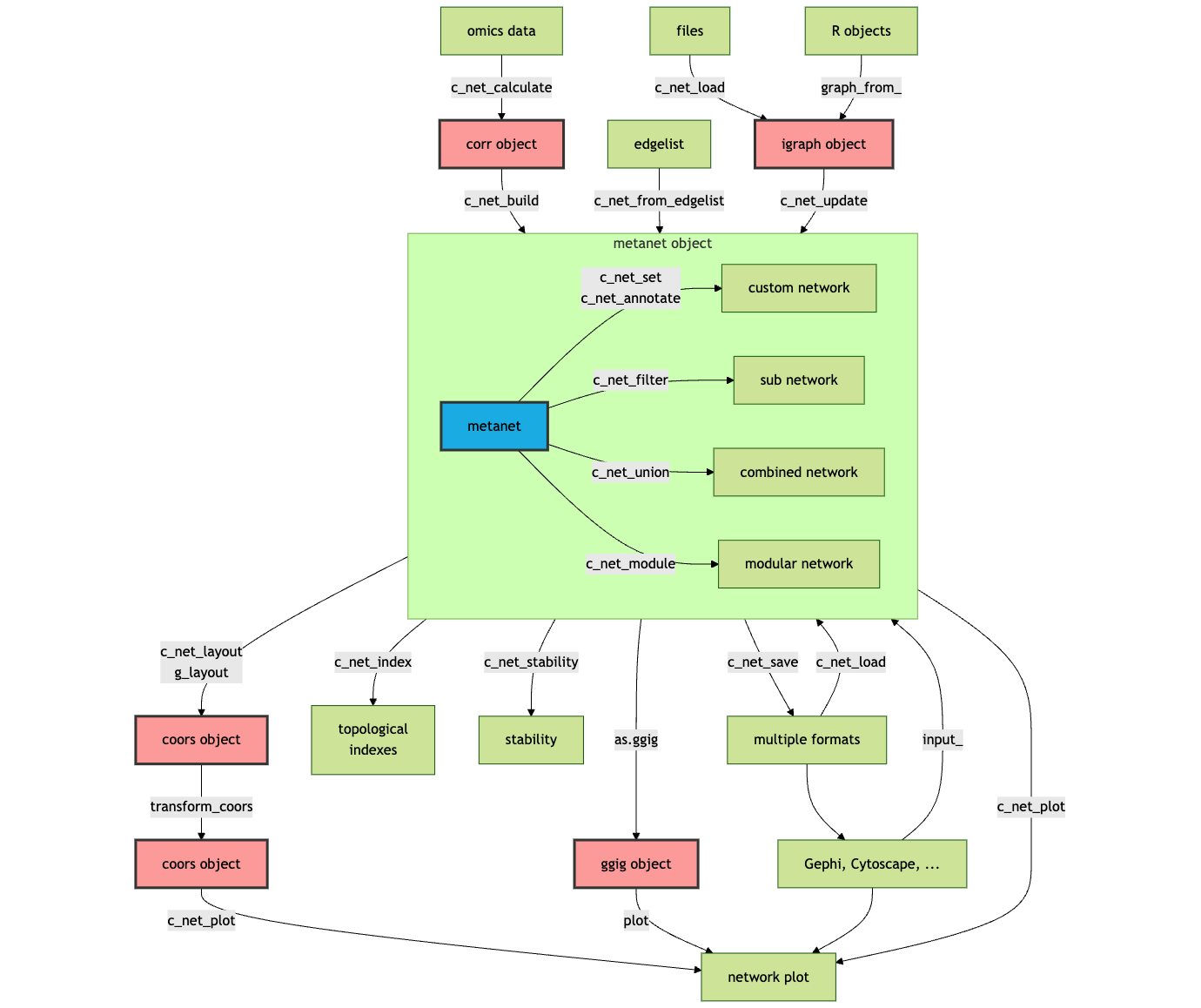
MetaNet
Simply build and draw a network plot, only need to use c_net_calculate(), c_net_build(), c_net_plot() three functions:
data("otutab", package = "pcutils")
# Inter-species correlation coefficients were calculated after transposition
totu <- t2(otutab[1:100, ])
cor <- c_net_calculate(totu)
net <- c_net_build(cor)
c_net_plot(net)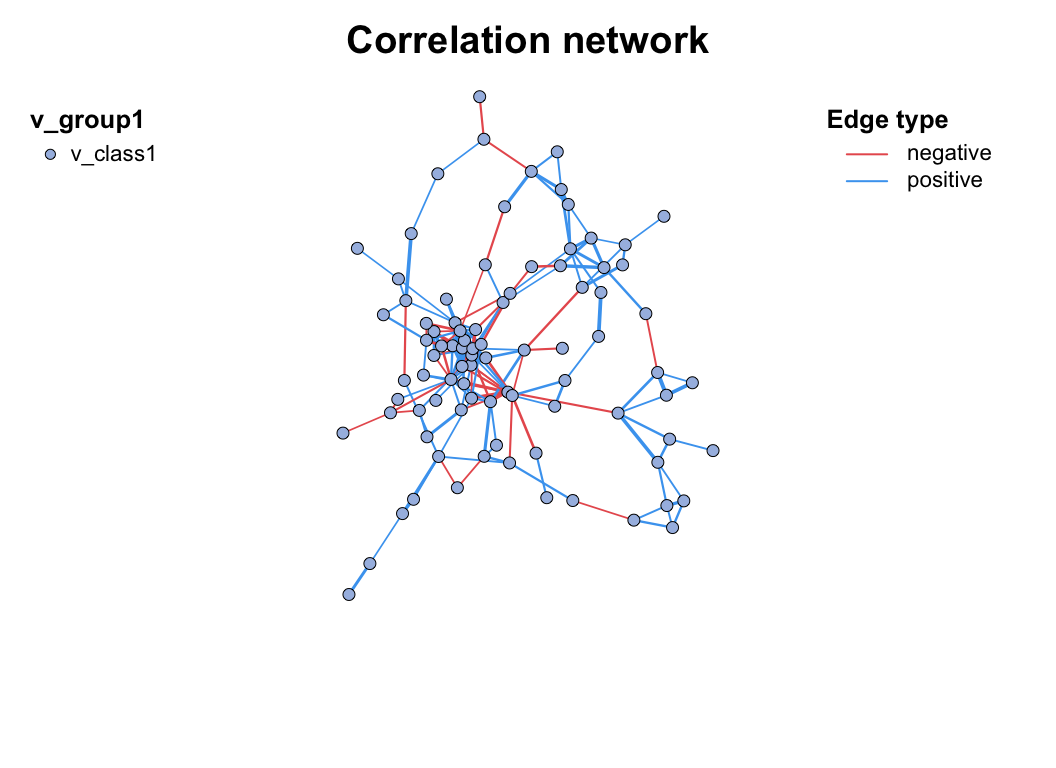
For more detailed usage, please refer to the sections below.
Session Info
sessionInfo()
## R version 4.2.2 (2022-10-31)
## Platform: aarch64-apple-darwin20 (64-bit)
## Running under: macOS 14.7
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/lib/libRblas.0.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.2-arm64/Resources/lib/libRlapack.dylib
##
## locale:
## [1] C/UTF-8/C/C/C/C
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] pcutils_0.2.8 dplyr_1.1.4 MetaNet_0.2.5 igraph_1.6.0
##
## loaded via a namespace (and not attached):
## [1] Rcpp_1.0.12 RColorBrewer_1.1-3 pillar_1.9.0 compiler_4.2.2
## [5] plyr_1.8.9 tools_4.2.2 digest_0.6.31 jsonlite_1.8.4
## [9] evaluate_0.19 lifecycle_1.0.4 tibble_3.2.1 gtable_0.3.4
## [13] pkgconfig_2.0.3 rlang_1.1.3 cli_3.6.2 yaml_2.3.6
## [17] xfun_0.49 fastmap_1.1.0 withr_3.0.0 stringr_1.5.1
## [21] knitr_1.45 generics_0.1.3 vctrs_0.6.5 htmlwidgets_1.6.1
## [25] grid_4.2.2 tidyselect_1.2.0 glue_1.7.0 R6_2.5.1
## [29] fansi_1.0.6 rmarkdown_2.25 ggplot2_3.5.1 reshape2_1.4.4
## [33] magrittr_2.0.3 scales_1.3.0 htmltools_0.5.4 colorspace_2.1-0
## [37] utf8_1.2.4 stringi_1.8.3 munsell_0.5.0