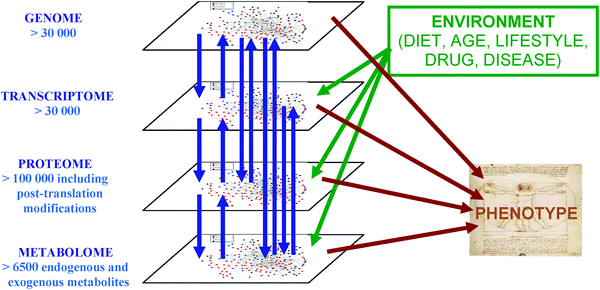
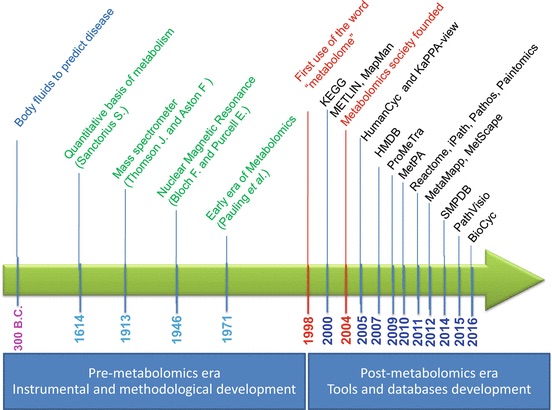
Ali, Ahmed, Yasmine Abouleila, Yoshihiro Shimizu, Eiso Hiyama, Samy Emara, Alireza Mashaghi, and Thomas Hankemeier. 2019.
“Single-Cell Metabolomics by Mass Spectrometry: Advances, Challenges, and Future Applications.” TrAC Trends in Analytical Chemistry 120 (November): 115436.
https://doi.org/10.1016/j.trac.2019.02.033.
Allam-Ndoul, Bénédicte, Frédéric Guénard, Véronique Garneau, Hubert Cormier, Olivier Barbier, Louis Pérusse, and Marie-Claude Vohl. 2016.
“Association Between Metabolite Profiles, Metabolic Syndrome and Obesity Status.” Nutrients 8 (6): 324.
https://doi.org/10.3390/nu8060324.
Alonso, Arnald, Sara Marsal, and Antonio Julià. 2015.
“Analytical Methods in Untargeted Metabolomics: State of the Art in 2015.” Frontiers in Bioengineering and Biotechnology 3 (March).
https://doi.org/10.3389/fbioe.2015.00023.
Baker, Monya. 2011.
“Metabolomics: From Small Molecules to Big Ideas.” Nature Methods 8 (2): 117–21.
https://doi.org/10.1038/nmeth0211-117.
Barnes, Stephen, H. Paul Benton, Krista Casazza, Sara J. Cooper, Xiangqin Cui, Xiuxia Du, Jeffrey Engler, et al. 2016a.
“Training in Metabolomics Research. I. Designing the Experiment, Collecting and Extracting Samples and Generating Metabolomics Data.” Journal of Mass Spectrometry 51 (7): 461–75.
https://doi.org/10.1002/jms.3782.
———, et al. 2016b.
“Training in Metabolomics Research. II. Processing and Statistical Analysis of Metabolomics Data, Metabolite Identification, Pathway Analysis, Applications of Metabolomics and Its Future.” Journal of Mass Spectrometry 51 (8): 535–48.
https://doi.org/10.1002/jms.3780.
Beale, David J., Farhana R. Pinu, Konstantinos A. Kouremenos, Mahesha M. Poojary, Vinod K. Narayana, Berin A. Boughton, Komal Kanojia, Saravanan Dayalan, Oliver A. H. Jones, and Daniel A. Dias. 2018.
“Review of Recent Developments in GC–MS Approaches to Metabolomics-Based Research.” Metabolomics 14 (11): 152.
https://doi.org/10.1007/s11306-018-1449-2.
Begou, O., H. G. Gika, I. D. Wilson, and G. Theodoridis. 2017.
“Hyphenated MS-based Targeted Approaches in Metabolomics.” Analyst 142 (17): 3079–3100.
https://doi.org/10.1039/C7AN00812K.
Bilbao, Aivett, Emmanuel Varesio, Jeremy Luban, Caterina Strambio-De-Castillia, Gérard Hopfgartner, Markus Müller, and Frédérique Lisacek. 2015.
“Processing Strategies and Software Solutions for Data-Independent Acquisition in Mass Spectrometry.” PROTEOMICS 15 (5-6): 964–80.
https://doi.org/10.1002/pmic.201400323.
Broeckling, Corey D., Richard D. Beger, Leo L. Cheng, Raquel Cumeras, Daniel J. Cuthbertson, Surendra Dasari, W. Clay Davis, et al. 2023.
“Current Practices in LC-MS Untargeted Metabolomics: A Scoping Review on the Use of Pooled Quality Control Samples.” Analytical Chemistry 95 (51): 18645–54.
https://doi.org/10.1021/acs.analchem.3c02924.
Bundy, Jacob G., Matthew P. Davey, and Mark R. Viant. 2009.
“Environmental Metabolomics: A Critical Review and Future Perspectives.” Metabolomics 5 (1): 3.
https://doi.org/10.1007/s11306-008-0152-0.
Cajka, Tomas, and Oliver Fiehn. 2016.
“Toward Merging Untargeted and Targeted Methods in Mass Spectrometry-Based Metabolomics and Lipidomics.” Analytical Chemistry 88 (1): 524–45.
https://doi.org/10.1021/acs.analchem.5b04491.
Castro-Puyana, María, Raquel Pérez-Míguez, Lidia Montero, and Miguel Herrero. 2017.
“Application of Mass Spectrometry-Based Metabolomics Approaches for Food Safety, Quality and Traceability.” TrAC Trends in Analytical Chemistry 93 (August): 102–18.
https://doi.org/10.1016/j.trac.2017.05.004.
Chang, Hui-Yin, Sean M. Colby, Xiuxia Du, Javier D. Gomez, Maximilian J. Helf, Katerina Kechris, Christine R. Kirkpatrick, et al. 2021.
“A Practical Guide to Metabolomics Software Development.” Analytical Chemistry 93 (4): 1912–23.
https://doi.org/10.1021/acs.analchem.0c03581.
Cheng, Susan, Svati H. Shah, Elizabeth J. Corwin, Oliver Fiehn, Robert L. Fitzgerald, Robert E. Gerszten, Thomas Illig, et al. 2017.
“Potential Impact and Study Considerations of Metabolomics in Cardiovascular Health and Disease: A Scientific Statement From the American Heart Association.” Circulation: Cardiovascular Genetics 10 (2): e000032.
https://doi.org/10.1161/HCG.0000000000000032.
Climaco Pinto, Rui, Ibrahim Karaman, Matthew R. Lewis, Jenny Hällqvist, Manuja Kaluarachchi, Gonçalo Graça, Elena Chekmeneva, et al. 2022.
“Finding Correspondence Between Metabolomic Features in Untargeted Liquid Chromatography–Mass Spectrometry Metabolomics Datasets.” Analytical Chemistry 94 (14): 5493–503.
https://doi.org/10.1021/acs.analchem.1c03592.
Considine, E. C., G. Thomas, A. L. Boulesteix, A. S. Khashan, and L. C. Kenny. 2017.
“Critical Review of Reporting of the Data Analysis Step in Metabolomics.” Metabolomics 14 (1): 7.
https://doi.org/10.1007/s11306-017-1299-3.
Domingo-Almenara, Xavier, J. Rafael Montenegro-Burke, H. Paul Benton, and Gary Siuzdak. 2018.
“Annotation: A Computational Solution for Streamlining Metabolomics Analysis.” Analytical Chemistry 90 (1): 480–89.
https://doi.org/10.1021/acs.analchem.7b03929.
Dryden, Michael D. M., Ryan Fobel, Christian Fobel, and Aaron R. Wheeler. 2017.
“Upon the Shoulders of Giants: Open-Source Hardware and Software in Analytical Chemistry.” Analytical Chemistry 89 (8): 4330–38.
https://doi.org/10.1021/acs.analchem.7b00485.
Du, Xinsong, Juan J. Aristizabal-Henao, Timothy J. Garrett, Mathias Brochhausen, William R. Hogan, and Dominick J. Lemas. 2022.
“A Checklist for Reproducible Computational Analysis in Clinical Metabolomics Research.” Metabolites 12 (1): 87.
https://doi.org/10.3390/metabo12010087.
Dudzik, Danuta, Cecilia Barbas-Bernardos, Antonia García, and Coral Barbas. 2018.
“Quality Assurance Procedures for Mass Spectrometry Untargeted Metabolomics. A Review.” Journal of Pharmaceutical and Biomedical Analysis, Review issue 2017, 147 (January): 149–73.
https://doi.org/10.1016/j.jpba.2017.07.044.
Fenaille, François, Pierre Barbier Saint-Hilaire, Kathleen Rousseau, and Christophe Junot. 2017.
“Data Acquisition Workflows in Liquid Chromatography Coupled to High Resolution Mass Spectrometry-Based Metabolomics: Where Do We Stand?” Journal of Chromatography A 1526 (Supplement C): 1–12.
https://doi.org/10.1016/j.chroma.2017.10.043.
Fessenden, Marissa. 2016.
“Metabolomics: Small Molecules, Single Cells.” Nature 540 (7631): 153–55.
https://doi.org/10.1038/540153a.
Fiehn, Oliver. 2002.
“Metabolomics – the Link Between Genotypes and Phenotypes.” Plant Molecular Biology 48 (1): 155–71.
https://doi.org/10.1023/A:1013713905833.
Gika, Helen G., Georgios A. Theodoridis, Robert S. Plumb, and Ian D. Wilson. 2014.
“Current Practice of Liquid Chromatography–Mass Spectrometry in Metabolomics and Metabonomics.” Journal of Pharmaceutical and Biomedical Analysis, Review
Papers on
Pharmaceutical and
Biomedical Analysis 2013, 87 (January): 12–25.
https://doi.org/10.1016/j.jpba.2013.06.032.
Goldansaz, Seyed Ali, An Chi Guo, Tanvir Sajed, Michael A. Steele, Graham S. Plastow, and David S. Wishart. 2017.
“Livestock Metabolomics and the Livestock Metabolome: A Systematic Review.” PLOS ONE 12 (5): e0177675.
https://doi.org/10.1371/journal.pone.0177675.
González, Oskar, Anne-Charlotte Dubbelman, and Thomas Hankemeier. 2022.
“Postcolumn Infusion as a Quality Control Tool for LC-MS-Based Analysis.” Postcolumn Infusion as a Quality Control Tool for LC-MS-Based Analysis, April.
https://doi.org/10.1021/jasms.2c00022.
González-Domínguez, Álvaro, Núria Estanyol-Torres, Carl Brunius, Rikard Landberg, and Raúl González-Domínguez. 2024.
“QComics: Recommendations and Guidelines for Robust, Easily Implementable and Reportable Quality Control of Metabolomics Data.” Analytical Chemistry 96 (3): 1064–72.
https://doi.org/10.1021/acs.analchem.3c03660.
González-Riano, Carolina, Danuta Dudzik, Antonia Garcia, Alberto Gil-de-la-Fuente, Ana Gradillas, Joanna Godzien, Ángeles López-Gonzálvez, et al. 2020.
“Recent Developments Along the Analytical Process for Metabolomics Workflows.” Analytical Chemistry 92 (1): 203–26.
https://doi.org/10.1021/acs.analchem.9b04553.
Griffiths, William J., Therese Koal, Yuqin Wang, Matthias Kohl, David P. Enot, and Hans-Peter Deigner. 2010.
“Targeted Metabolomics for Biomarker Discovery.” Angewandte Chemie International Edition 49 (32): 5426–45.
https://doi.org/10.1002/anie.200905579.
Gromski, Piotr S., Howbeer Muhamadali, David I. Ellis, Yun Xu, Elon Correa, Michael L. Turner, and Royston Goodacre. 2015.
“A Tutorial Review: Metabolomics and Partial Least Squares-Discriminant Analysis – a Marriage of Convenience or a Shotgun Wedding.” Analytica Chimica Acta 879 (June): 10–23.
https://doi.org/10.1016/j.aca.2015.02.012.
Hansen, Rebecca L., and Young Jin Lee. 2018.
“High-Spatial Resolution Mass Spectrometry Imaging: Toward Single Cell Metabolomics in Plant Tissues.” The Chemical Record 18 (1): 65–77.
https://doi.org/10.1002/tcr.201700027.
Hites, Ronald A., and Karl J. Jobst. 2018.
“Is Nontargeted Screening Reproducible?” Environmental Science & Technology 52 (21): 11975–76.
https://doi.org/10.1021/acs.est.8b05671.
Jones, Dean P., Youngja Park, and Thomas R. Ziegler. 2012.
“Nutritional Metabolomics: Progress in Addressing Complexity in Diet and Health.” Annual Review of Nutrition 32 (1): 183–202.
https://doi.org/10.1146/annurev-nutr-072610-145159.
Jorge, Tiago F., Ana T. Mata, and Carla António. 2016.
“Mass Spectrometry as a Quantitative Tool in Plant Metabolomics.” Phil. Trans. R. Soc. A 374 (2079): 20150370.
https://doi.org/10.1098/rsta.2015.0370.
Kapoore, Rahul Vijay, and Seetharaman Vaidyanathan. 2016.
“Towards Quantitative Mass Spectrometry-Based Metabolomics in Microbial and Mammalian Systems.” Phil. Trans. R. Soc. A 374 (2079): 20150363.
https://doi.org/10.1098/rsta.2015.0363.
Kennedy, Adam D., Bryan M. Wittmann, Anne M. Evans, Luke A. D. Miller, Douglas R. Toal, Shaun Lonergan, Sarah H. Elsea, and Kirk L. Pappan. 2018.
“Metabolomics in the Clinic: A Review of the Shared and Unique Features of Untargeted Metabolomics for Clinical Research and Clinical Testing.” Journal of Mass Spectrometry 53 (11): 1143–54.
https://doi.org/10.1002/jms.4292.
Kusonmano, Kanthida, Wanwipa Vongsangnak, and Pramote Chumnanpuen. 2016.
“Informatics for Metabolomics.” In
Translational Biomedical Informatics, 91–115. Advances in
Experimental Medicine and
Biology. Springer, Singapore.
https://doi.org/10.1007/978-981-10-1503-8_5.
Levy, Allison J., Nicholas R. Oranzi, Atiye Ahmadireskety, Robin H. J. Kemperman, Michael S. Wei, and Richard A. Yost. 2019.
“Recent Progress in Metabolomics Using Ion Mobility-Mass Spectrometry.” TrAC Trends in Analytical Chemistry 116 (July): 274–81.
https://doi.org/10.1016/j.trac.2019.05.001.
Liu, Xinyu, Lina Zhou, Xianzhe Shi, and Guowang Xu. 2019.
“New Advances in Analytical Methods for Mass Spectrometry-Based Large-Scale Metabolomics Study.” TrAC Trends in Analytical Chemistry 121 (December): 115665.
https://doi.org/10.1016/j.trac.2019.115665.
Lu, Wenyun, Bryson D. Bennett, and Joshua D. Rabinowitz. 2008.
“Analytical Strategies for LC–MS-based Targeted Metabolomics.” Journal of Chromatography B, Hyphenated
Techniques for
Global Metabolite Profiling, 871 (2): 236–42.
https://doi.org/10.1016/j.jchromb.2008.04.031.
Lu, Xin, and Guowang Xu. 2008.
“LC-MS Metabonomics Methodology in Biomarker Discovery.” In
Biomarker Methods in Drug Discovery and Development, edited by Feng Wang, 291–315. Methods in
Pharmacology and
Toxicology™. Humana Press.
https://doi.org/10.1007/978-1-59745-463-6_14.
Lv, Wangjie, Zhongda Zeng, Yuqing Zhang, Qingqing Wang, Lichao Wang, Zhaoxuan Zhang, Xianzhe Shi, Xinjie Zhao, and Guowang Xu. 2022.
“Comprehensive Metabolite Quantitative Assay Based on Alternate Metabolomics and Lipidomics Analyses.” Analytica Chimica Acta 1215 (July): 339979.
https://doi.org/10.1016/j.aca.2022.339979.
Madsen, Rasmus, Torbjörn Lundstedt, and Johan Trygg. 2010.
“Chemometrics in Metabolomics—A Review in Human Disease Diagnosis.” Analytica Chimica Acta 659 (1): 23–33.
https://doi.org/10.1016/j.aca.2009.11.042.
Mangul, Serghei, Thiago Mosqueiro, Richard J. Abdill, Dat Duong, Keith Mitchell, Varuni Sarwal, Brian Hill, et al. 2019.
“Challenges and Recommendations to Improve the Installability and Archival Stability of Omics Computational Tools.” PLOS Biology 17 (6): e3000333.
https://doi.org/10.1371/journal.pbio.3000333.
Martens, Jonathan, Giel Berden, Rianne E. van Outersterp, Leo A. J. Kluijtmans, Udo F. Engelke, Clara D. M. van Karnebeek, Ron A. Wevers, and Jos Oomens. 2017.
“Molecular Identification in Metabolomics Using Infrared Ion Spectroscopy.” Scientific Reports 7 (June).
https://doi.org/10.1038/s41598-017-03387-4.
Matich, Eryn K., Nita G. Chavez Soria, Diana S. Aga, and G. Ekin Atilla-Gokcumen. 2019.
“Applications of Metabolomics in Assessing Ecological Effects of Emerging Contaminants and Pollutants on Plants.” Journal of Hazardous Materials 373 (July): 527–35.
https://doi.org/10.1016/j.jhazmat.2019.02.084.
Miggiels, Paul, Bert Wouters, Gerard J. P. van Westen, Anne-Charlotte Dubbelman, and Thomas Hankemeier. 2019.
“Novel Technologies for Metabolomics: More for Less.” TrAC Trends in Analytical Chemistry 120 (November): 115323.
https://doi.org/10.1016/j.trac.2018.11.021.
Misra, Biswapriya B. 2018.
“New Tools and Resources in Metabolomics: 2016–2017.” ELECTROPHORESIS 39 (7): 909–23.
https://doi.org/10.1002/elps.201700441.
Misra, Biswapriya B., Johannes F. Fahrmann, and Dmitry Grapov. 2017.
“Review of Emerging Metabolomic Tools and Resources: 2015–2016.” ELECTROPHORESIS 38 (18): 2257–74.
https://doi.org/10.1002/elps.201700110.
Misra, Biswapriya B., and Justin J. J. van der Hooft. 2016.
“Updates in Metabolomics Tools and Resources: 2014–2015.” ELECTROPHORESIS 37 (1): 86–110.
https://doi.org/10.1002/elps.201500417.
Müller, Manfred J., and Anja Bosy-Westphal. 2020.
“From a ‘Metabolomics Fashion’ to a Sound Application of Metabolomics in Research on Human Nutrition.” European Journal of Clinical Nutrition 74 (12): 1619–29.
https://doi.org/10.1038/s41430-020-00781-6.
Ni, Zhixu, Michele Wölk, Geoff Jukes, Karla Mendivelso Espinosa, Robert Ahrends, Lucila Aimo, Jorge Alvarez-Jarreta, et al. 2022.
“Guiding the Choice of Informatics Software and Tools for Lipidomics Research Applications.” Nature Methods, December, 1–12.
https://doi.org/10.1038/s41592-022-01710-0.
Pezzatti, Julian, Julien Boccard, Santiago Codesido, Yoric Gagnebin, Abhinav Joshi, Didier Picard, Víctor González-Ruiz, and Serge Rudaz. 2020.
“Implementation of Liquid Chromatography–High Resolution Mass Spectrometry Methods for Untargeted Metabolomic Analyses of Biological Samples: A Tutorial.” Analytica Chimica Acta 1105 (April): 28–44.
https://doi.org/10.1016/j.aca.2019.12.062.
Place, Benjamin J., Elin M. Ulrich, Jonathan K. Challis, Alex Chao, Bowen Du, Kristin Favela, Yong-Lai Feng, et al. 2021.
“An Introduction to the Benchmarking and Publications for Non-Targeted Analysis Working Group.” Analytical Chemistry 93 (49): 16289–96.
https://doi.org/10.1021/acs.analchem.1c02660.
Rey-Stolle, Fernanda, Danuta Dudzik, Carolina Gonzalez-Riano, Miguel Fernández-García, Vanesa Alonso-Herranz, David Rojo, Coral Barbas, and Antonia García. 2022.
“Low and High Resolution Gas Chromatography-Mass Spectrometry for Untargeted Metabolomics: A Tutorial.” Analytica Chimica Acta 1210 (June): 339043.
https://doi.org/10.1016/j.aca.2021.339043.
Sarpe, Vladimir, and David C Schriemer. 2017.
“Supporting Metabolomics with Adaptable Software: Design Architectures for the End-User.” Current Opinion in Biotechnology, Analytical biotechnology, 43 (February): 110–17.
https://doi.org/10.1016/j.copbio.2016.11.001.
Schrimpe-Rutledge, Alexandra C., Simona G. Codreanu, Stacy D. Sherrod, and John A. McLean. 2016.
“Untargeted Metabolomics Strategies—Challenges and Emerging Directions.” Journal of The American Society for Mass Spectrometry 27 (12): 1897–1905.
https://doi.org/10.1007/s13361-016-1469-y.
Schymanski, Emma L., and Antony J. Williams. 2017.
“Open Science for Identifying ‘Known Unknown’ Chemicals.” Environmental Science & Technology 51 (10): 5357–59.
https://doi.org/10.1021/acs.est.7b01908.
Siskos, Alexandros P., Pooja Jain, Werner Römisch-Margl, Mark Bennett, David Achaintre, Yasmin Asad, Luke Marney, et al. 2017.
“Interlaboratory Reproducibility of a Targeted Metabolomics Platform for Analysis of Human Serum and Plasma.” Analytical Chemistry 89 (1): 656–65.
https://doi.org/10.1021/acs.analchem.6b02930.
Smirnov, Kirill S., Tanja V. Maier, Alesia Walker, Silke S. Heinzmann, Sara Forcisi, Inés Martinez, Jens Walter, and Philippe Schmitt-Kopplin. 2016.
“Challenges of Metabolomics in Human Gut Microbiota Research.” International Journal of Medical Microbiology, Intestinal microbiota - a microbial ecosystem at the edge between immune homeostasis and inflammation, 306 (5): 266–79.
https://doi.org/10.1016/j.ijmm.2016.03.006.
Spicer, Rachel, Reza M. Salek, Pablo Moreno, Daniel Cañueto, and Christoph Steinbeck. 2017.
“Navigating Freely-Available Software Tools for Metabolomics Analysis.” Metabolomics 13 (9).
https://doi.org/10.1007/s11306-017-1242-7.
Spratlin, Jennifer L., Natalie J. Serkova, and S. Gail Eckhardt. 2009.
“Clinical Applications of Metabolomics in Oncology: A Review.” Clinical Cancer Research 15 (2): 431–40.
https://doi.org/10.1158/1078-0432.CCR-08-1059.
Sumner, Lloyd W., Alexander Amberg, Dave Barrett, Michael H. Beale, Richard Beger, Clare A. Daykin, Teresa W.-M. Fan, et al. 2007.
“Proposed Minimum Reporting Standards for Chemical Analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI).” Metabolomics : Official Journal of the Metabolomic Society 3 (3): 211–21.
https://doi.org/10.1007/s11306-007-0082-2.
Sumner, Lloyd W, Pedro Mendes, and Richard A Dixon. 2003.
“Plant Metabolomics: Large-Scale Phytochemistry in the Functional Genomics Era.” Phytochemistry, Plant
Metabolomics, 62 (6): 817–36.
https://doi.org/10.1016/S0031-9422(02)00708-2.
Tang, Yanan, Caley B. Craven, Nicholas J. P. Wawryk, Junlang Qiu, Feng Li, and Xing-Fang Li. 2020.
“Advances in Mass Spectrometry-Based Omics Analysis of Trace Organics in Water.” TrAC Trends in Analytical Chemistry 128 (July): 115918.
https://doi.org/10.1016/j.trac.2020.115918.
Theodoridis, Georgios A., Helen G. Gika, Elizabeth J. Want, and Ian D. Wilson. 2012.
“Liquid Chromatography–Mass Spectrometry Based Global Metabolite Profiling: A Review.” Analytica Chimica Acta 711 (January): 7–16.
https://doi.org/10.1016/j.aca.2011.09.042.
Tian, Tze-Feng, San-Yuan Wang, Tien-Chueh Kuo, Cheng-En Tan, Guan-Yuan Chen, Ching-Hua Kuo, Chi-Hsin Sally Chen, Chang-Chuan Chan, Olivia A. Lin, and Y. Jane Tseng. 2016.
“Web Server for Peak Detection, Baseline Correction, and Alignment in Two-Dimensional Gas Chromatography Mass Spectrometry-Based Metabolomics Data.” Analytical Chemistry 88 (21): 10395–403.
https://doi.org/10.1021/acs.analchem.6b00755.
Uppal, Karan, Douglas I. Walker, Ken Liu, Shuzhao Li, Young-Mi Go, and Dean P. Jones. 2016.
“Computational Metabolomics: A Framework for the Million Metabolome.” Chemical Research in Toxicology 29 (12): 1956–75.
https://doi.org/10.1021/acs.chemrestox.6b00179.
Verhoeven, Aswin, Martin Giera, and Oleg A. Mayboroda. 2020.
“Scientific Workflow Managers in Metabolomics: An Overview.” Analyst 145 (11): 3801–8.
https://doi.org/10.1039/D0AN00272K.
Viant, Mark R., Timothy M. D. Ebbels, Richard D. Beger, Drew R. Ekman, David J. T. Epps, Hennicke Kamp, Pim E. G. Leonards, et al. 2019.
“Use Cases, Best Practice and Reporting Standards for Metabolomics in Regulatory Toxicology.” Nature Communications 10 (1): 3041.
https://doi.org/10.1038/s41467-019-10900-y.
Vinaixa, Maria, Emma L. Schymanski, Steffen Neumann, Miriam Navarro, Reza M. Salek, and Oscar Yanes. 2016.
“Mass Spectral Databases for LC/MS- and GC/MS-based Metabolomics: State of the Field and Future Prospects.” TrAC Trends in Analytical Chemistry 78 (April): 23–35.
https://doi.org/10.1016/j.trac.2015.09.005.
Vitale, Chiara Maria, Arjen Lommen, Carolin Huber, Kevin Wagner, Borja Garlito Molina, Rosalie Nijssen, Elliott James Price, et al. 2022.
“Harmonized Quality Assurance/Quality Control Provisions for Nontargeted Measurement of Urinary Pesticide Biomarkers in the HBM4EU Multisite SPECIMEn Study.” Analytical Chemistry 94 (22): 7833–43.
https://doi.org/10.1021/acs.analchem.2c00061.
Wallach, Joshua D., Kevin W. Boyack, and John P. A. Ioannidis. 2018.
“Reproducible Research Practices, Transparency, and Open Access Data in the Biomedical Literature, 2015–2017.” PLOS Biology 16 (11): e2006930.
https://doi.org/10.1371/journal.pbio.2006930.
Warth, Benedikt, Scott Spangler, Mingliang Fang, Caroline H. Johnson, Erica M. Forsberg, Ana Granados, Richard L. Martin, et al. 2017.
“Exposome-Scale Investigations Guided by Global Metabolomics, Pathway Analysis, and Cognitive Computing.” Analytical Chemistry 89 (21): 11505–13.
https://doi.org/10.1021/acs.analchem.7b02759.
Weljie, Aalim M., Jack Newton, Pascal Mercier, Erin Carlson, and Carolyn M. Slupsky. 2006.
“Targeted Profiling: Quantitative Analysis of 1H NMR Metabolomics Data.” Analytical Chemistry 78 (13): 4430–42.
https://doi.org/10.1021/ac060209g.
Wishart, David S. 2016.
“Emerging Applications of Metabolomics in Drug Discovery and Precision Medicine.” Nature Reviews Drug Discovery 15 (7): 473–84.
https://doi.org/10.1038/nrd.2016.32.
Wolfender, Jean-Luc, Guillaume Marti, Aurélien Thomas, and Samuel Bertrand. 2015.
“Current Approaches and Challenges for the Metabolite Profiling of Complex Natural Extracts.” Journal of Chromatography A, Editors’
Choice IX, 1382 (February): 136–64.
https://doi.org/10.1016/j.chroma.2014.10.091.
Yates Iii, John R. 2011.
“A Century of Mass Spectrometry: From Atoms to Proteomes.” Nature Methods 8 (8): 633–37.
https://doi.org/10.1038/nmeth.1659.
Yuan, Min, Susanne B. Breitkopf, Xuemei Yang, and John M. Asara. 2012.
“A Positive/Negative Ion–Switching, Targeted Mass Spectrometry–Based Metabolomics Platform for Bodily Fluids, Cells, and Fresh and Fixed Tissue.” Nature Protocols 7 (5): 872–81.
https://doi.org/10.1038/nprot.2012.024.
Zenobi, R. 2013.
“Single-Cell Metabolomics: Analytical and Biological Perspectives.” Science 342 (6163): 1243259.
https://doi.org/10.1126/science.1243259.
Zhang, Aihua, Hui Sun, Ping Wang, Ying Han, and Xijun Wang. 2012.
“Modern Analytical Techniques in Metabolomics Analysis.” The Analyst 137 (2): 293–300.
https://doi.org/10.1039/C1AN15605E.
Zhou, Juntuo, and Yuxin Yin. 2016.
“Strategies for Large-Scale Targeted Metabolomics Quantification by Liquid Chromatography-Mass Spectrometry.” Analyst 141 (23): 6362–73.
https://doi.org/10.1039/C6AN01753C.