Chapter 2 Part two of An Example
2.1 Analysis
The Main effects of depression, general anxiety, perfectionism, OCD symptoms, self esteem and stress were tested as precursors of group classification using multinomial logistic analysis modelling. The main effects and 2-way interactions were examined. Stepwise regression was also performed using the R stepAIC function on main effects and interactions model.
The Data was partioned 80:20 through random sampling, for developing a predictive model and validation respectively. Observations with missing values were ommited (74 omitted) before proceeding with model building.
The models were tested by calculating the classification matrix using the validation data and finding the misclassification proportion; the goal of lowering the misclassification compared to the Main effects model and Main effects including interactions.2.2 Main Effects Model
The Main Effects were examined, Classification as either Normal, Depressive or having an Eating Disorder (DiaGroup).
set.seed(3137924)
CogData=na.omit(CogData)
ind=sample(2,nrow(CogData),replace=TRUE,prob=c(0.8,0.2))
CogDataSample=CogData[ind==1,]
CogDataValidation=CogData[ind==2,]
#CogDataSample=CogData[sample(nrow(CogData), 400), ]
mod.fit=multinom(formula = DiagGroup ~ .,data=CogDataSample)
mod.fit1b=stepAIC(mod.fit)#Anova(mod.fit)
#### Main Effects FUll Model
cm=table(predict(mod.fit, CogDataValidation),CogDataValidation$DiagGroup)
#print(cm)
Full.Model.misclassification=(1-sum(diag(cm))/sum(cm))*100
#print(Full.Model.misclassification)
confusion = data.frame(cm)
confusion = ddply(confusion, "Var1", transform, Proportion = Freq / sum(Freq))
Plot1 = ggplot() + geom_tile(aes(x=Var1, y=Var2, fill=Proportion),
data=confusion, color="black",size=0.1) +
labs(x="Actual",y="Predicted") # rotate axis ticks text
Plot1 = Plot1 + geom_text(aes(x=Var1,y=Var2, label=sprintf("%.3f", Proportion)),
data=confusion, size=3, colour="black") +
scale_fill_gradient(low="yellow",high="purple") +theme_bw()+ theme(panel.border = element_blank())
Plot1 = Plot1 + geom_tile(aes(x=Var1,y=Var2),
data=subset(confusion, as.character(Var1)==as.character(Var2)),
color="black", size=0.3, fill="black", alpha=0)+ggtitle("Diagnosis Group Confusion Matrix(Main Effects)")+
ylim(rev(levels(confusion$Var2))) + xlim(levels(confusion$Var1))
### Reduced Main Effects Model
## Correlation between variables test
##DataCov = do.call( rbind, lapply( split(CogData, CogData$DiagGroup),
## function(CogData) data.frame(group=CogData$DiagGroup[1], mCov=cor(CogData$Depression_Total,CogData$GlobalEDEQ)) ##) )
#Anova(mod.fit1b)
cm1b=table(predict(mod.fit1b, CogDataValidation),CogDataValidation$DiagGroup)
#print(cm1b)
Full1b.Model.misclassification=(1-sum(diag(cm1b))/sum(cm1b))*100
#print(Full1b.Model.misclassification)
confusion1b = data.frame(cm1b)
confusion1b = ddply(confusion1b, "Var1", transform, Proportion = Freq / sum(Freq))
Plot1b = ggplot() + geom_tile(aes(x=Var1, y=Var2, fill=Proportion),
data=confusion1b, color="black",size=0.1) +
labs(x="Actual",y="Predicted") # rotate axis ticks text
Plot1b = Plot1b + geom_text(aes(x=Var1,y=Var2, label=sprintf("%.3f", Proportion)),
data=confusion1b, size=3, colour="black")+
scale_fill_gradient(low="yellow",high="purple") +theme_bw()+ theme(panel.border = element_blank())
Plot1b = Plot1b + geom_tile(aes(x=Var1,y=Var2),
data=subset(confusion1b, as.character(Var1)==as.character(Var2)),
color="black", size=0.3, fill="black", alpha=0)+ggtitle("Diagnosis Group Confusion Matrix(Reduced Main Effects)")+
ylim(rev(levels(confusion1b$Var2))) + xlim(levels(confusion1b$Var1))
library(gridExtra)
grid.arrange(Plot1,Plot1b,ncol=1)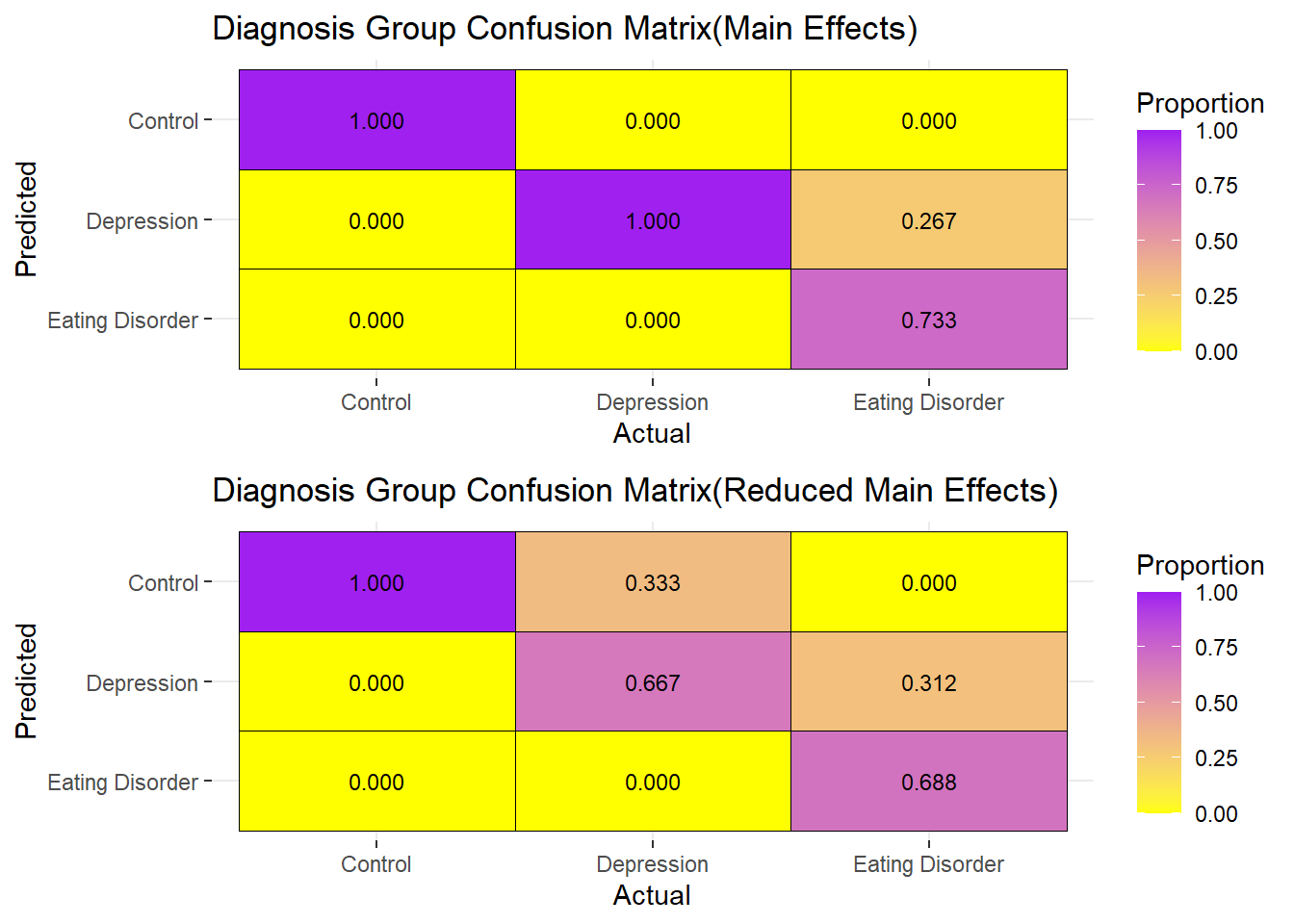
Both the GlobalEDEQ and Depression_Total are used to diagnose having an Eating Disorder and Depression respectively, the main effects model shows that Stress_Total scores and Self_Esteem scores are also important. The Main Effects Confusion Matrix correctly classifies all those as normal to be normal, as well as those with depression are classified as having depression.
73.3% of those classified as having an eating disorder are correctly classified, while 26.7% of those having an eating disorder are classified as having depression.
The misclassification using the full main effects is 5.2632%.
The stepwise reduced model, which only has and , performs relatively poorly in comparison to the Full Main effects model, having a relatively higher misclassification error of .
2.3 Main Effects and Interactions
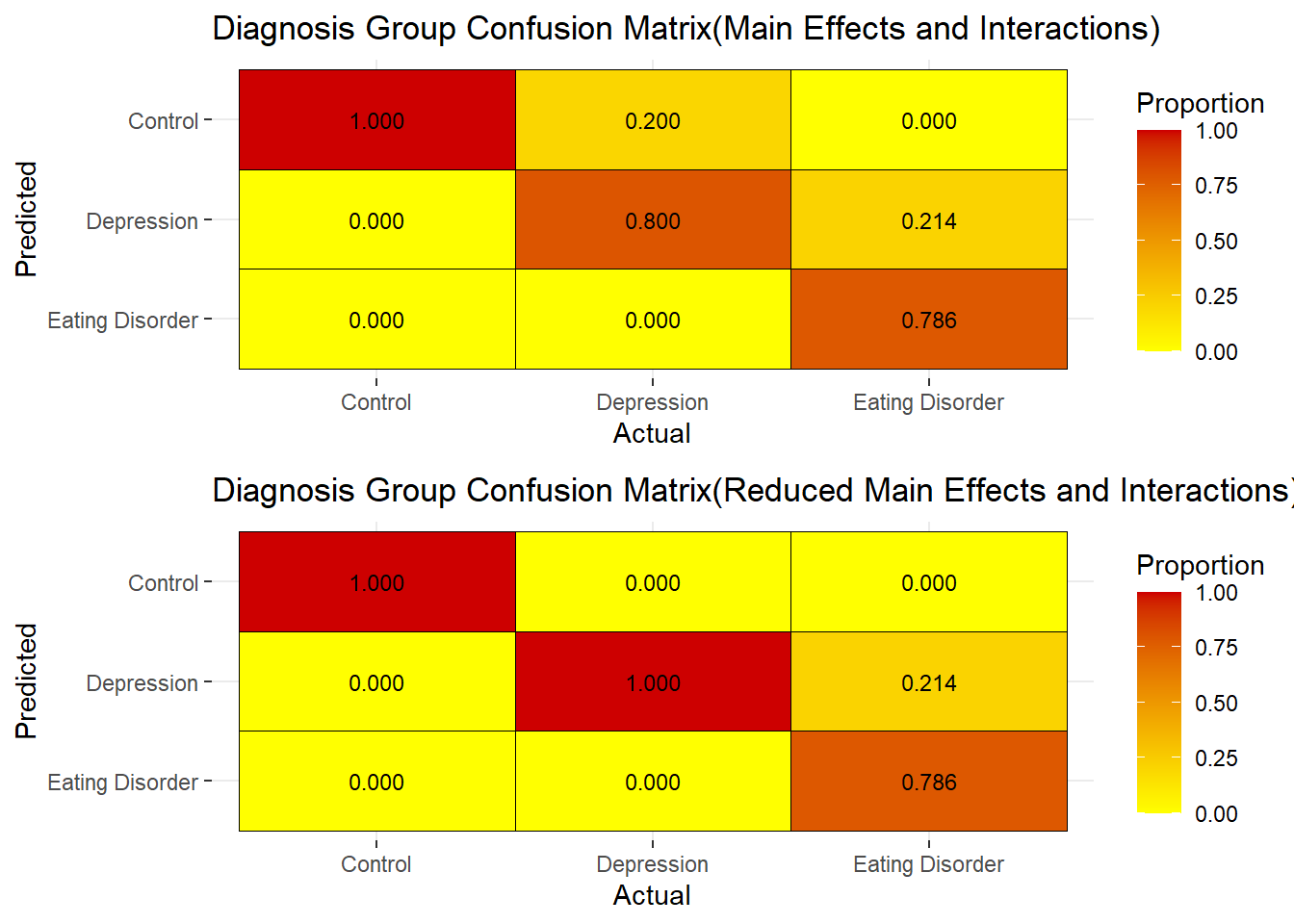 GlobalEDEQ, Depression_Total,Anxiety_Total,OCD_Symptoms:Depression_Total and Depression_Total:Stress_Total were statistically significant with the Main effects with Interactions full model. Achieving a 5.2632% misclassification error, which is the same as the main effects model.
Analyzing the Reduced Main Effects Model with Interactions the following variables are statistically significant.
GlobalEDEQ, Depression_Total,Anxiety_Total,OCD_Symptoms:Depression_Total and Depression_Total:Stress_Total were statistically significant with the Main effects with Interactions full model. Achieving a 5.2632% misclassification error, which is the same as the main effects model.
Analyzing the Reduced Main Effects Model with Interactions the following variables are statistically significant.
The Reduced Main Effects with Interactions Confusion Matrix correctly classifies all those as normal to be normal, as well as those with depression are classified as having depression.
78.6% of those classified as having an eating disorder are correctly classified,while 21.4% of those having an eating disorder are classified as having depression.
The misclassification using the full main effects and interactions is 3.9474%, which is the lowest of all models.
2.4 Discussion
In addition to Main Effects and Main Effects with Interactions, stepwise reduced models were also analyzed and their misclassification proportions were calculated for comparison. As can be seen in Table 1.0 .
The Reduced Main Effects with Interactions achieved the lowest misclassification error of 3.9474%.
Identifying characteristics that are prevalent for women whom either have Depression or an Eating Disorder as well as those with the potentional for co-morbidity. Characteristics which where significant were the Depression score component which is usually administered to patients to determine whether they have depression as well as the GlobalEDEQ score, which is used to determine whether a person has an eating disorder.
On all tested models, there were classifications of those having an eating disorder, as having depression however the reverse is not seen. This may suggest that depression may be co-morbid with having an eating disorder.