Exercise Solutions
Section 1.1.1
- You should expect these variables to be very closely correlated, with the points clustered very close to a line.
ggplot(data = mpg) +
geom_point(mapping = aes(x = cty, y = hwy))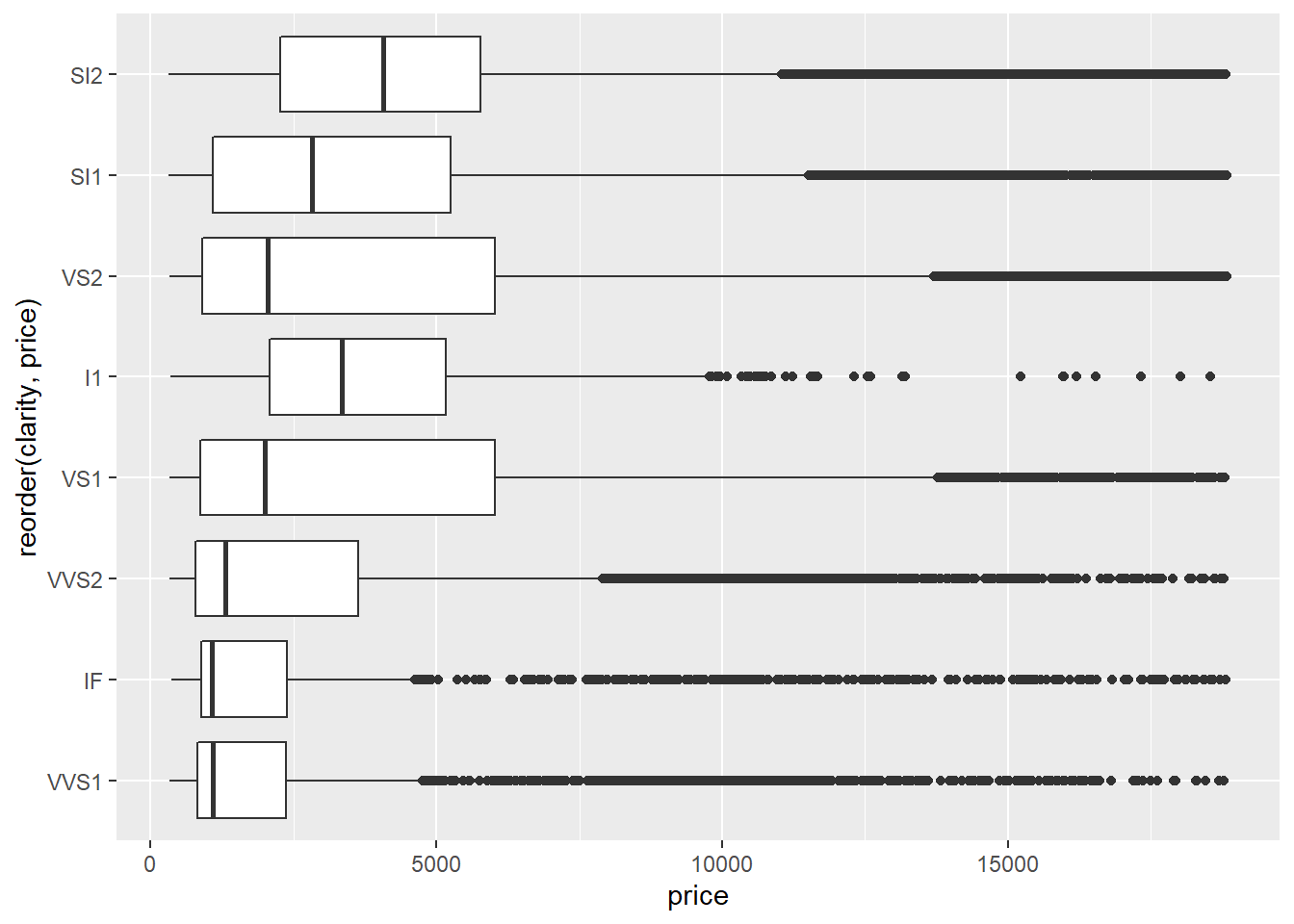
manufacturer,model,year,cyl,trans,drv,fl, andclassare categorical. The others are continuous. (You could make an argument foryearandcylbeing continuous as well, but because each only has a few values and the possible values cannot occur across a continuous spectrum, they are more appropriately classified as categorical.)You could use either
hwyorctyto measure fuel efficiency. The scatter plot below would indicate that front-wheel drives generally have the best fuel efficiency but exhibit the most variation. Rear-wheel drives get the worst fuel efficiency and exhibit the least variation.
ggplot(data = mpg) +
geom_point(mapping = aes(x = drv, y = hwy))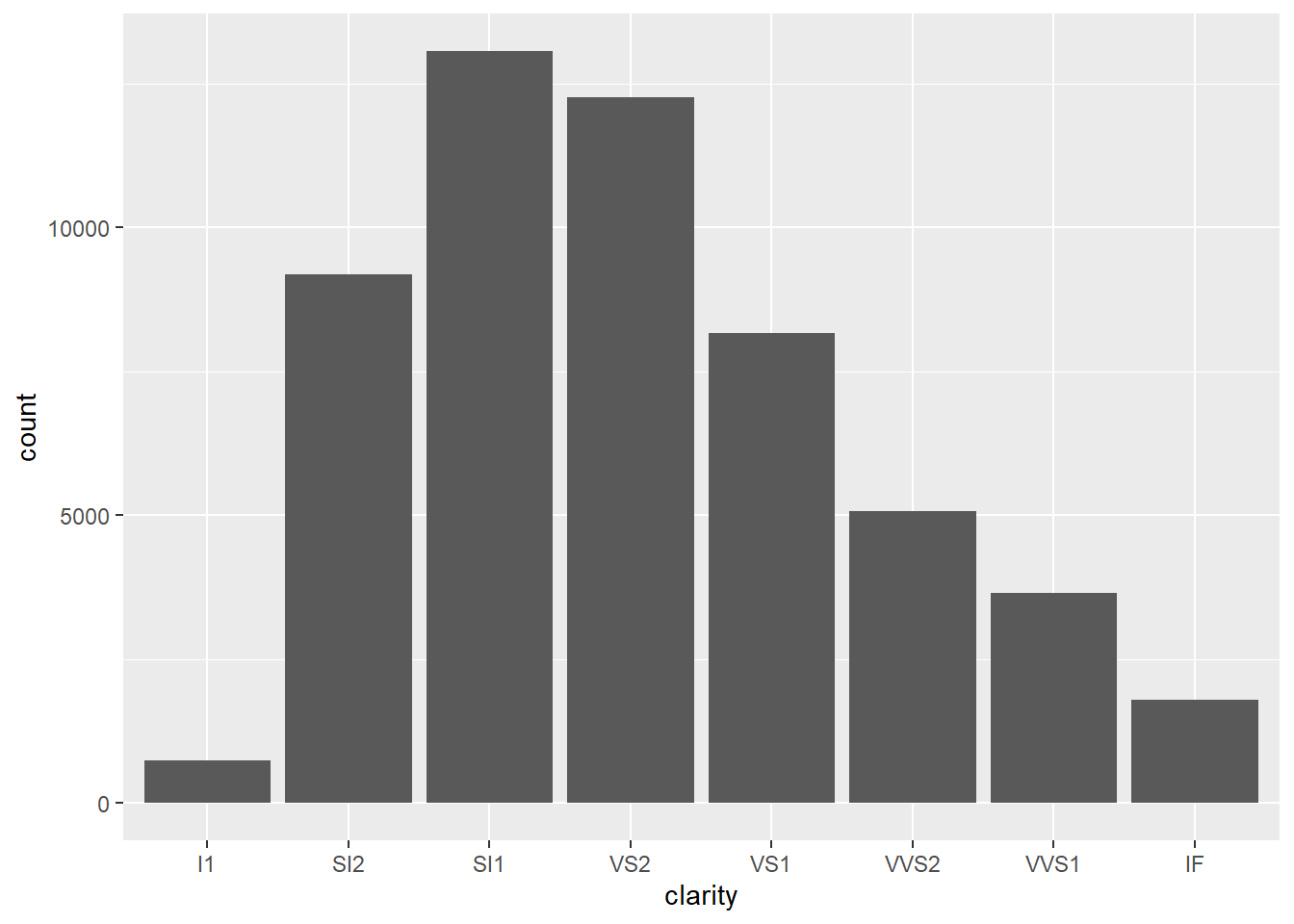
The points are stacked vertically. This happens because
drvis categorical. (We’ll see in Section 1.6 that box plots are better ways to visualize a continuous vs. categorical relationship.)In the scatter plot below, a dot just indicates that a car with that
class-drvcombination exists in the data set. For example, there’s a car withclass = 2seateranddrv = r. While this is somewhat helpful, it doesn’t convey a trend, which is what scatter plots are supposed to do.
ggplot(data = mpg) +
geom_point(mapping = aes(x = class, y = drv))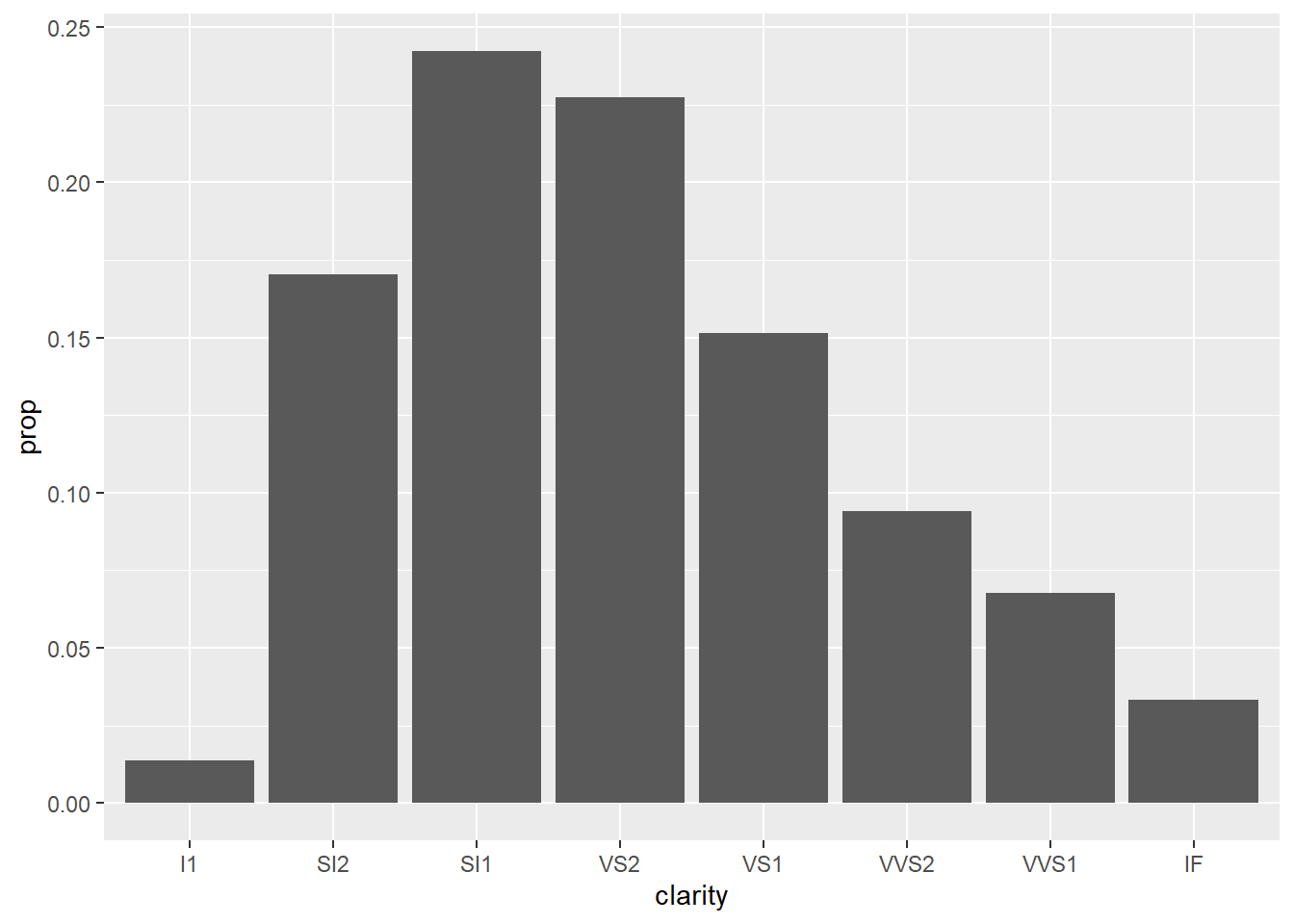
- Scatter plots are most useful when they illustrate a trend in the data, i.e., a way that a change in one variable brings about a change in a related variable. Changes in variables are easier to think about and visualize when the variables are continuous.
Section 1.2.1
- The warning message indicates that you can only map a variable to
shapeif it has 6 or fewer values since looking at more than 6 shapes can be visually confusing. The 62 rows that were removed must be those for whichclass = SUVsinceSUVis the seventh value ofclasswhen listed alphabetically.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, shape = class))## Warning: The shape palette can deal with a maximum of 6 discrete values because more than 6 becomes
## difficult to discriminate; you have 7. Consider specifying shapes manually if you must have
## them.## Warning: Removed 62 rows containing missing values (geom_point).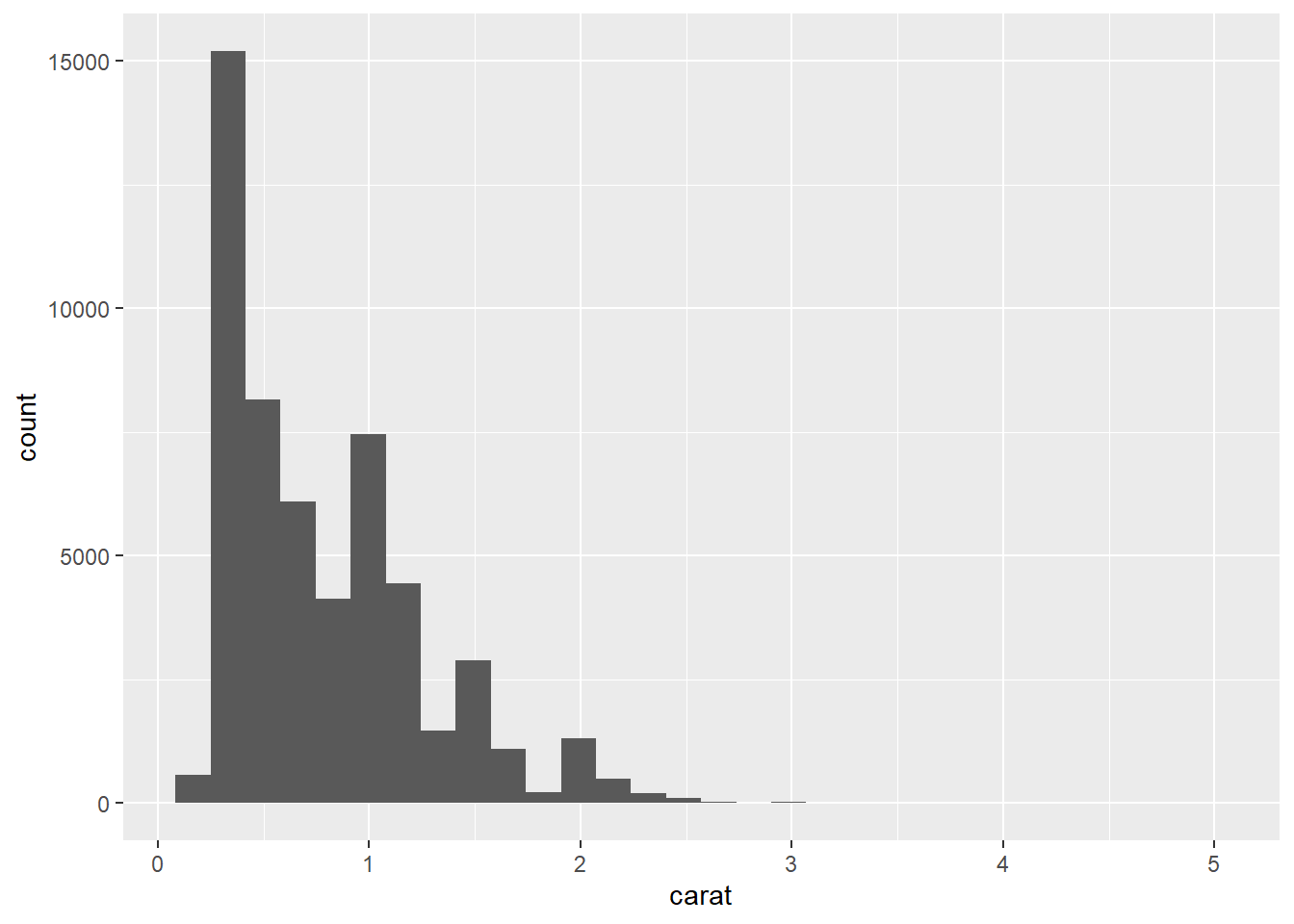
- One example would be to use
drvinstead ofclasssince it only has 3 values:
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, shape = drv))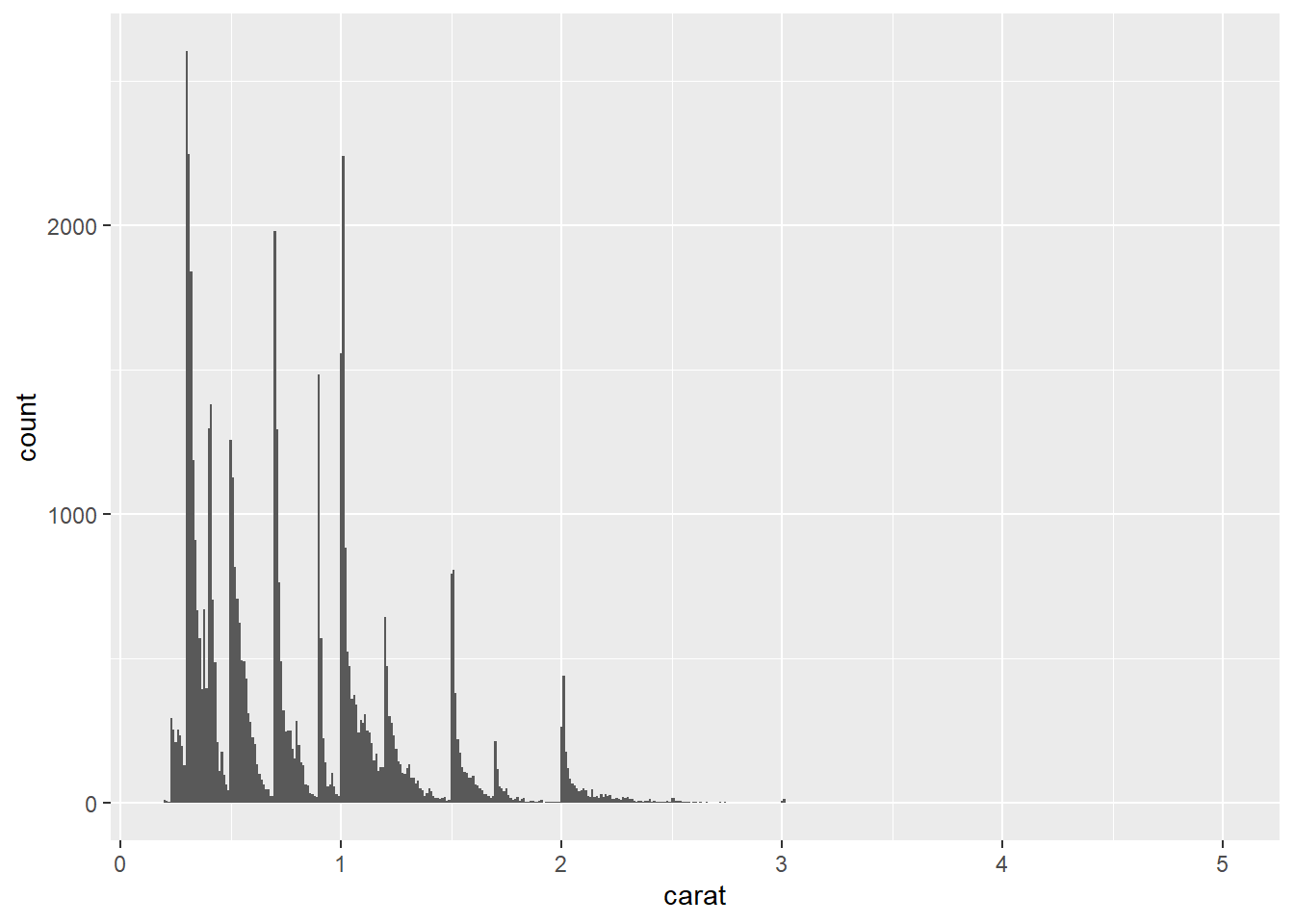
- The scatter plot below shows what happens when the continuous variable
ctyis mapped toshape. The error message indicates that this is not an option.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, shape = cty))## Error in `scale_f()`:
## ! A continuous variable can not be mapped to shape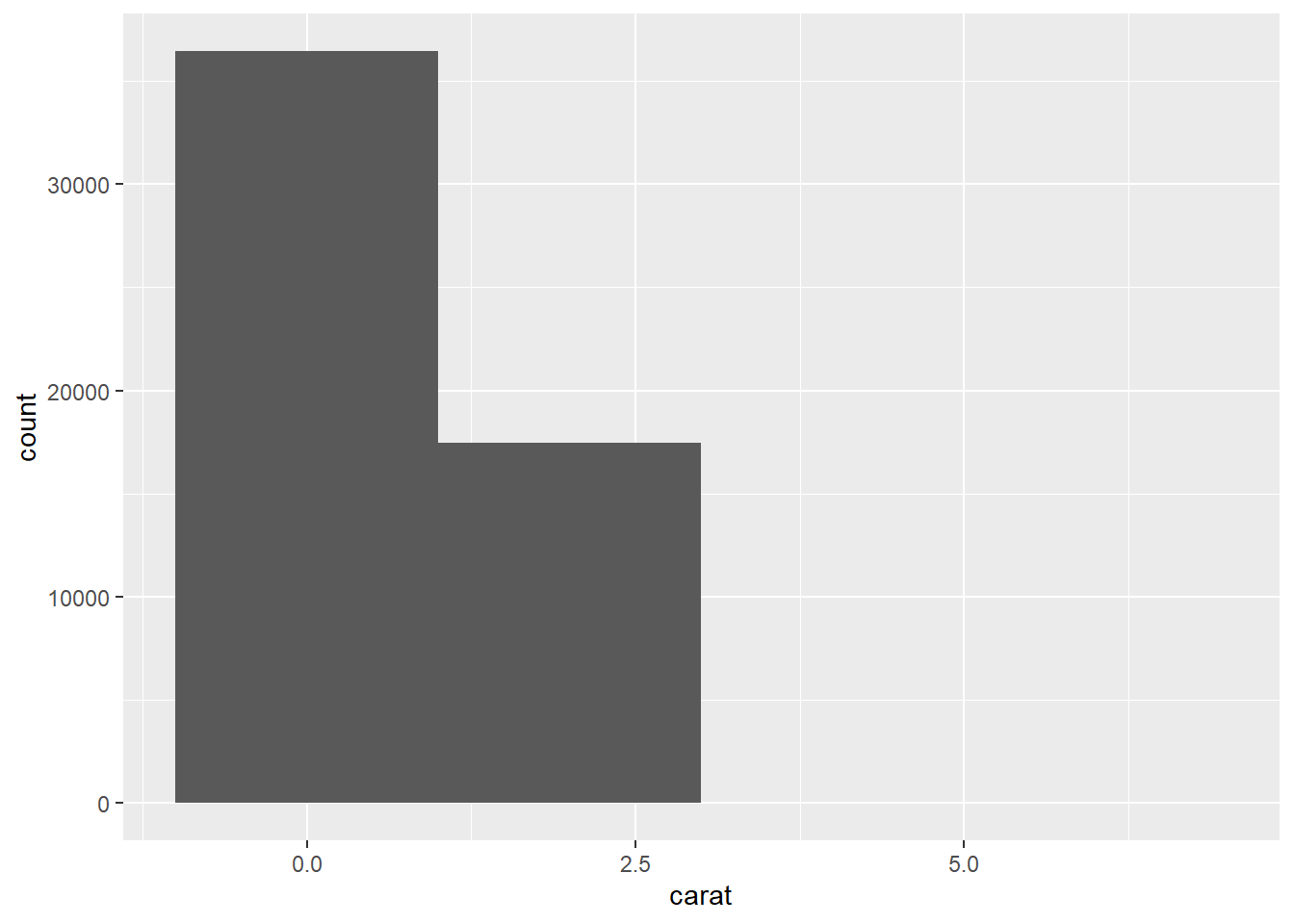
- First, let’s map the continuous
ctytosize. We see that the size of the dots corresponds to the magnitude of the values ofcty. The legend displays representative dot sizes.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, size = cty))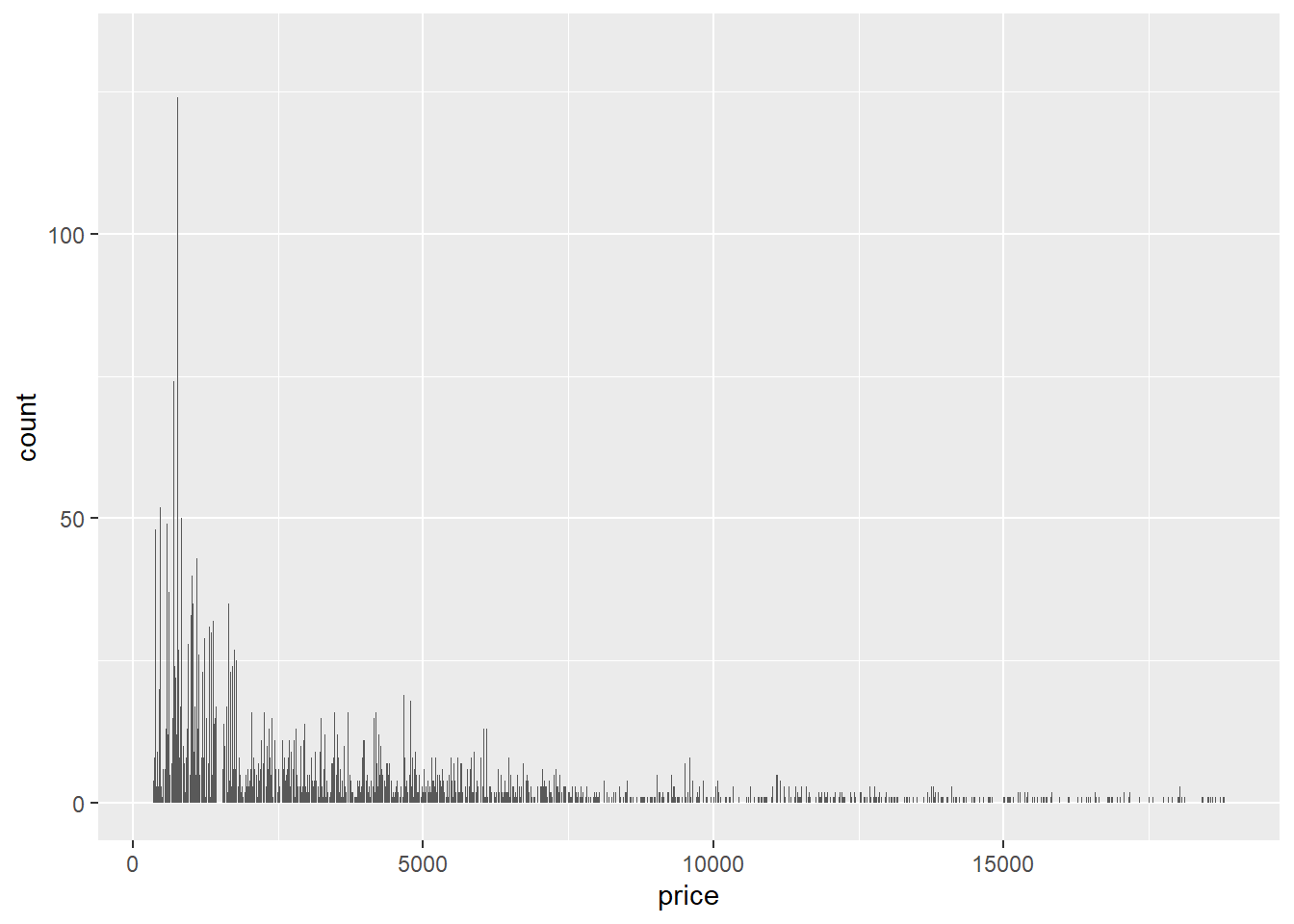
If we map the categorical drv to size, we get a warning message that indicates that size is not an appropriate aesthetic for categorical variables. This is because such variables usually have no meaningful notion of magnitude, so the size aesthetic can be misleading. For example, in the plot below, one might get the idea that rear-wheel drives are somehow “bigger” than front-wheel and 4-wheel drives.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, size = drv))## Warning: Using size for a discrete variable is not advised.
- Mapping the continuous
ctytocolor, we see that a gradation of colors is assigned to the continuous interval ofctyvalues.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = cty))
- The scatter plot below maps
classtocoloranddrvtoshape. The result is a visualization which is somewhat hard to read. Generally speaking, mapping to more than one extra aesthetic crams a little too much information into a visualization.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = class, shape = drv))
- The scatter plot below maps the
drvvariable to bothcolorandshape. Mappingdrvto two different aesthetics is a good way to further differentiate the scatter plot points that represent the differentdrvvalues.
ggplot(data = mpg) +
geom_point(mapping = aes(x = displ, y = hwy, color = drv, shape = drv))
- This technique gives us a way to visually separate a scatter plot according to a given condition. Below, we can visualize the relationship between
displandhwyseparately for cars with fewer than 6 cylinders and those with 6 or more cylinders.
ggplot(mpg) +
geom_point(aes(displ, hwy, color = cyl<6))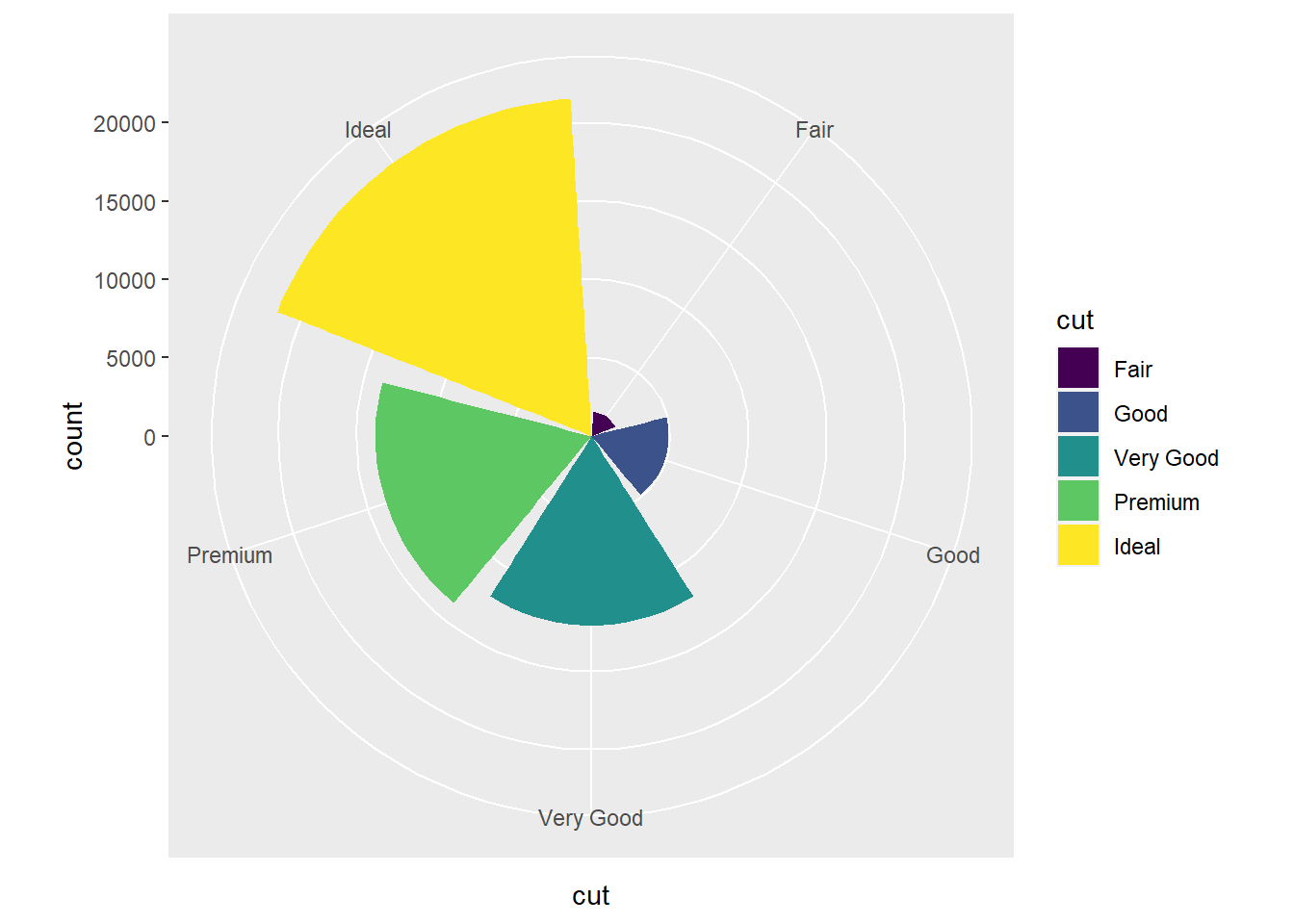
Section 1.4.1
- The available aesthetics are found in the documentation by entering
?geom_smooth. The visualizations below mapdrvtolinetypeandsize.
ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy, linetype = drv))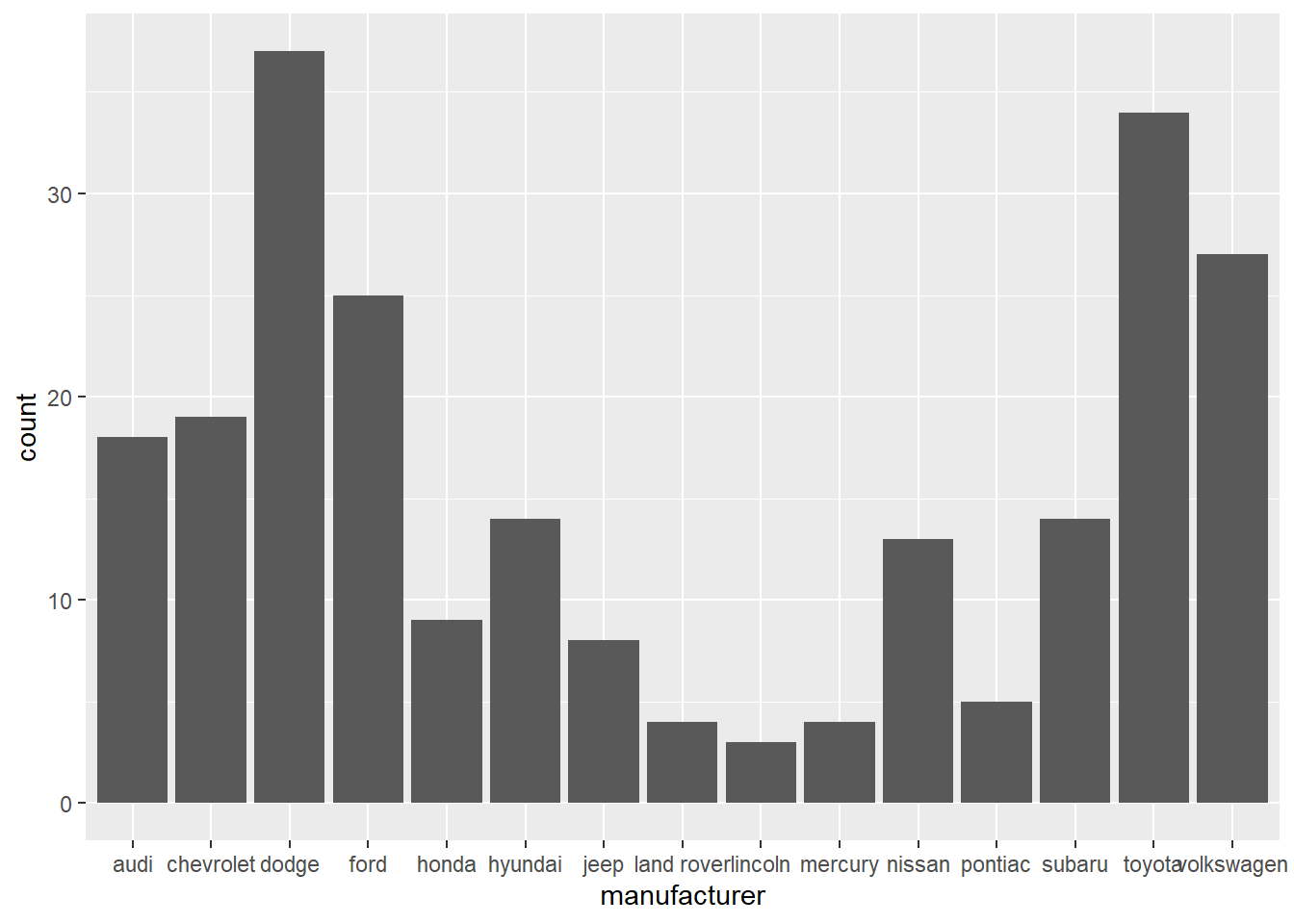
ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy, size = drv))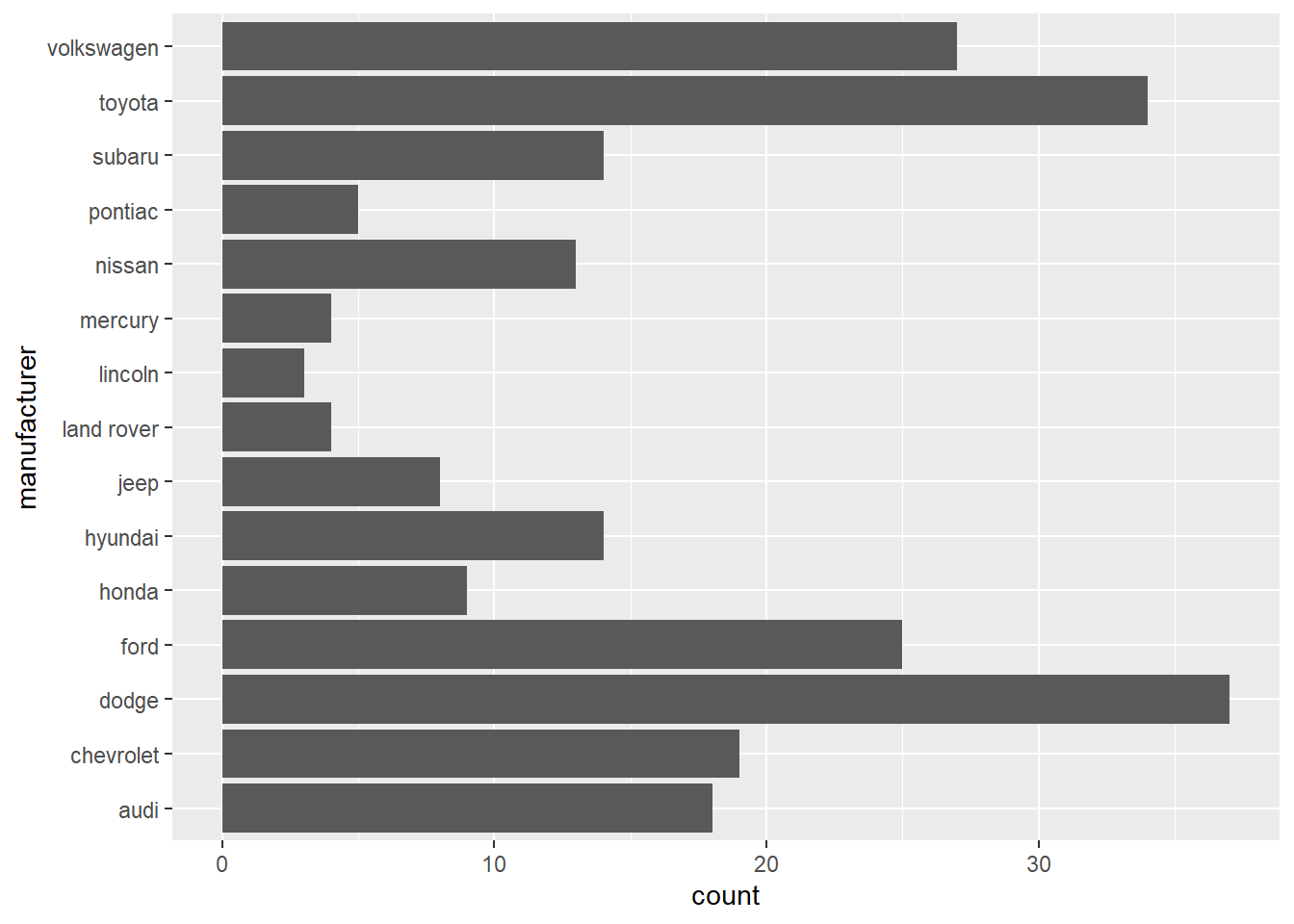
ggplot(data = mpg, mapping = aes(x = cty, y = hwy)) +
geom_point(mapping = aes(color = drv)) +
geom_smooth(se = FALSE) +
labs(title = "The Relationship Between Highway and City Fuel Efficiency",
x = "City Fuel Efficiency (miles per gallon)",
y = "Highway Fuel Efficiency (miles per gallon)",
color = "Drive Train Type")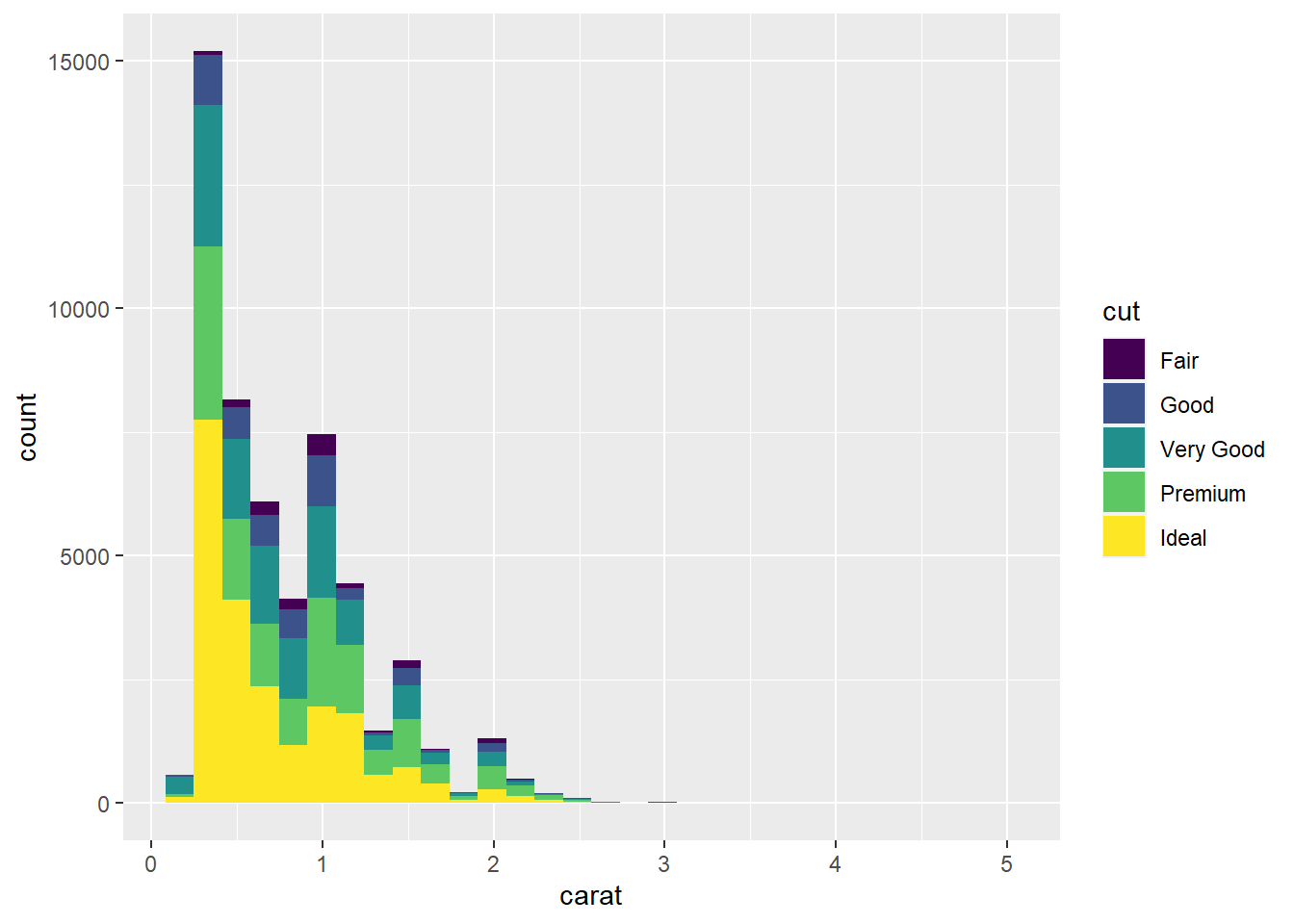
- The visualization below shows that the
coloraesthetic should probably not be used with a categorical variable with several values since it leads to a very cluttered plot.
ggplot(data = mpg) +
geom_smooth(mapping = aes(x = displ, y = hwy, color = class))
Section 1.6.1
- Since
caratandpriceare both continuous, a scatter plot is appropriate. Notice below thatpriceis the y-axis variable, andcaratis the x-axis variable. This is becausecaratis an independent variable (meaning that it’s not affected by the other variables) andpriceis a dependent variable (meaning that it is affected by the other variables).
ggplot(data = diamonds) +
geom_point(mapping = aes(x = carat, y = price))
- Since
clarityis categorical, a better way to visualize its affect onpriceis to use a box plot.
ggplot(data = diamonds) +
geom_boxplot(mapping = aes(x = clarity, y = price))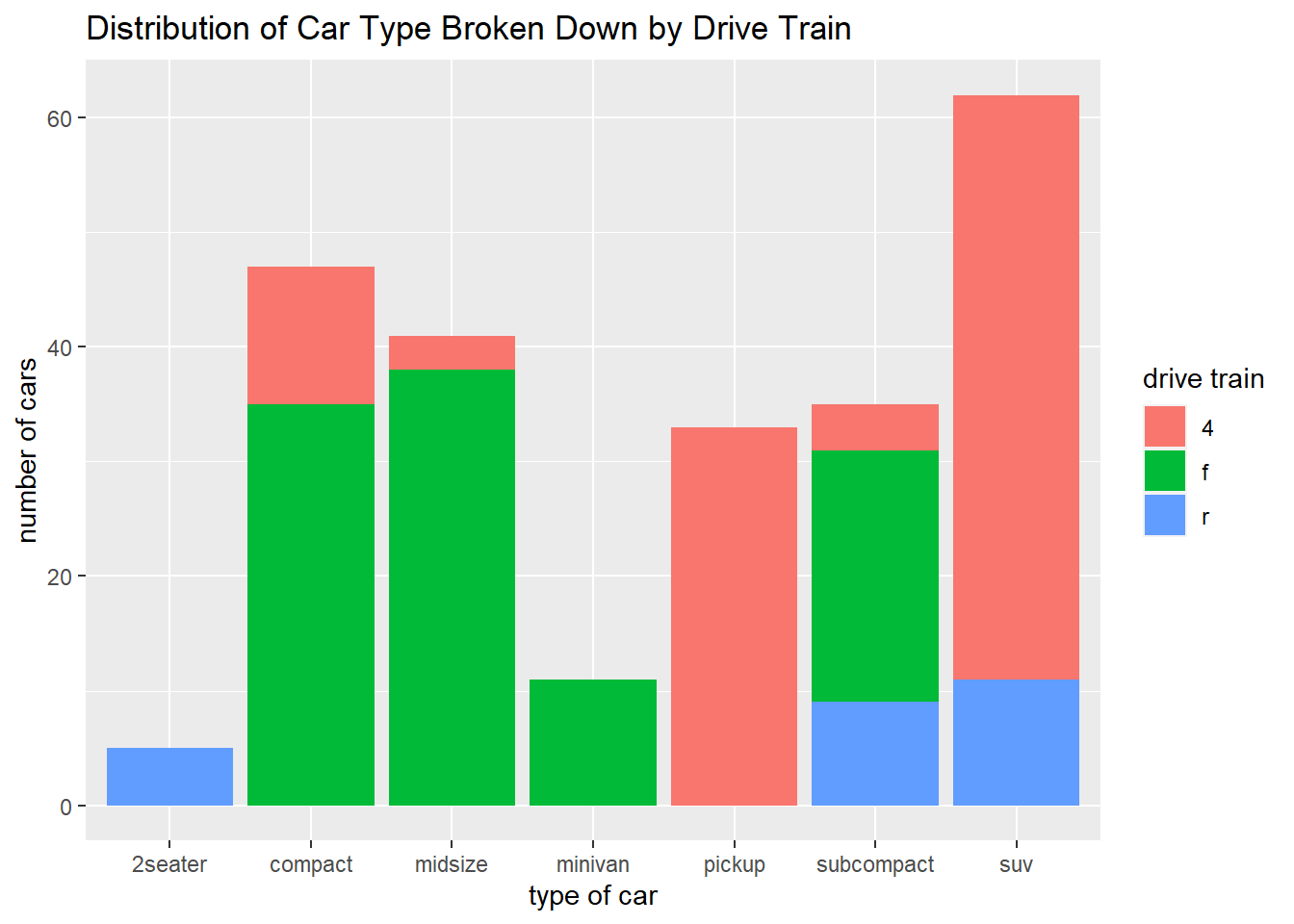
Judging by the median value of
price, it looks likeSI2is the highest priced clarity value. You could also make an argument forVS2orVS1since they have the highest values for Q3.VS2andVS1clarities also seem to have the most variation inpricesince they have the largest interquartile ranges.The fact that, for any given
clarityvalue, all of the outliers lie above the main cluster indicates that the data set contains several diamonds with extremely high prices that are much larger than the median prices. This is often the case with valuable commodities - there are always several items whose values are extremely high compared to the median. (Think houses, baseball cards, cars, antiques, etc.)The
fillaesthetic seems to best among the three at visually distinguishing the values ofcut.
ggplot(data = diamonds) +
geom_boxplot(mapping = aes(x = color, y = price, color = cut))
ggplot(data = diamonds) +
geom_boxplot(mapping = aes(x = color, y = price, fill = cut))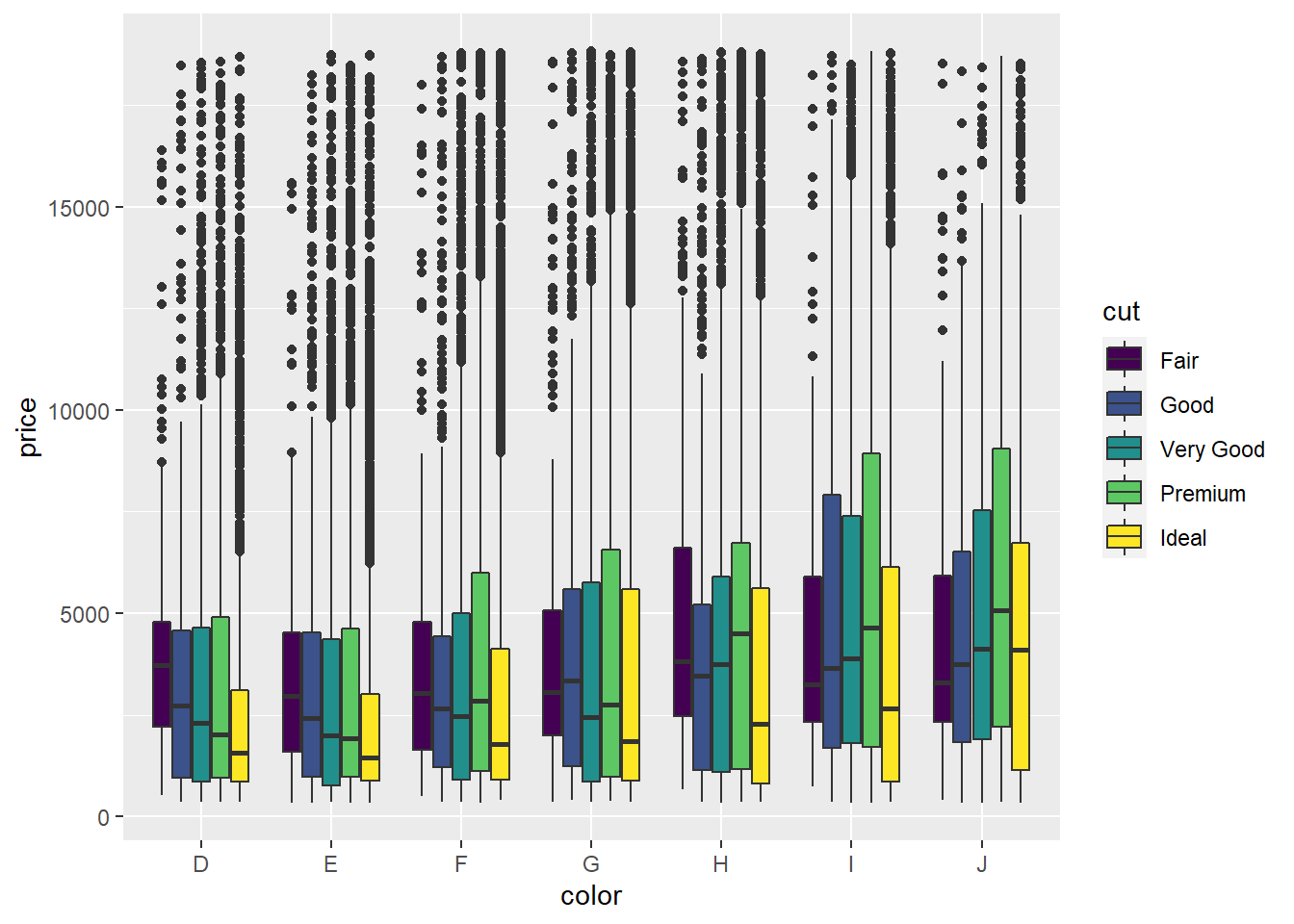
ggplot(data = diamonds) +
geom_boxplot(mapping = aes(x = color, y = price, linetype = cut))
ggplot(data = diamonds) +
geom_boxplot(mapping = aes(x = reorder(clarity, price), y = price)) +
coord_flip()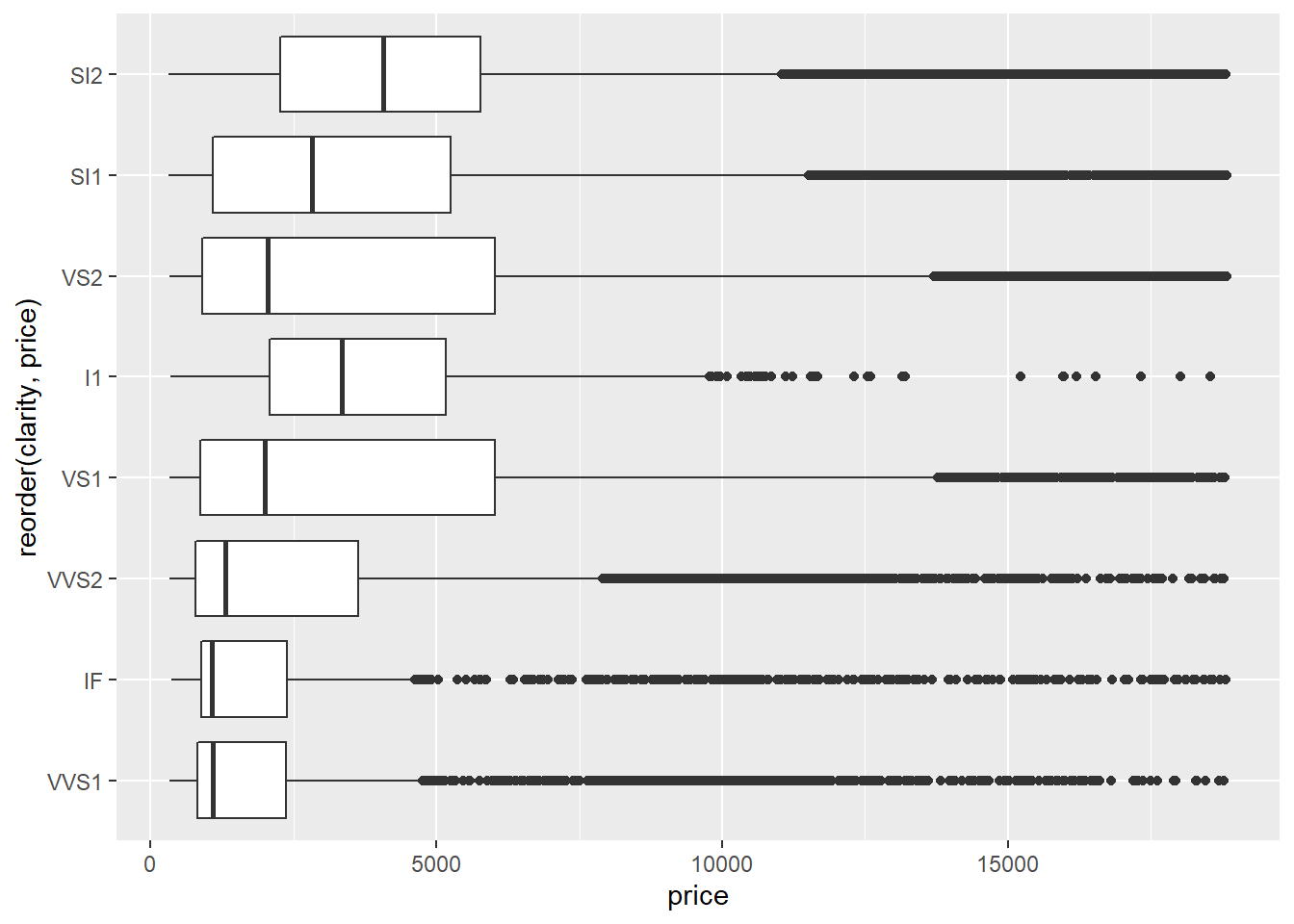
Section 1.7.1
- Since
clarityis categorical, we should use a bar graph.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = clarity))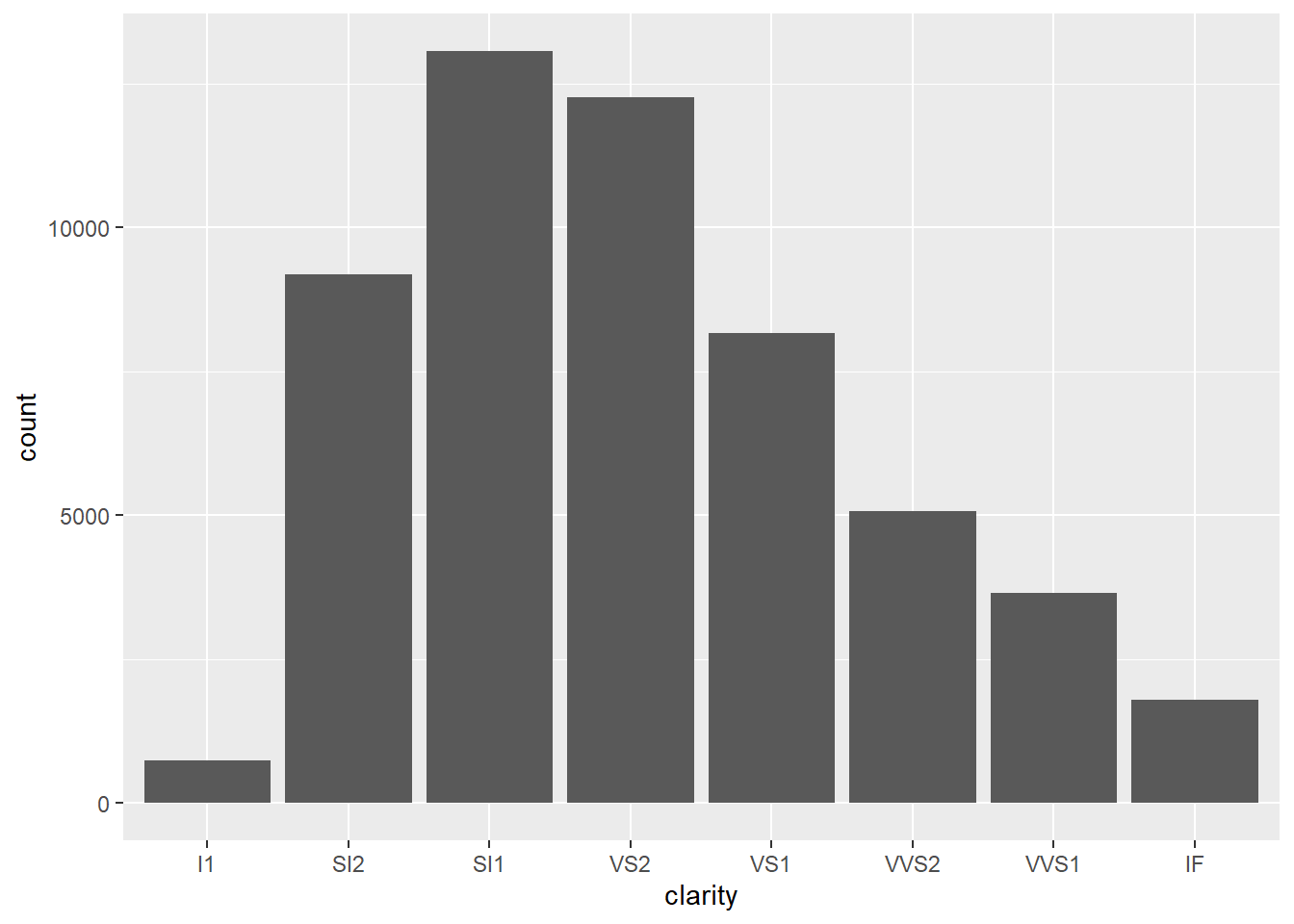
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = clarity, y = stat(prop), group = 1))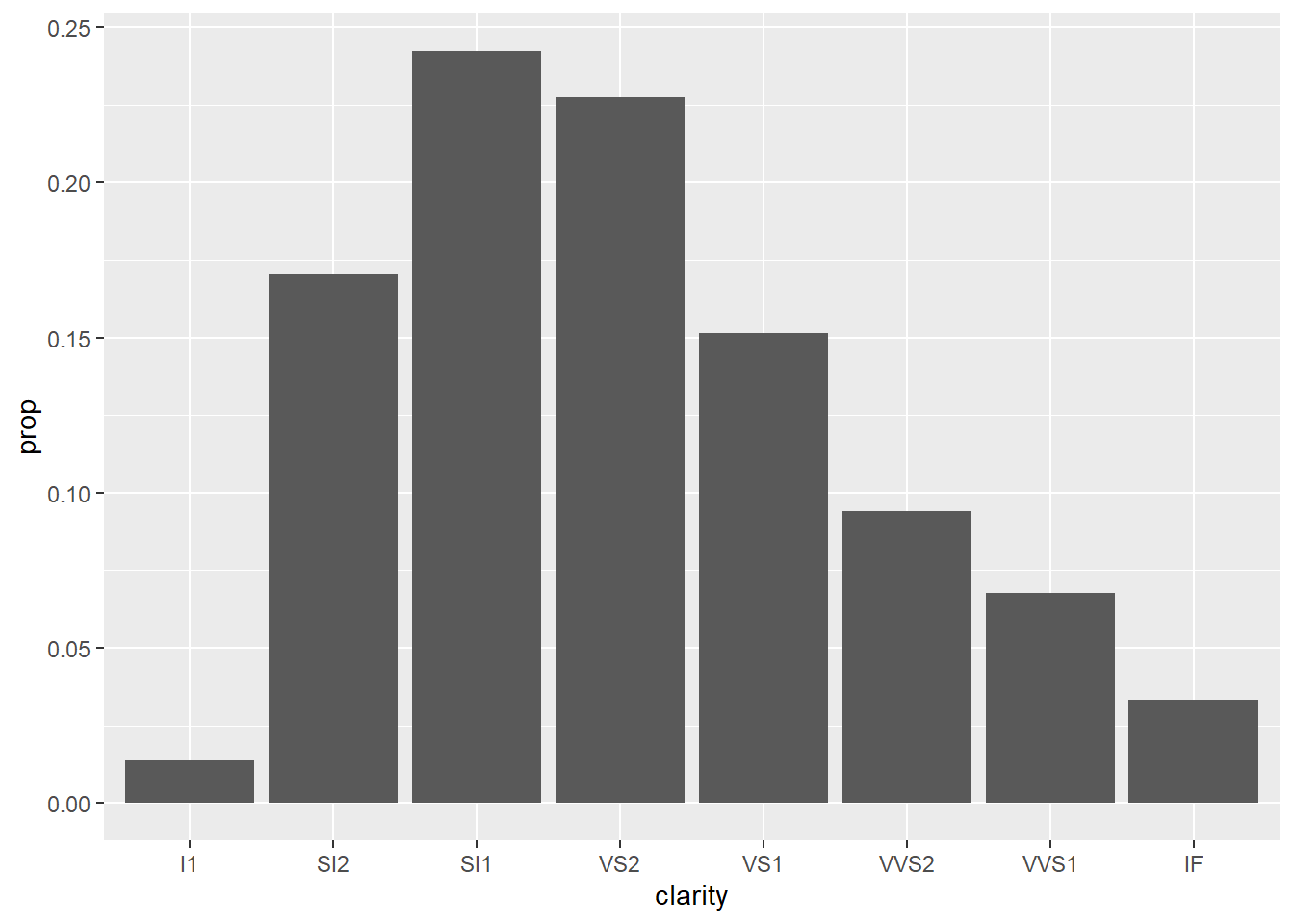
- Since
caratis continuous, we should now use a histogram.
ggplot(data = diamonds) +
geom_histogram(mapping = aes(x = carat))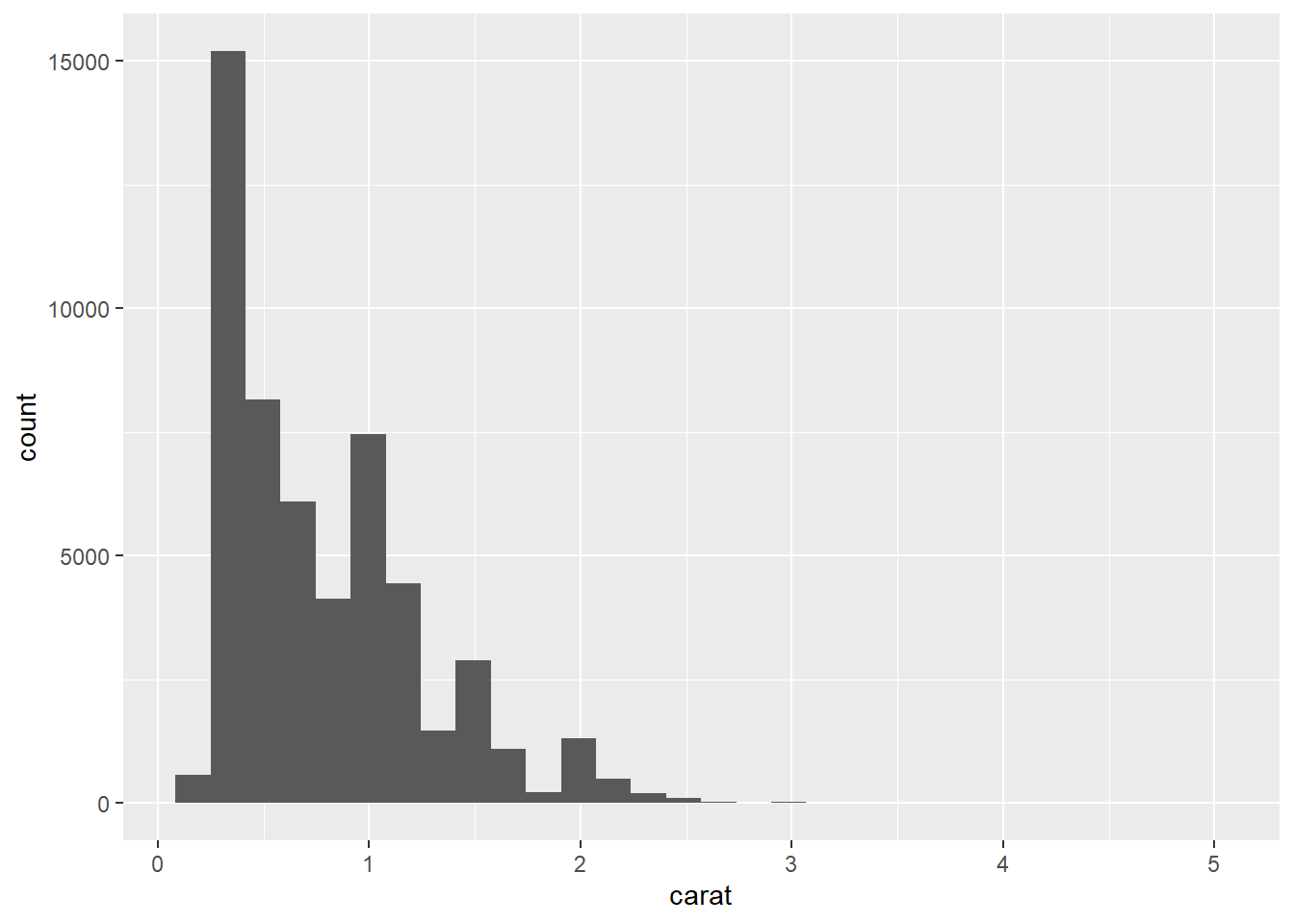
- The first histogram below has bins that are way too narrow. The histogram is too granular to visualize the true distribution. The second one has bins which are way too wide, producing a histogram that is too coarse.
ggplot(data = diamonds) +
geom_histogram(mapping = aes(x = carat), binwidth = 0.01)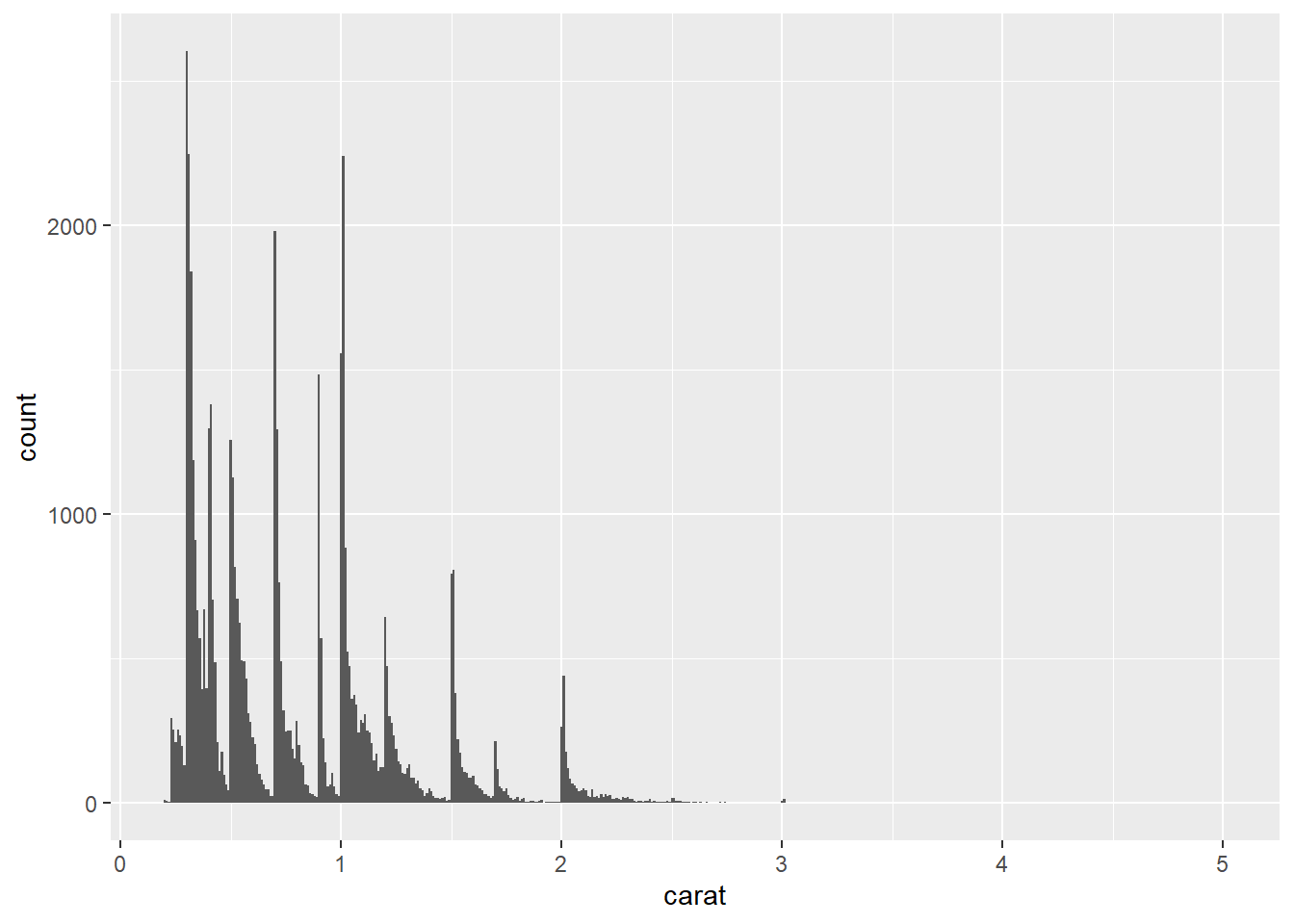
ggplot(data = diamonds) +
geom_histogram(mapping = aes(x = carat), binwidth = 2)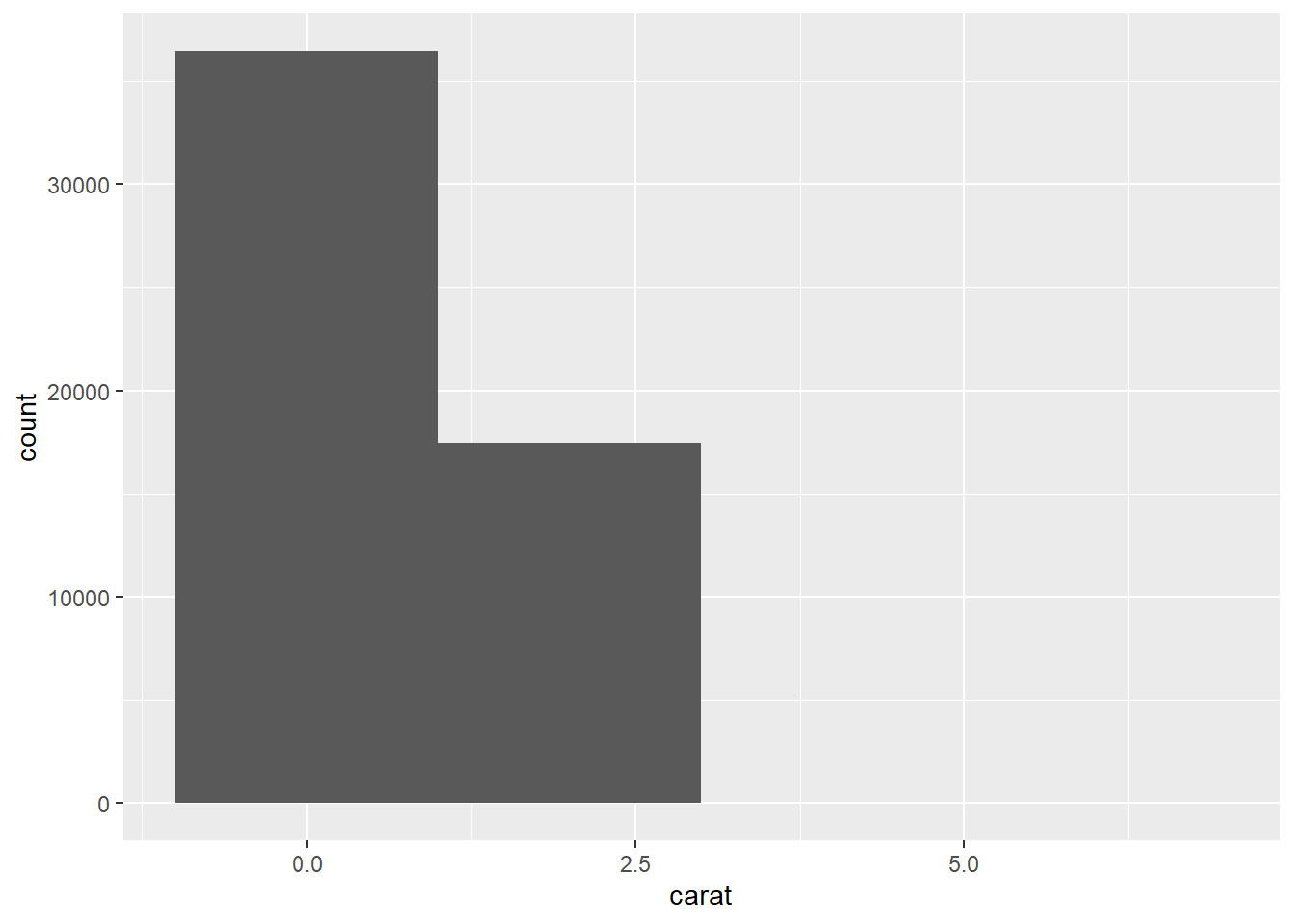
- The following bar graph looks so spiky because it’s producing a bar for every single value of
pricein the data set, of which there are hundreds, at least. Sincepriceis continuous, a histogram would have been the way to visualize its distribution.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = price))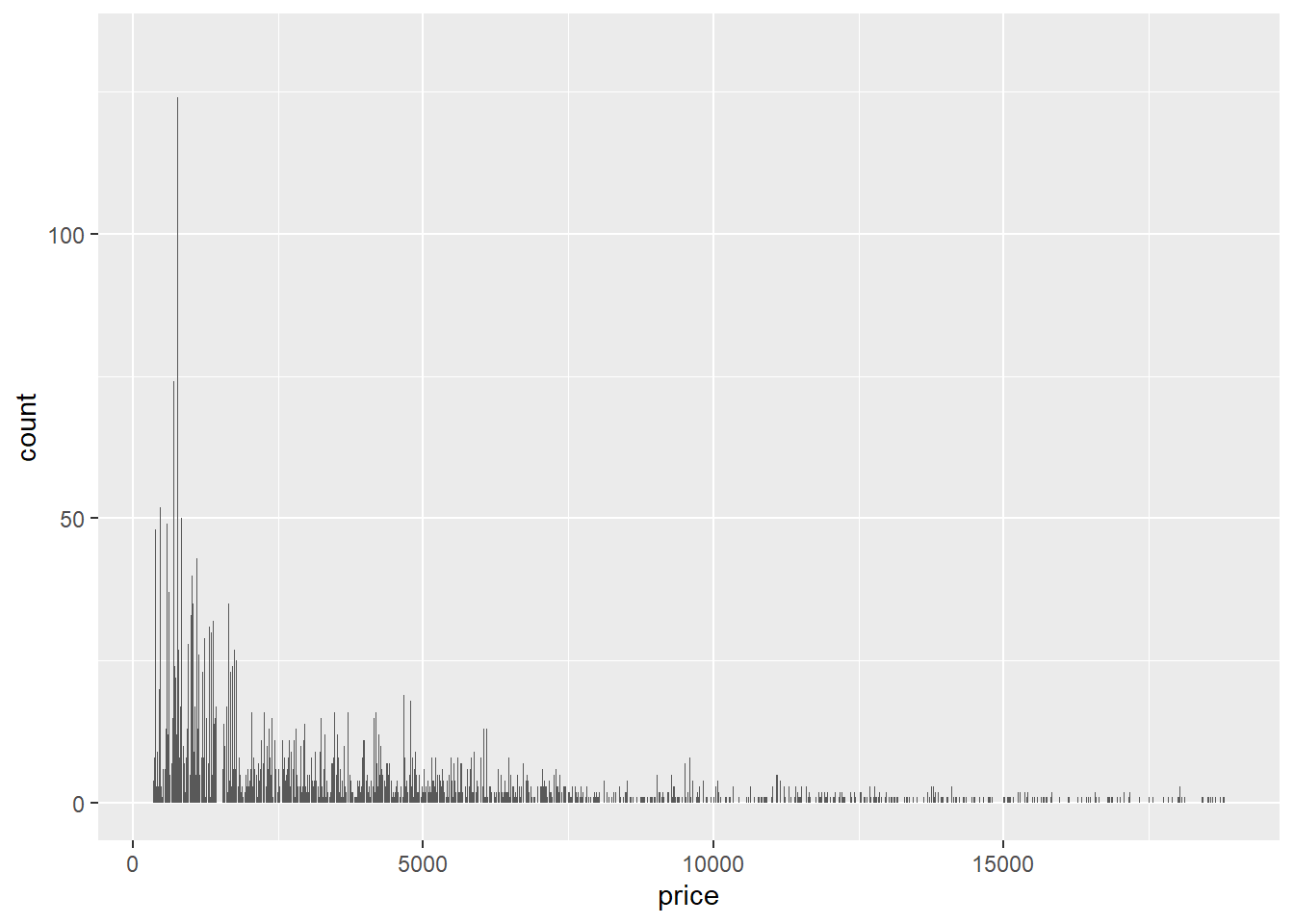
- Mapping
cuttofillproduces a much better result than mapping tocolor.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = clarity, color = cut))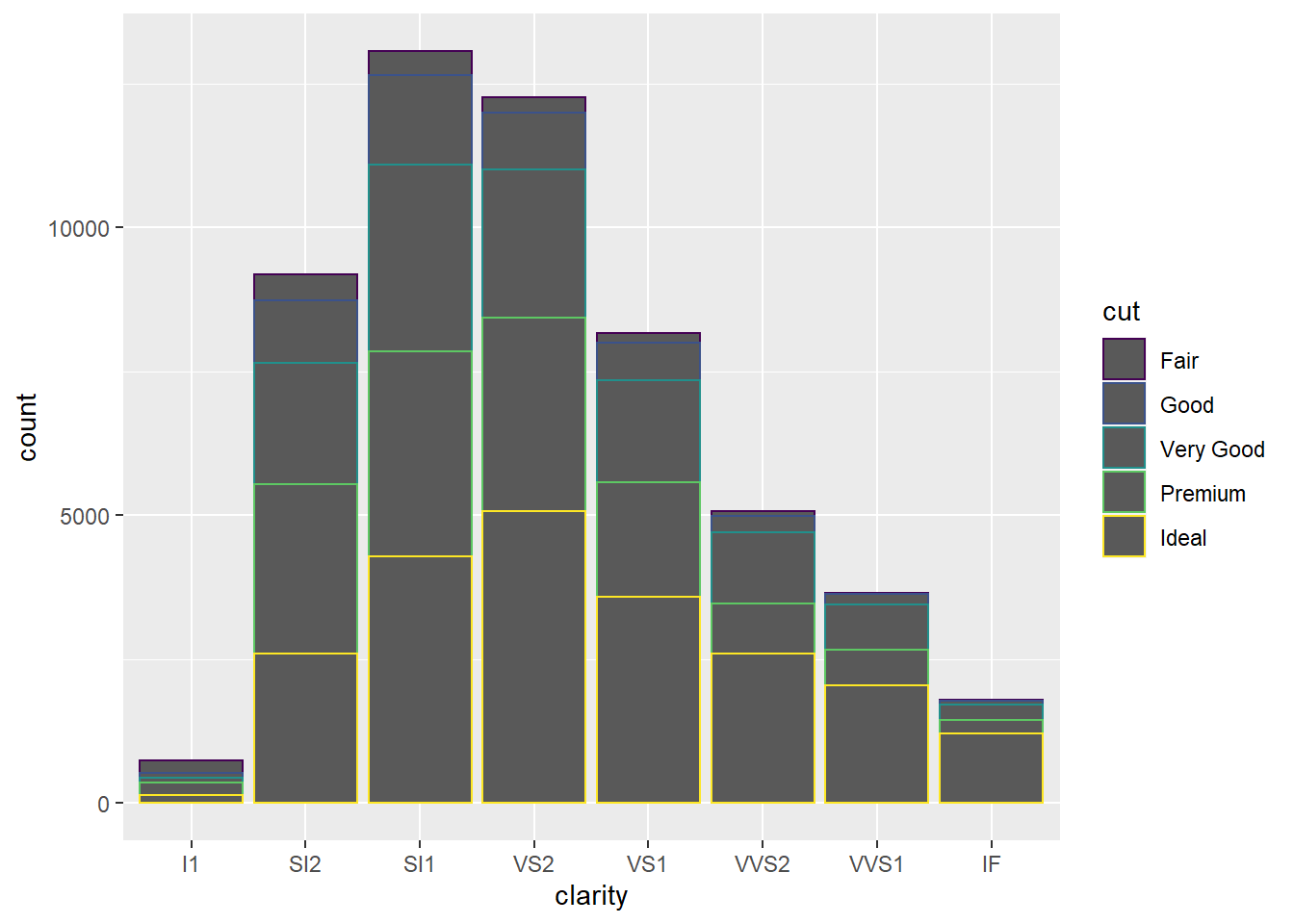
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = clarity, fill = cut))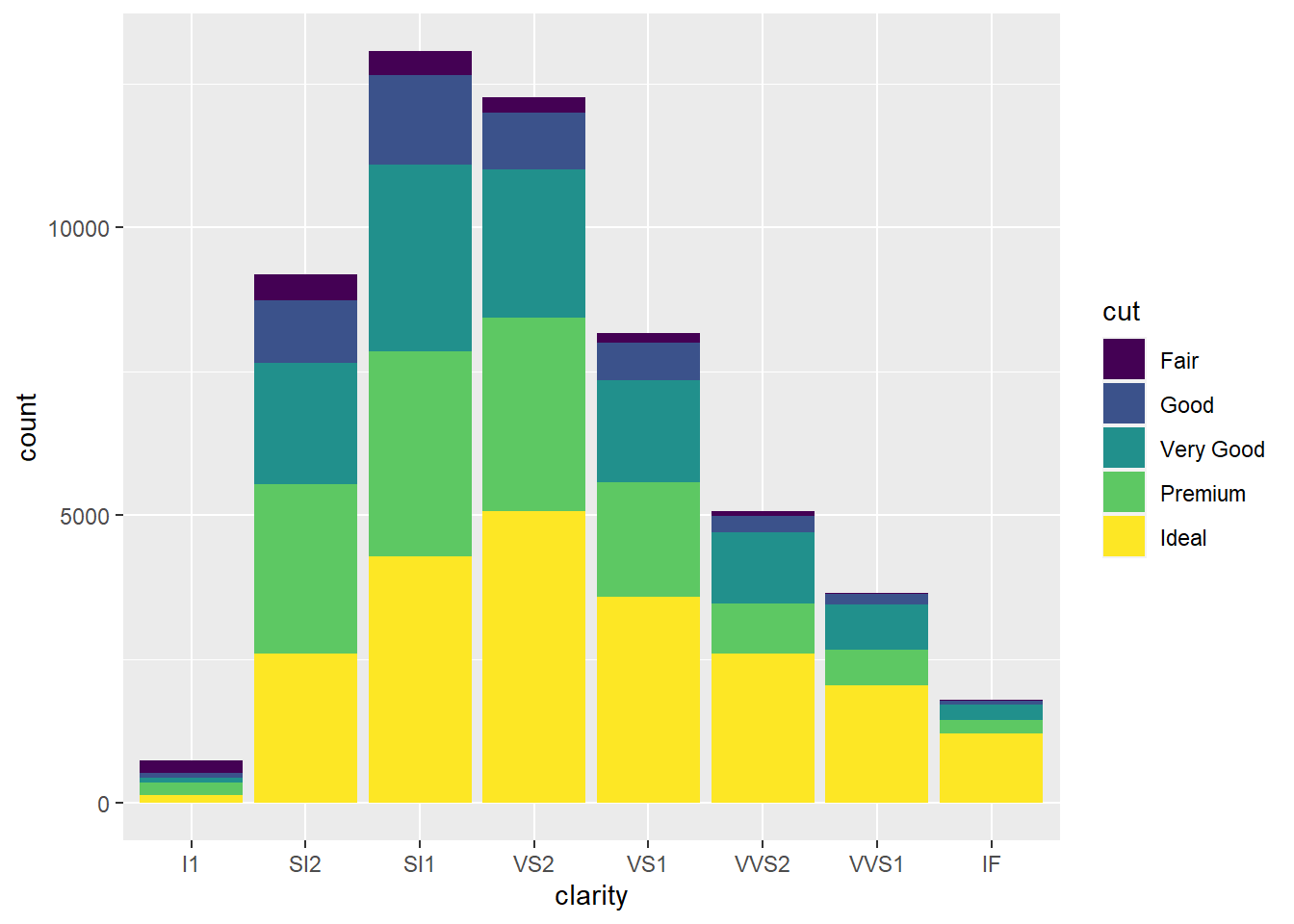
- Since using the optional
position = "dodge"argument produces a bar for eachcutvalue within a givenclarityvalue, it’s easier to see howcutvalues are distributed within eachclarityvalue since it allows you to compare relative counts more easily.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = clarity, fill = cut), position = "dodge")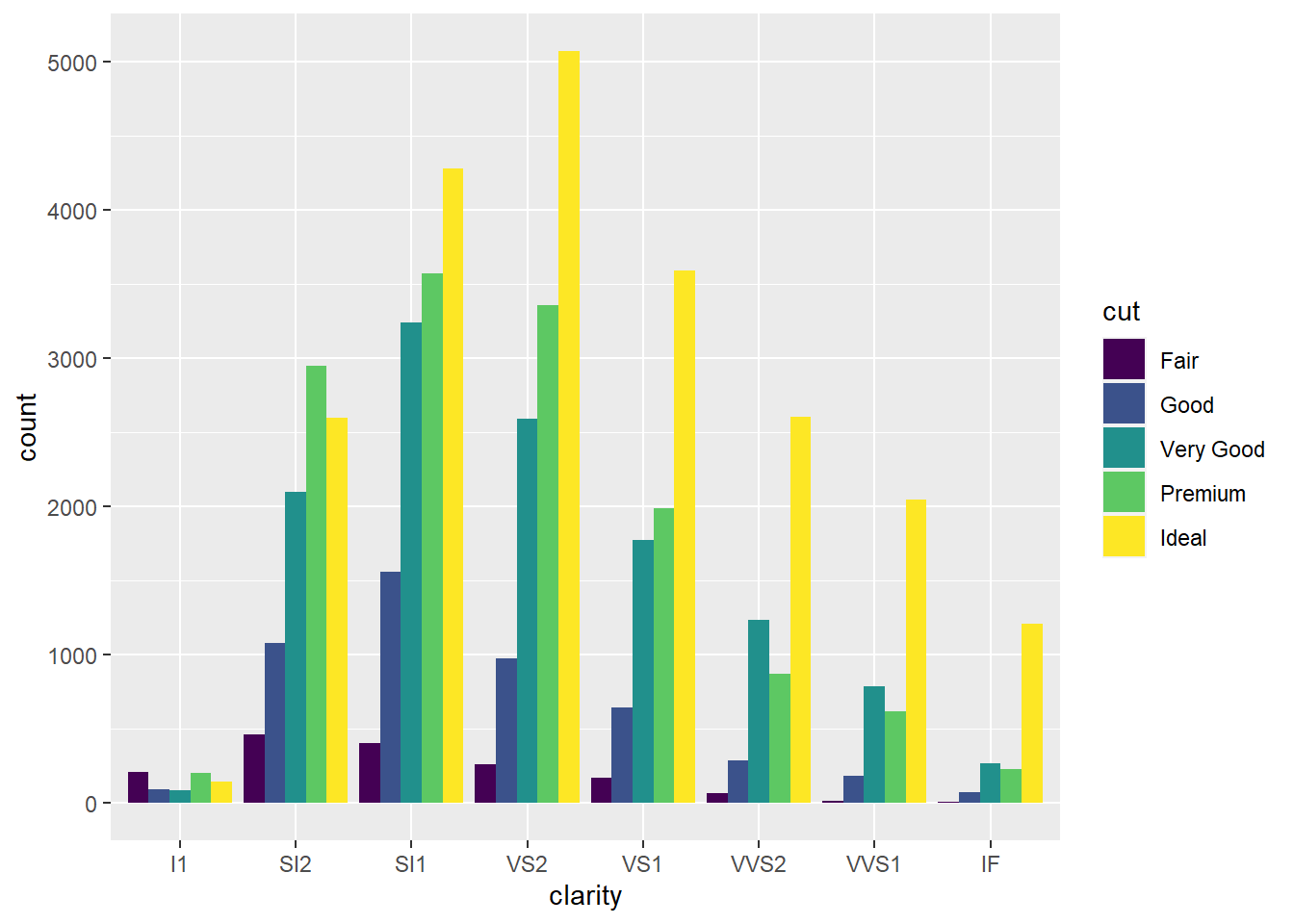
- Mapping
cutto two different aesthetics is redundant, but doing so makes it easier to distinguish the bars for the variouscutvalues. (See Exercise 7 from Section 1.2.1.)
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, fill = cut))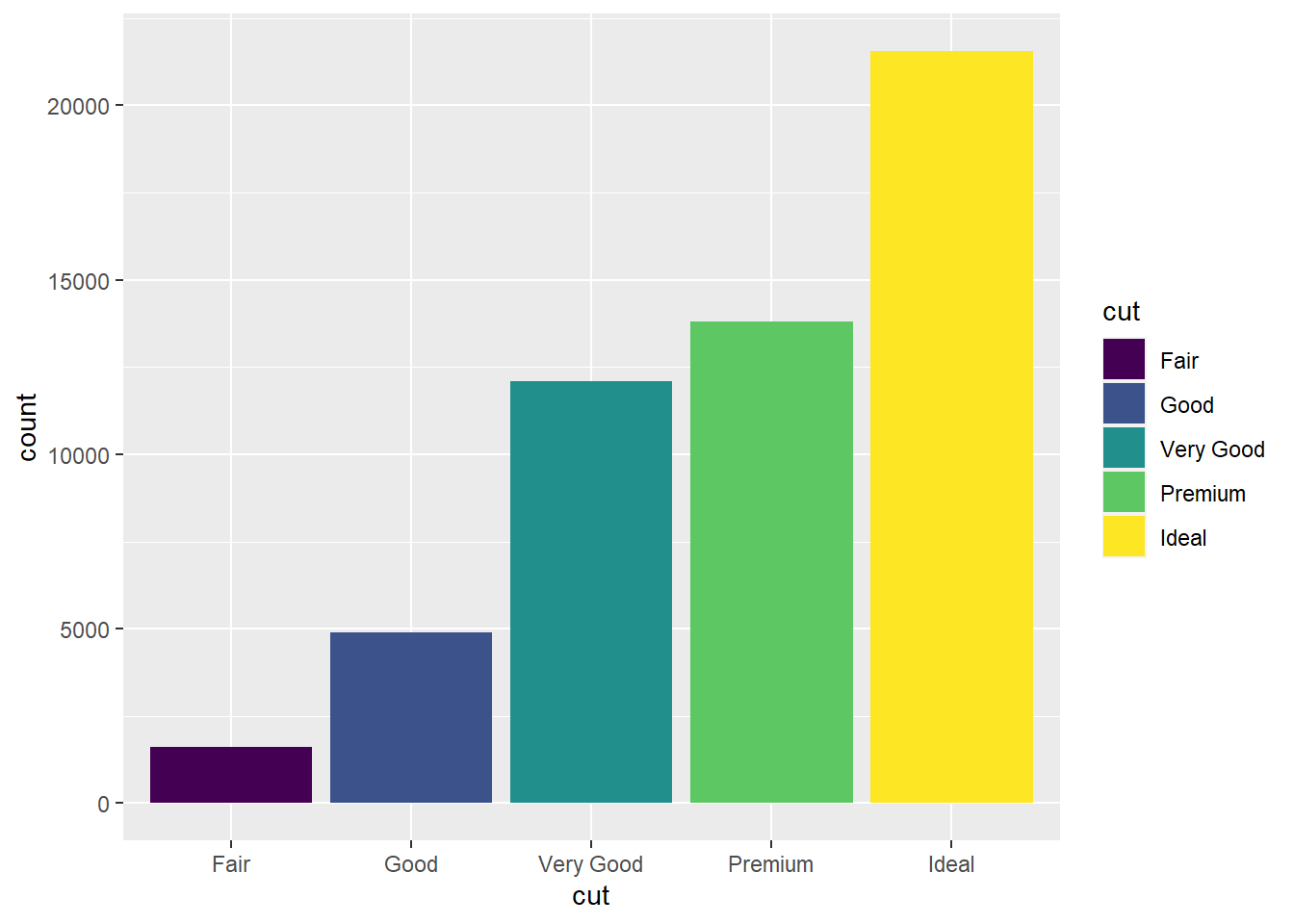
- You get a pie chart, where the radius of each piece of pie corresponds to the count for that
cutvalue.
ggplot(data = diamonds) +
geom_bar(mapping = aes(x = cut, fill = cut)) +
coord_polar()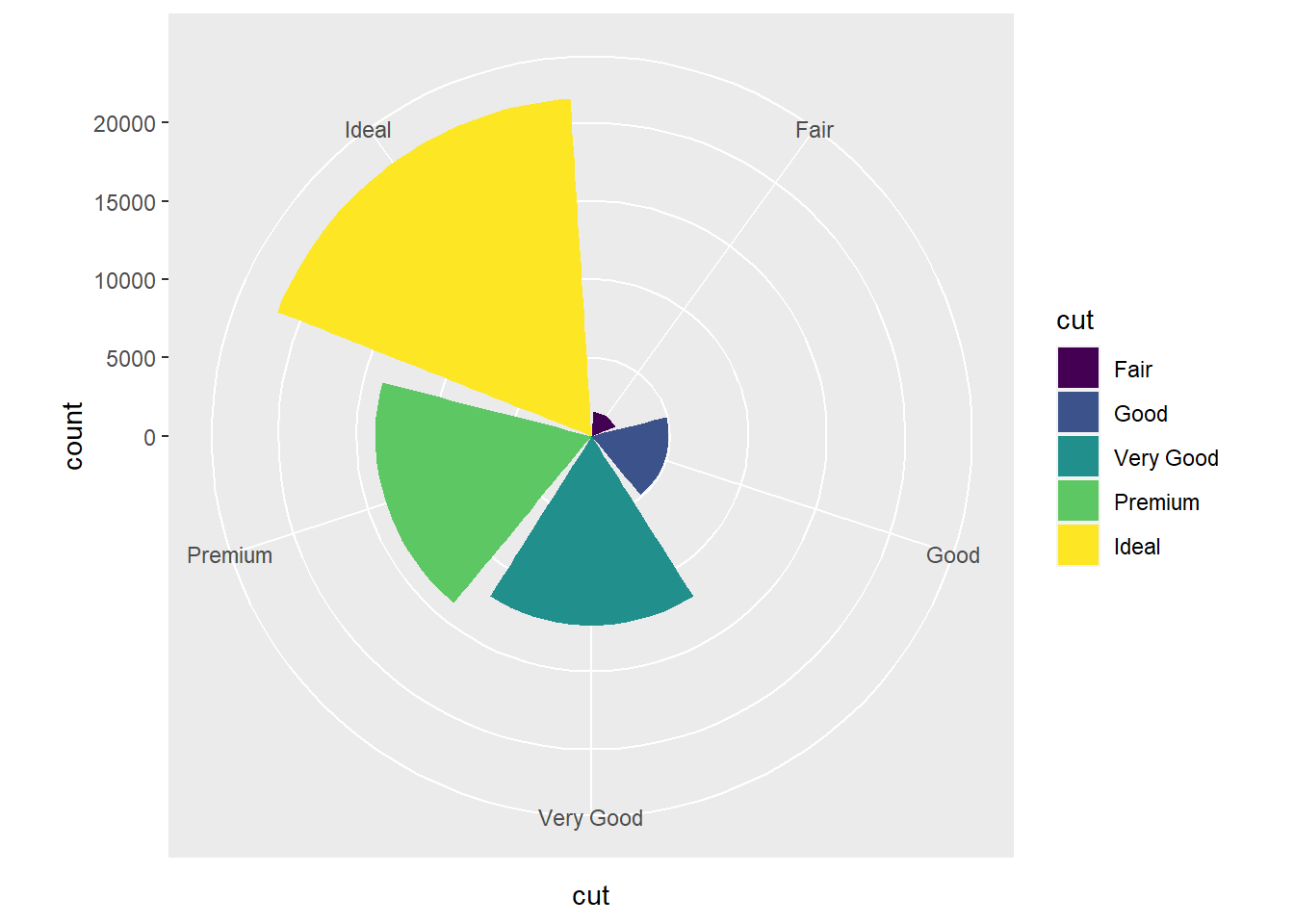
- In the first bar graph below, the names of the
manufacturervalues are crowded together on the x-axis, making them hard to read. The second bar graph fixes this problems by adding acoord_flip()layer.
ggplot(data = mpg) +
geom_bar(mapping = aes(x = manufacturer))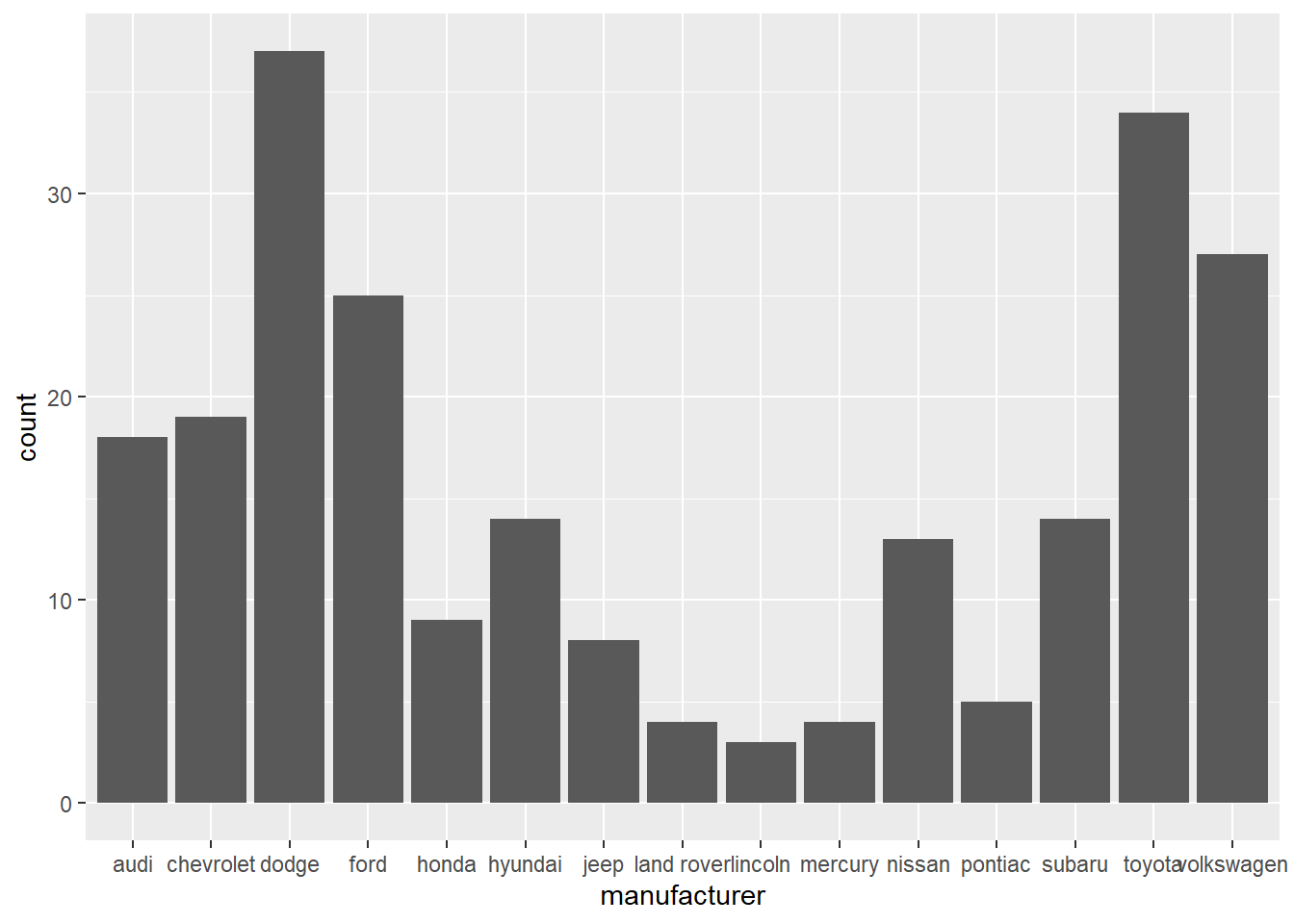
ggplot(data = mpg) +
geom_bar(mapping = aes(x = manufacturer)) +
coord_flip()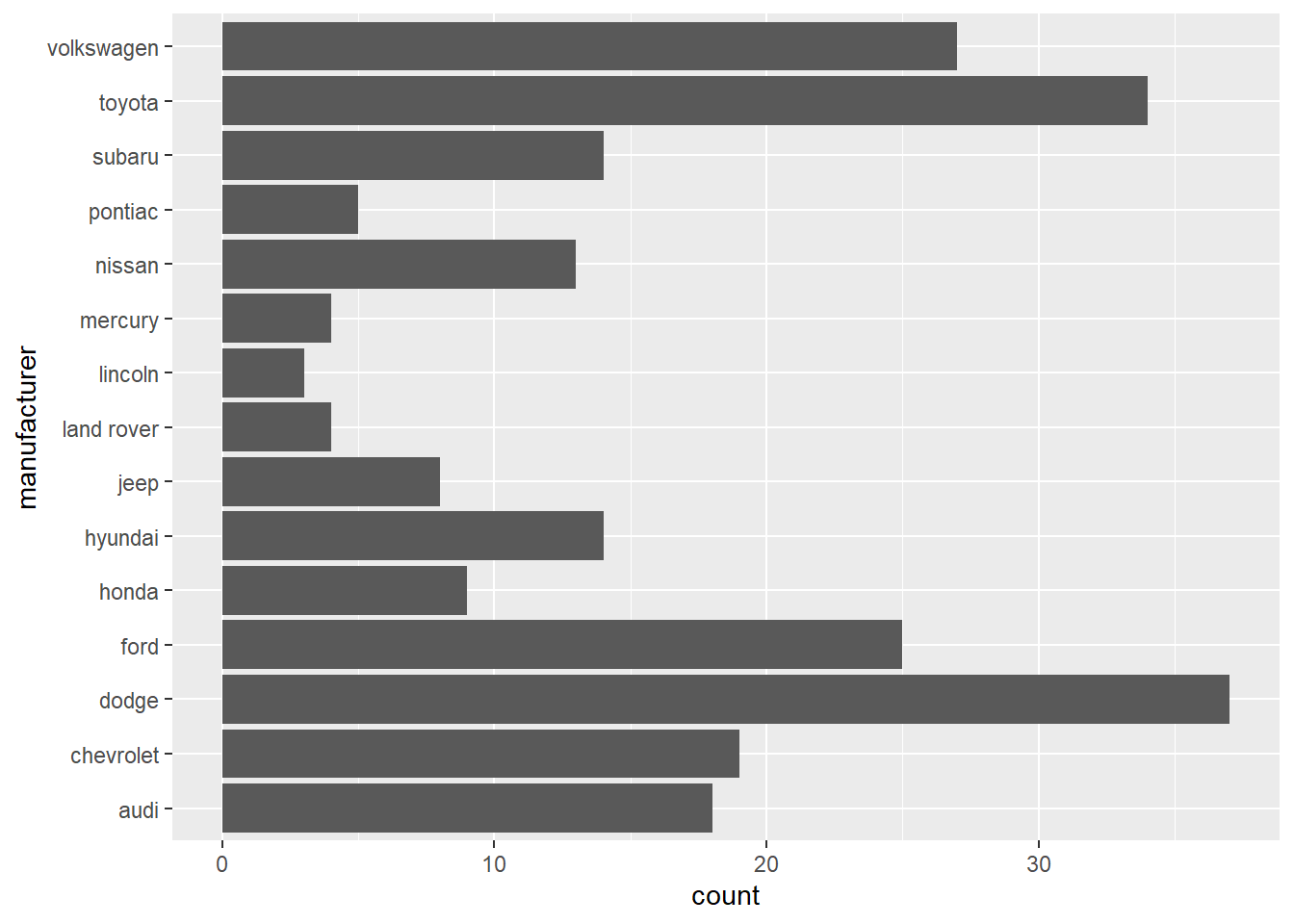
ggplot(data = diamonds) +
geom_histogram(mapping = aes(x = carat, fill = cut))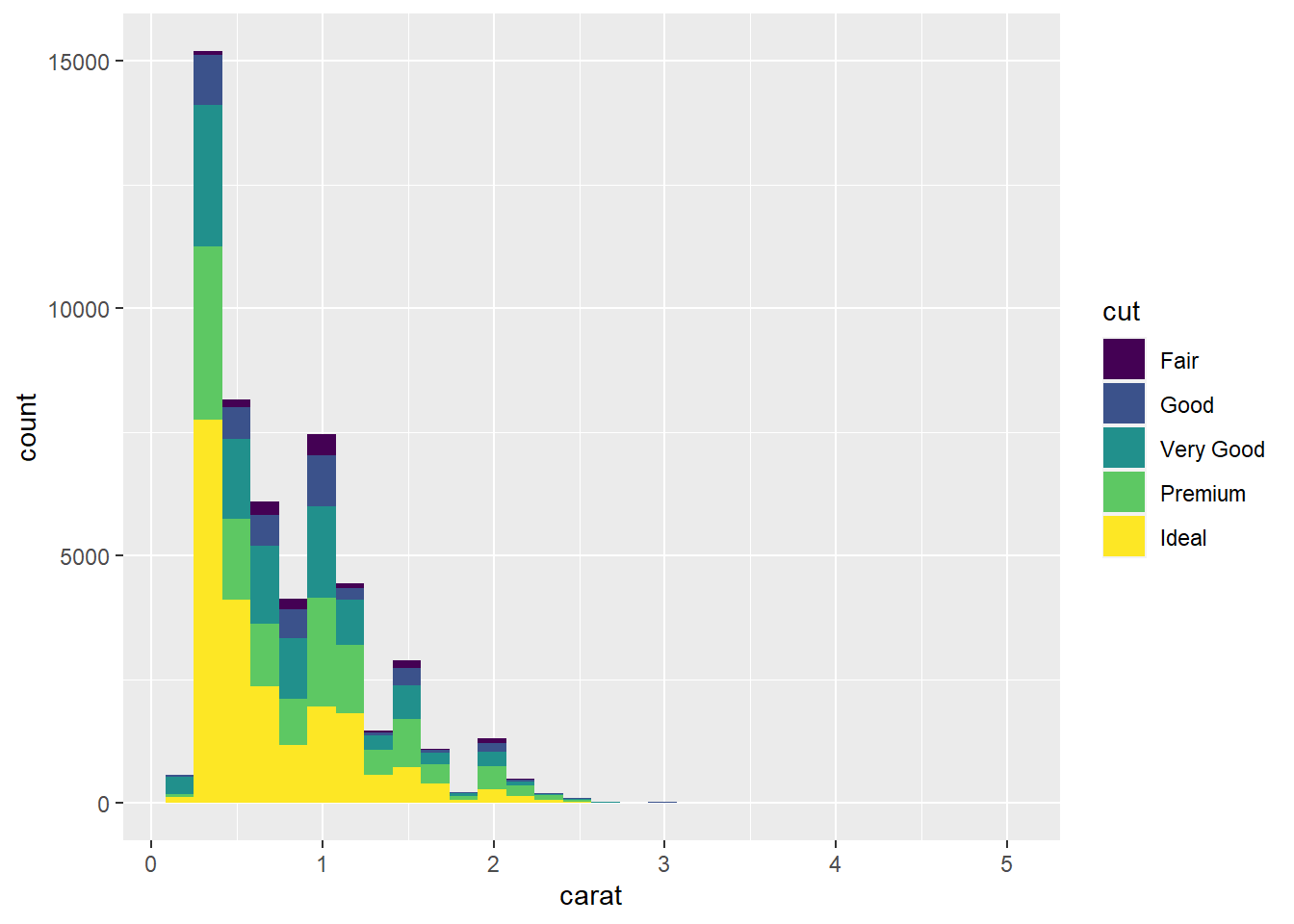
- A frequency polynomial contains the same information as a histogram but displays it in a more minimal way.
ggplot(data = diamonds) +
geom_freqpoly(mapping = aes(x = price))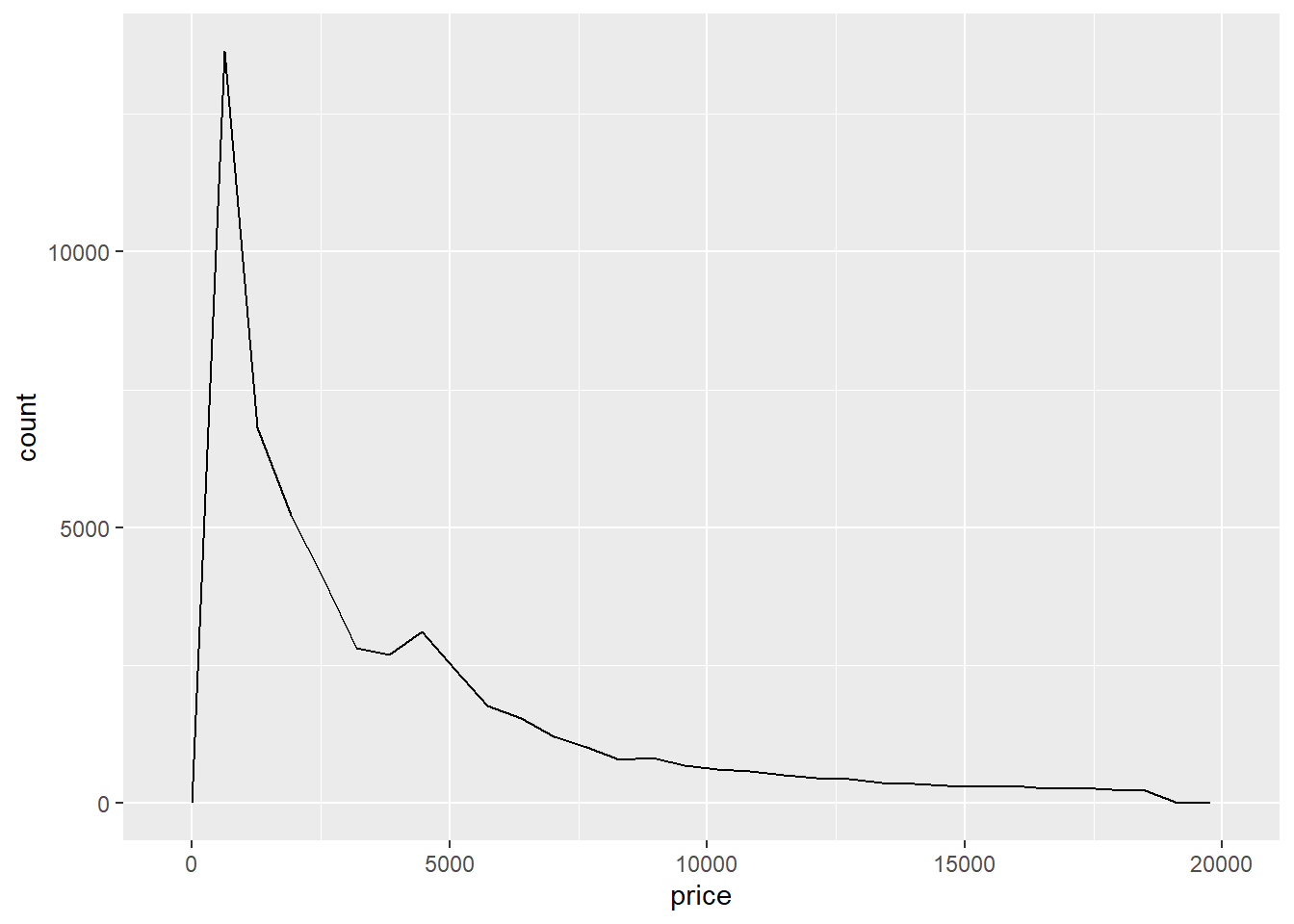
- First, you may have noticed that mapping
cutto fill, as was done in Exercise 11, is not as helpful ingeom_freqpoly. Mapping tocolorproduces a nice plot. It seems a little better than the histogram in Exercise 11 since it avoids the stacking of the colored segments of the bars.
ggplot(data = diamonds) +
geom_freqpoly(mapping = aes(x = carat, color = cut))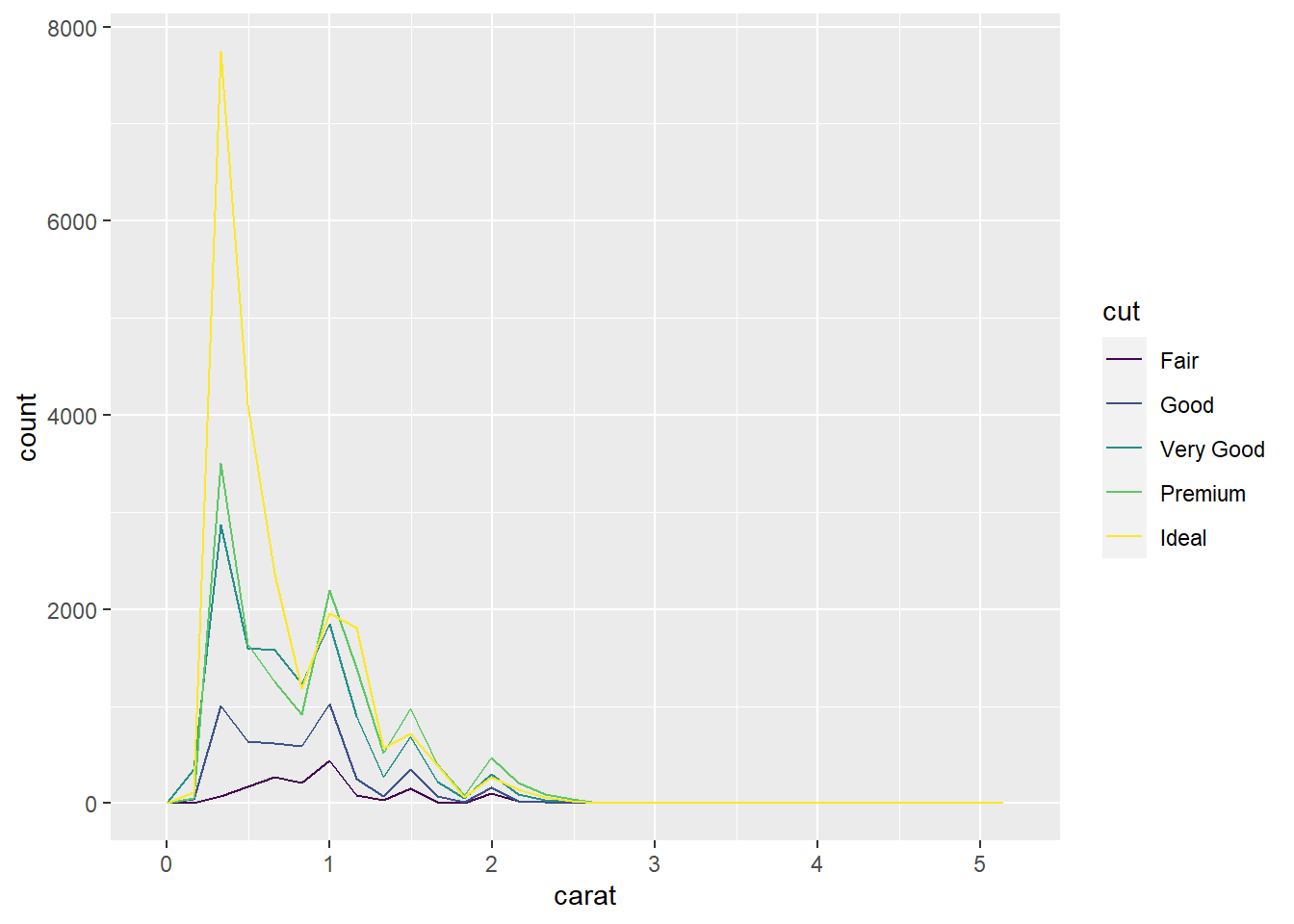
ggplot(data = mpg) +
geom_bar(mapping = aes(x = class, fill = drv)) +
labs(x = "type of car",
y = "number of cars",
fill = "drive train",
title = "Distribution of Car Type Broken Down by Drive Train")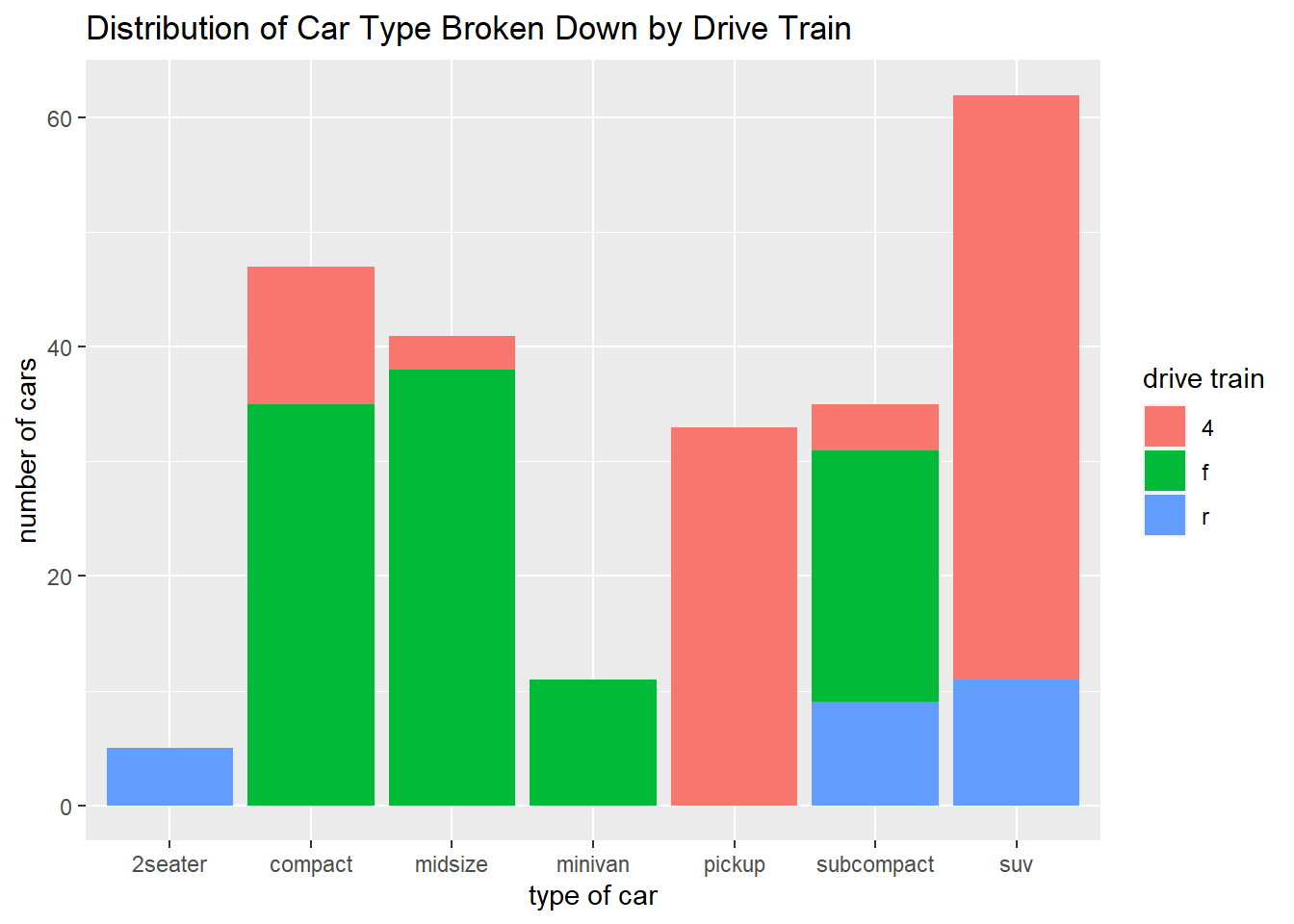
Section 2.1.1
There are 336,776 observations (rows) and 19 variables (columns).
filter(flights, month == 2)filter(flights, carrier == "UA" | carrier == "AA")filter(flights, month >= 6 & month <= 8)filter(flights, arr_delay > 120 & dep_delay <= 0)filter(flights, dep_delay > 60 & dep_delay - arr_delay > 30)filter(flights, dep_time < 600)A canceled flight would have an
NAfor its departure time, so we can just count the number ofNAs in this column as shown below:
sum(is.na(flights$dep_time))## [1] 8255- We would have to find the number of flights with a 0 or negative
arr_delayvalue and then divide that number by the total number of arriving flights Delta flew. The following shows a way to do this. (Recall that when you compute the mean of a column ofTRUE/FALSEvalues, you’re computing the percentage ofTRUEs in the column.)
# First create a data set containing the Delta flights that arrived at their destinations:
Delta_arr <- filter(flights, carrier == "DL" & !is.na(arr_delay))
# Now find the percentage of rows of this filtered data set with `arr_delay <= 0`:
mean(Delta_arr$arr_delay <= 0)## [1] 0.6556087So Delta’s on-time arrival rate in 2013 was about 66%. For the winter on-time arrival rate, we can repeat this calculation after filtering Delta_arr down to just the winter flights.
winter_Delta_arr <- filter(Delta_arr, month == 12 | month <= 2)
mean(winter_Delta_arr$arr_delay <= 0)## [1] 0.6546032The on-time arrival rate is about 65%.
Section 2.2.1
- Let’s sort the
msleepdata set byconservationand thenViewit:
View(arrange(msleep, conservation))Scrolling through the sorted data set, we see the rows with NA in the conservation column appear at the end. To move them to the top, we can do the following, remembering that is.na(conservation) will return a 1 for the rows with an NA in the conservation column and a 0 otherwise.
arrange(msleep, desc(is.na(conservation)))- The longest delay was 1301 minutes.
arrange(flights, desc(dep_delay))## # A tibble: 336,776 x 19
## year month day dep_time sched_d~1 dep_d~2 arr_t~3 sched~4 arr_d~5 carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 9 641 900 1301 1242 1530 1272 HA 51 N384HA JFK HNL
## 2 2013 6 15 1432 1935 1137 1607 2120 1127 MQ 3535 N504MQ JFK CMH
## 3 2013 1 10 1121 1635 1126 1239 1810 1109 MQ 3695 N517MQ EWR ORD
## 4 2013 9 20 1139 1845 1014 1457 2210 1007 AA 177 N338AA JFK SFO
## 5 2013 7 22 845 1600 1005 1044 1815 989 MQ 3075 N665MQ JFK CVG
## 6 2013 4 10 1100 1900 960 1342 2211 931 DL 2391 N959DL JFK TPA
## 7 2013 3 17 2321 810 911 135 1020 915 DL 2119 N927DA LGA MSP
## 8 2013 6 27 959 1900 899 1236 2226 850 DL 2007 N3762Y JFK PDX
## 9 2013 7 22 2257 759 898 121 1026 895 DL 2047 N6716C LGA ATL
## 10 2013 12 5 756 1700 896 1058 2020 878 AA 172 N5DMAA EWR MIA
## # ... with 336,766 more rows, 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>,
## # time_hour <dttm>, and abbreviated variable names 1: sched_dep_time, 2: dep_delay, 3: arr_time,
## # 4: sched_arr_time, 5: arr_delay
## # i Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names- It looks like several flights departed at 12:01am.
arrange(flights, dep_time)## # A tibble: 336,776 x 19
## year month day dep_time sched_d~1 dep_d~2 arr_t~3 sched~4 arr_d~5 carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 13 1 2249 72 108 2357 71 B6 22 N206JB JFK SYR
## 2 2013 1 31 1 2100 181 124 2225 179 WN 530 N550WN LGA MDW
## 3 2013 11 13 1 2359 2 442 440 2 B6 1503 N627JB JFK SJU
## 4 2013 12 16 1 2359 2 447 437 10 B6 839 N607JB JFK BQN
## 5 2013 12 20 1 2359 2 430 440 -10 B6 1503 N608JB JFK SJU
## 6 2013 12 26 1 2359 2 437 440 -3 B6 1503 N527JB JFK SJU
## 7 2013 12 30 1 2359 2 441 437 4 B6 839 N508JB JFK BQN
## 8 2013 2 11 1 2100 181 111 2225 166 WN 530 N231WN LGA MDW
## 9 2013 2 24 1 2245 76 121 2354 87 B6 608 N216JB JFK PWM
## 10 2013 3 8 1 2355 6 431 440 -9 B6 739 N586JB JFK PSE
## # ... with 336,766 more rows, 5 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>,
## # time_hour <dttm>, and abbreviated variable names 1: sched_dep_time, 2: dep_delay, 3: arr_time,
## # 4: sched_arr_time, 5: arr_delay
## # i Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names- The fastest average speed was Delta flight 1499 from LGA to ATL on May 25.
arrange(flights, desc(distance/air_time))The slowest average speed was US Airways flight 1860 from LGA to PHL on January 28.
arrange(flights, distance/air_time)- The Hawaiian Airlines flights from JFK to HNL flew the farthest distance of 4983 miles.
arrange(flights, desc(distance))The shortest flight in the data set was US Airways flight 1632 from EWR to LGA on July 27. (It was canceled.)
arrange(flights, distance)Section 2.3.1
- Checking the
destcolumn of the transformed data set created below, we see that the most distant American Airlines destination is SFO.
flights %>%
filter(carrier == "AA") %>%
arrange(desc(distance))- We can refine the filter from the previous exercise to include only flights with LGA as the origin airport. The most distant destination is DFW.
flights %>%
filter(carrier == "AA" & origin == "LGA") %>%
arrange(desc(distance))- The most delayed winter flight was Hawaiian Airlines flight 51 from JFK to HNL on January 9.
flights %>%
filter(month == 12 | month <= 2) %>%
arrange(desc(dep_delay))The most delayed summer flight was Envoy Air flight 3535 from JFK to CMH on June 15.
flights %>%
filter(month >= 6 & month <= 8) %>%
arrange(desc(dep_delay))Section 2.5.1
- The fastest flight was Delta flight 1499 from LGA to ATL on May 25. It’s average air speed was about 703 mph.
flights %>%
filter(!is.na(dep_time)) %>% # <---- Filters out canceled flights
mutate(air_speed = distance/air_time * 60) %>%
arrange(desc(air_speed)) %>%
select(month, day, carrier, flight, origin, dest, air_speed)The slowest flight was US Airways flight 1860 from LGA to PHL on January 28. It’s average air speed was about 77 mph.
flights %>%
filter(!is.na(dep_time)) %>% # <---- Filters out canceled flights
mutate(air_speed = distance/air_time * 60) %>%
arrange(air_speed) %>%
select(month, day, carrier, flight, origin, dest, air_speed)- The
differencecolumn below shows the values of the the newly createdflight_minutesvariable minus theair_timevariable already inflights. A little digging on the internet could tell us thatair_timeonly includes the times during which the plane is actually in the air and does not include taxiing on the runway, etc.dep_timeandarr_timeare the times when the plane departs from and arrives at the gate, soflight_minutesincludes time spent on the runway. This would meanflight_minutesshould be consistently larger thanair_time, producing all positive values in the difference column.
However, this is not what we observe. If we look carefully at the flights with a negative difference value, we might notice that the destination airport is not in the Eastern Time Zone. The arr_time values in the Central Time Zone are thus 60 minutes behind Eastern, Mountain Time Zone arr_times are 120 minutes behind, and Pacific Time Zones are 180 minutes behind. This explains the negative difference values and also why you would see, for example, a flight from Newark to San Francisco with a flight_minutes value of only 205.
flights %>%
mutate(dep_mins_midnight = (dep_time %/% 100)*60 + (dep_time %% 100),
arr_mins_midnight = (arr_time %/% 100)*60 + (arr_time %% 100),
flight_minutes = arr_mins_midnight - dep_mins_midnight,
difference = flight_minutes - air_time) %>%
select(month, day, origin, dest, air_time, flight_minutes, difference)## # A tibble: 336,776 x 7
## month day origin dest air_time flight_minutes difference
## <int> <int> <chr> <chr> <dbl> <dbl> <dbl>
## 1 1 1 EWR IAH 227 193 -34
## 2 1 1 LGA IAH 227 197 -30
## 3 1 1 JFK MIA 160 221 61
## 4 1 1 JFK BQN 183 260 77
## 5 1 1 LGA ATL 116 138 22
## 6 1 1 EWR ORD 150 106 -44
## 7 1 1 EWR FLL 158 198 40
## 8 1 1 LGA IAD 53 72 19
## 9 1 1 JFK MCO 140 161 21
## 10 1 1 LGA ORD 138 115 -23
## # ... with 336,766 more rows
## # i Use `print(n = ...)` to see more rows- The first day of the year with a canceled flight was January 1.
flights %>%
mutate(status = ifelse(is.na(dep_time), "canceled", "not canceled")) %>%
arrange(status)## # A tibble: 336,776 x 20
## year month day dep_time sched_d~1 dep_d~2 arr_t~3 sched~4 arr_d~5 carrier flight tailnum origin dest
## <int> <int> <int> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr>
## 1 2013 1 1 NA 1630 NA NA 1815 NA EV 4308 N18120 EWR RDU
## 2 2013 1 1 NA 1935 NA NA 2240 NA AA 791 N3EHAA LGA DFW
## 3 2013 1 1 NA 1500 NA NA 1825 NA AA 1925 N3EVAA LGA MIA
## 4 2013 1 1 NA 600 NA NA 901 NA B6 125 N618JB JFK FLL
## 5 2013 1 2 NA 1540 NA NA 1747 NA EV 4352 N10575 EWR CVG
## 6 2013 1 2 NA 1620 NA NA 1746 NA EV 4406 N13949 EWR PIT
## 7 2013 1 2 NA 1355 NA NA 1459 NA EV 4434 N10575 EWR MHT
## 8 2013 1 2 NA 1420 NA NA 1644 NA EV 4935 N759EV EWR ATL
## 9 2013 1 2 NA 1321 NA NA 1536 NA EV 3849 N13550 EWR IND
## 10 2013 1 2 NA 1545 NA NA 1910 NA AA 133 <NA> JFK LAX
## # ... with 336,766 more rows, 6 more variables: air_time <dbl>, distance <dbl>, hour <dbl>, minute <dbl>,
## # time_hour <dttm>, status <chr>, and abbreviated variable names 1: sched_dep_time, 2: dep_delay,
## # 3: arr_time, 4: sched_arr_time, 5: arr_delay
## # i Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names- There were 9430 flights for which
arr_statusisNA. The reason thatNAwas assigned toarr_statusfor these flights is that thearr_delayvalue for these flights isNA, so the condition in theifelsewould be checking whetherNA <= 0. The result of this condition can be neitherTRUEnorFALSE, it will instead just beNA.
flights_w_arr_status <- flights %>%
mutate(arr_status = ifelse(arr_delay <= 0,
"on time",
"late"))
sum(is.na(flights_w_arr_status$arr_status))## [1] 9430- We will rearrange the conditions so that the first one checked is
arr_delay <= 0and the second one isis.na(arr_delay). We can then find a row with anNAin thearr_delaycolumn by sorting byis.na(arr_delay)in descending order.
The results below show that arr_status was not correctly assigned a “canceled” value for these flights. This is because the first condition in the nested ifelse is arr_delay <= 0. As we saw in the previous exercise, when the arr_delay value is NA, this first condition is neither TRUE nor FALSE, so neither of the conditional statements will be executed. Instead, a value of NA will be assigned to arr_status.
flights_w_arr_status2 <- flights %>%
mutate(arr_status = ifelse(arr_delay <= 0,
"on time",
ifelse(is.na(arr_delay),
"canceled",
"late"))) %>%
arrange(desc(is.na(arr_delay))) %>%
select(arr_delay, arr_status)
flights_w_arr_status2## # A tibble: 336,776 x 2
## arr_delay arr_status
## <dbl> <chr>
## 1 NA <NA>
## 2 NA <NA>
## 3 NA <NA>
## 4 NA <NA>
## 5 NA <NA>
## 6 NA <NA>
## 7 NA <NA>
## 8 NA <NA>
## 9 NA <NA>
## 10 NA <NA>
## # ... with 336,766 more rows
## # i Use `print(n = ...)` to see more rows- We’ll interchange the first two conditions as we did in the previous exercise and then again sort by
is.na(arr_status)in descending order to find the flights for whicharr_delayisNA. We can see thatarr_statusis correctly assigned acanceledvalue. This shows that the order in which the conditions are listed does not matter in acase_whenstatement. The previous exercise shows that the order does matter in a nestedifelse, which means thatcase_whenstatements are more robust ways to handle conditional statements.
flights_w_arr_status3 <- flights %>%
mutate(arr_status = case_when(arr_delay <= 0 ~ "on time",
is.na(arr_delay) ~ "canceled",
arr_delay >0 ~ "delayed")) %>%
arrange(desc(is.na(arr_delay))) %>%
select(arr_delay, arr_status)
flights_w_arr_status3## # A tibble: 336,776 x 2
## arr_delay arr_status
## <dbl> <chr>
## 1 NA canceled
## 2 NA canceled
## 3 NA canceled
## 4 NA canceled
## 5 NA canceled
## 6 NA canceled
## 7 NA canceled
## 8 NA canceled
## 9 NA canceled
## 10 NA canceled
## # ... with 336,766 more rows
## # i Use `print(n = ...)` to see more rowsflights %>%
mutate(season = case_when(month >= 3 & month <= 5 ~ "spring",
month >= 6 & month <= 8 ~ "summer",
month >= 9 & month <= 11 ~ "fall",
TRUE ~ "winter"))- It might help to recall the distribution of
priceindiamondsby looking at its histogram shown below.
ggplot(data = diamonds) +
geom_histogram(mapping = aes(x = price))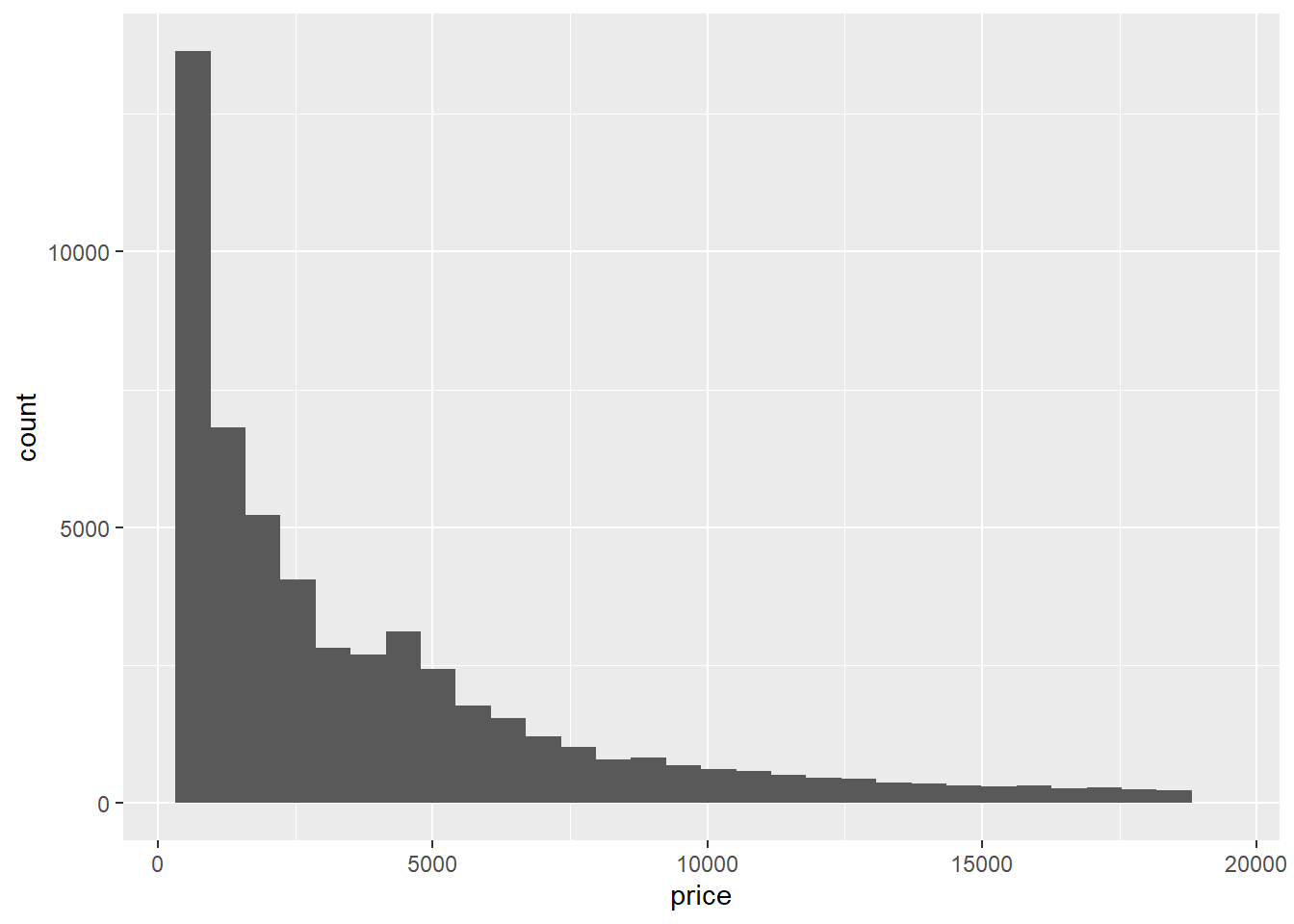
The histogram might suggest the following cutoffs: expensive for price < 5000, very expensive for 5000 <= price < 10000, insanely expensive for 10000 <= price < 15000 and priceless for price >= 15000. We can assign these labels with the following case_when:
diamonds2 <- diamonds %>%
mutate(category = case_when(price < 5000 ~ "expensive",
price >= 5000 & price < 10000 ~ "very expensive",
price >= 10000 & price < 15000 ~ "insanely expensive",
TRUE ~ "priceless"))Finally, we can visualize the distribution with a bar graph:
ggplot(data = diamonds2) +
geom_bar(mapping = aes(x = category))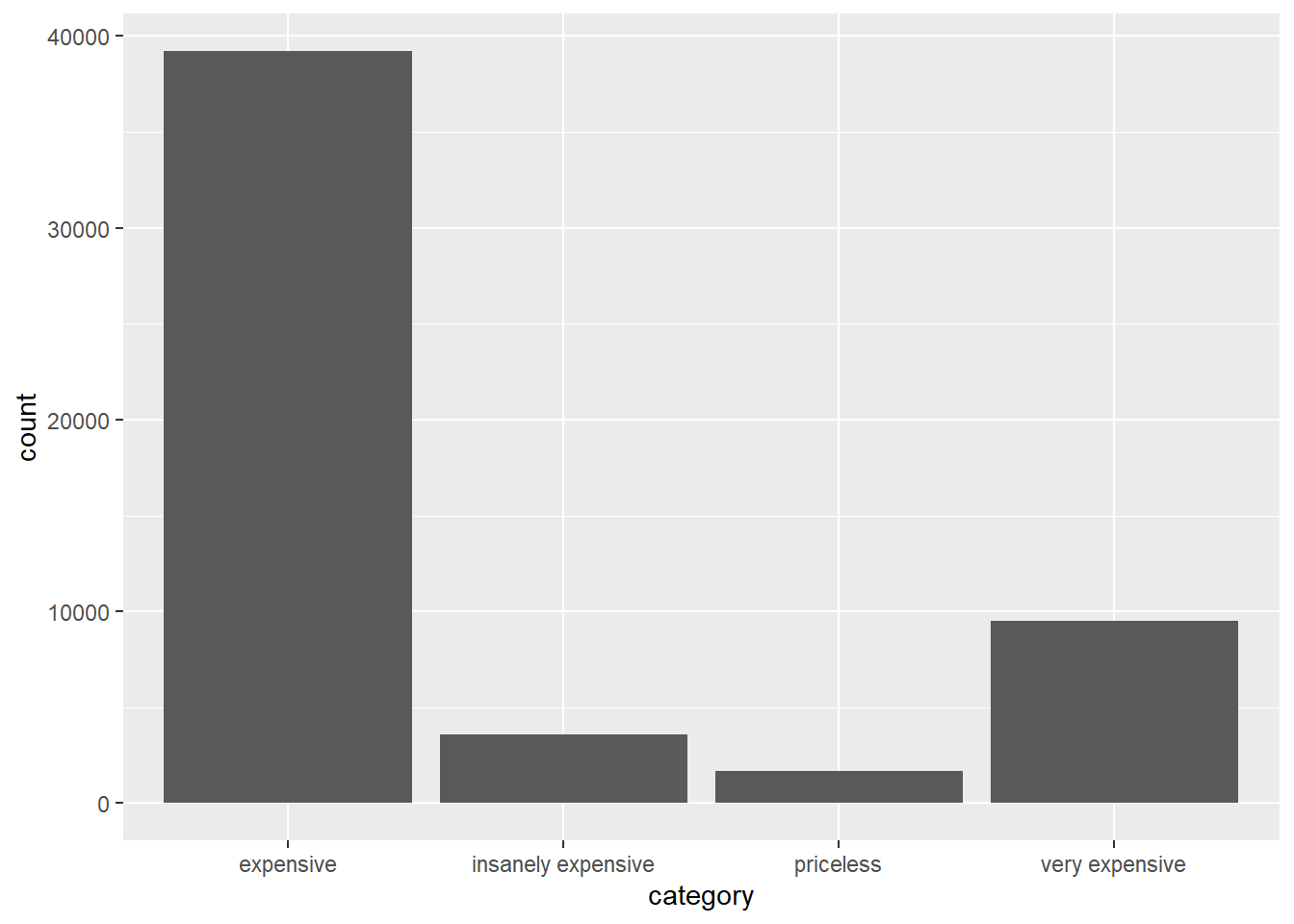
Battinghas 110,495 observations and 22 variables.Our results are surprising because there are several players with perfect batting averages. This is explained by the fact that these players had very few at bats.
Batting %>%
mutate(batting_average = H/AB) %>%
arrange(desc(batting_average))- The highest single season batting average of 0.440 belonged to Hugh Duffy in 1894.
Batting %>%
filter(AB >= 350) %>%
mutate(batting_average = H/AB) %>%
arrange(desc(batting_average))- The highest batting average of the modern era belonged to Tony Gwynn, who hit 0.394 in 1994.
Batting %>%
filter(yearID >= 1947,
AB >= 350) %>%
mutate(batting_average = H/AB) %>%
arrange(desc(batting_average))- The last player to hit 0.400 was Ted Williams in 1941.
Batting %>%
filter(AB >= 350) %>%
mutate(batting_average = H/AB) %>%
filter(batting_average >= 0.4) %>%
arrange(desc(yearID))Section 2.6.1
- Be sure to remove the
NAvalues when computing the mean.
flights %>%
group_by(month, day) %>%
summarize(avg_dep_delay = mean(dep_delay, na.rm = TRUE))- The carrier with the best on-time rate was AS (Alaska Airlines) at about 73%. However, American Airlines and Delta Airlines should be recognized as well since they maintained comparably high on-time rates while flying tens of thousands more flights than Alaska Airlines.
The carrier with the worst on-time rate was FL (Airtran Airways) at about 40%. It should also be mentioned that Express Jet (EV) flew over 50,000 flights while compiling an on-time rate of just 52%.
arr_rate <- flights %>%
group_by(carrier) %>%
summarize(on_time_rate = mean(arr_delay <= 0, na.rm = TRUE),
count = n())
arrange(arr_rate, desc(on_time_rate))## # A tibble: 16 x 3
## carrier on_time_rate count
## <chr> <dbl> <int>
## 1 AS 0.733 714
## 2 HA 0.716 342
## 3 AA 0.665 32729
## 4 VX 0.659 5162
## 5 DL 0.656 48110
## 6 OO 0.655 32
## 7 US 0.629 20536
## 8 9E 0.616 18460
## 9 UA 0.615 58665
## 10 B6 0.563 54635
## 11 WN 0.560 12275
## 12 MQ 0.533 26397
## 13 YV 0.526 601
## 14 EV 0.521 54173
## 15 F9 0.424 685
## 16 FL 0.403 3260arrange(arr_rate, on_time_rate)## # A tibble: 16 x 3
## carrier on_time_rate count
## <chr> <dbl> <int>
## 1 FL 0.403 3260
## 2 F9 0.424 685
## 3 EV 0.521 54173
## 4 YV 0.526 601
## 5 MQ 0.533 26397
## 6 WN 0.560 12275
## 7 B6 0.563 54635
## 8 UA 0.615 58665
## 9 9E 0.616 18460
## 10 US 0.629 20536
## 11 OO 0.655 32
## 12 DL 0.656 48110
## 13 VX 0.659 5162
## 14 AA 0.665 32729
## 15 HA 0.716 342
## 16 AS 0.733 714- One way we can answer this question by comparing the minimum
air_timevalue to the averageair_timevalue at each destination and sorting by the difference to find destinations with minimums way smaller than averages.
It looks like there was a flight to Minneapolis-St. Paul that arrived almost 1 hour faster than the average flight to that destination. (It looks like this flight left the origin gate at 3:58PM EST and arrived at the destination gate at 5:45PM CST, which would mean 107 minutes gate-to-gate. The air_time value of 93 is thus probably not an entry error. The flight had a departure delay of 45 minutes and may have been trying to make up time in the air.)
flights %>%
filter(!is.na(air_time)) %>%
group_by(dest) %>%
summarize(min_air_time = min(air_time),
avg_air_time = mean(air_time)) %>%
arrange(desc(avg_air_time - min_air_time))- Express Jet flew to 61 different destinations.
flights %>%
group_by(carrier) %>%
summarize(dist_dest = n_distinct(dest)) %>%
arrange(desc(dist_dest))- Standard deviation is the statistic most often used to measure variation in a variable. The plane with tail number N76062 had the highest standard deviation is distances flown at about 1796 miles.
flights %>%
filter(!is.na(tailnum)) %>%
group_by(tailnum) %>%
summarize(variation = sd(distance, na.rm = TRUE),
count = n()) %>%
arrange(desc(variation))- February 8 had the most cancellations at 472. There were 7 days with no cancellations, including, luckily, Thanksgiving and the day after Thanksgiving.
cancellations <- flights %>%
group_by(month, day) %>%
summarize(flights_canceled = sum(is.na(dep_time)))
arrange(cancellations, desc(flights_canceled))## # A tibble: 365 x 3
## # Groups: month [12]
## month day flights_canceled
## <int> <int> <int>
## 1 2 8 472
## 2 2 9 393
## 3 5 23 221
## 4 12 10 204
## 5 9 12 192
## 6 3 6 180
## 7 3 8 180
## 8 12 5 158
## 9 12 14 125
## 10 6 28 123
## # ... with 355 more rows
## # i Use `print(n = ...)` to see more rowsarrange(cancellations, flights_canceled)## # A tibble: 365 x 3
## # Groups: month [12]
## month day flights_canceled
## <int> <int> <int>
## 1 4 21 0
## 2 5 17 0
## 3 5 26 0
## 4 10 5 0
## 5 10 20 0
## 6 11 28 0
## 7 11 29 0
## 8 1 6 1
## 9 1 19 1
## 10 2 16 1
## # ... with 355 more rows
## # i Use `print(n = ...)` to see more rowsHR_by_year <- Batting %>%
group_by(yearID) %>%
summarize(max_HR = max(HR))- The huge jump around 1920 is explained by the arrival of Babe Ruth. Home runs totals remained very high in his aftermath, showing how dramatically he changed the game. You might also recognize some high spikes during the PED-fueled years of Mark McGwire, Sammy Sosa, and Barry Bonds, and some low spikes during strike- or COVID- shortened seasons.
ggplot(data = HR_by_year, mapping = aes(x = yearID, y = max_HR)) +
geom_point() +
geom_line()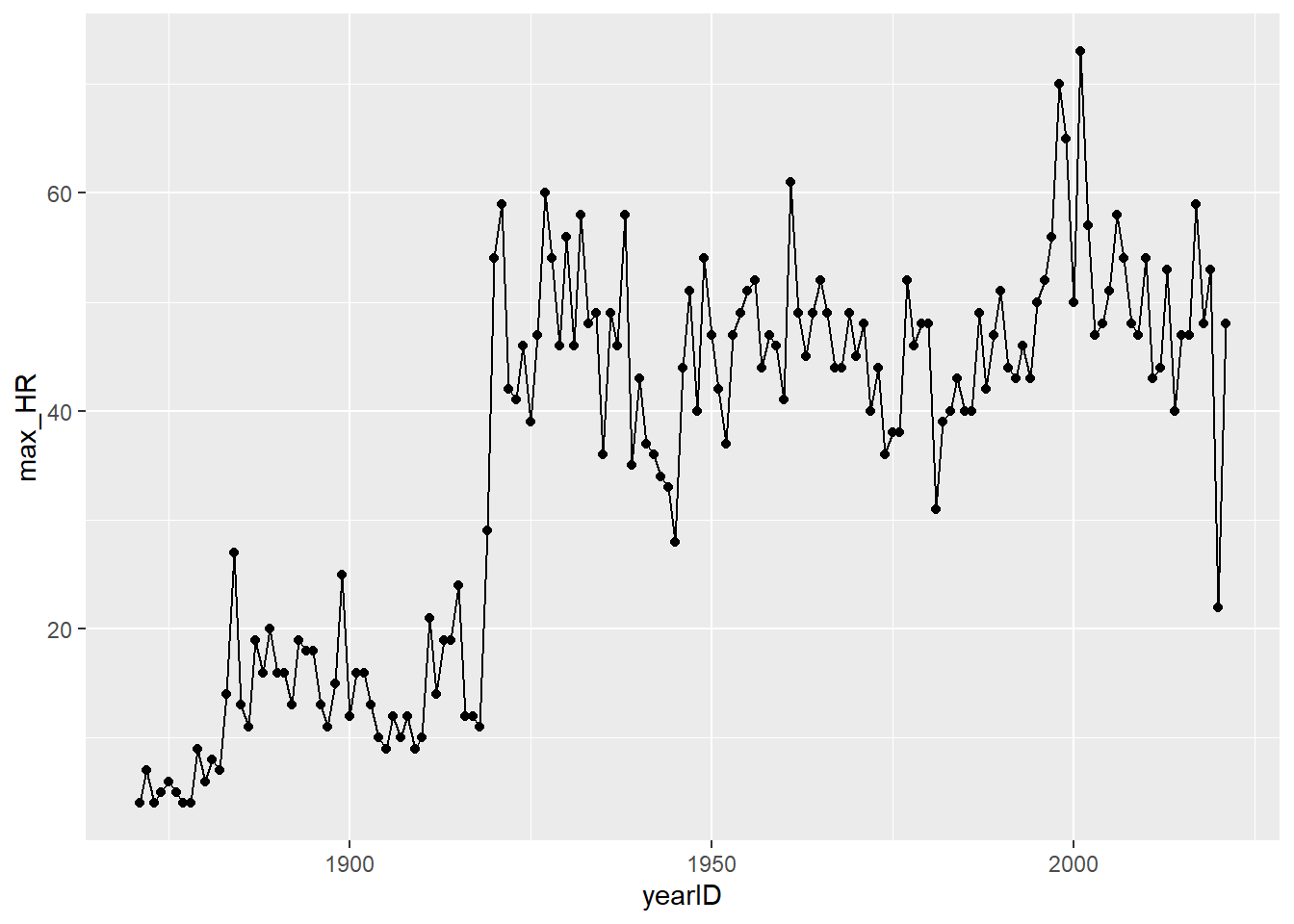
Batting %>%
group_by(playerID) %>%
summarize(at_bats = sum(AB, na.rm = TRUE),
hits = sum(H, na.rm = TRUE),
batting_avg = hits/at_bats) %>%
filter(at_bats >= 3000) %>%
arrange(desc(batting_avg))## # A tibble: 1,774 x 4
## playerID at_bats hits batting_avg
## <chr> <int> <int> <dbl>
## 1 cobbty01 11436 4189 0.366
## 2 hornsro01 8173 2930 0.358
## 3 jacksjo01 4981 1772 0.356
## 4 odoulle01 3264 1140 0.349
## 5 delahed01 7510 2597 0.346
## 6 speaktr01 10195 3514 0.345
## 7 hamilbi01 6283 2164 0.344
## 8 willite01 7706 2654 0.344
## 9 broutda01 6726 2303 0.342
## 10 ruthba01 8398 2873 0.342
## # ... with 1,764 more rows
## # i Use `print(n = ...)` to see more rowsSection 3.1.1
The code to import the file is
sample_xlsx <- read_excel("sample_xlsx.xlsx"). You are also required to load the readxl library. This is different from the previous imports in that the file is an Excel spreadsheet, and thus theread_excelfunction is required instead ofread_csv.A more descriptive name for this data set would be
colleges. We should also uncheck the box that says, “First Row as Names” on the import screen. The code to import it with these adjustments iscolleges <- read_excel("sample_2.xlsx", col_names = FALSE).The columns name assigned were
...1,...2, and...3. You can specify your own column names during the import as follows:
colleges <- read_excel("sample_2.xlsx", col_names = c("college", "city", "state"))schedule <- tribble(
~course, ~title, ~credits,
"MS 121", "Calculus I", 4,
"MS 231", "Multivariable Calculus", 3,
"MS 282", "Applied Statistics with R", 3
)Section 3.2.1
The variable
time_hourhas a type of “dttm.” This stands for “date-time” and is a combination of a date and a time.
mpg2 <- mpg %>%
mutate(hwy = as.double(hwy),
cty = as.double(cty))- Coercing an integer into a character returns a one-character string consisting of the symbol for the integer. It will no longer have a numberical value. For example,
as.character(7L)returns"7". - The same thing happens as in part (a). For example,
as.character(2.8)returns"2.8". - Coercing a double into an integer returns the integer part of the number. For example,
as.integer(2.8)returns 2. - Coercing a character value containing letters into either a double or an integer returns
NA. You can try this on a specific example such asas.integer("q")oras.double("q"). However, if the character is actually a number, then coercing it into an integer or double restores its numerical value. For example,as.integer("7")returns the number 7, andas.double("7.7")returns the number 7.7.
- Coercing an integer into a character returns a one-character string consisting of the symbol for the integer. It will no longer have a numberical value. For example,
dayis a double, but it should be an integer.- We’ll have to change
monthto a factor where the levels are the months in chronological order.
# part (a)
holidays <- read_csv("holidays.csv")
holidays <- holidays %>%
mutate(day = as.integer(day))# part (b)
month_levels <- c("Jan", "Feb", "Mar", "Apr", "May", "Jun",
"Jul", "Aug", "Sep", "Oct", "Nov", "Dec")
holidays <- holidays %>%
mutate(month = factor(month, levels = month_levels)) %>%
arrange(month, day)
holidays## # A tibble: 9 x 3
## month day holiday
## <fct> <int> <chr>
## 1 Jan 1 New Year's Day
## 2 Jan 18 Martin Luther King, Jr. Day
## 3 Feb 14 Valentine's Day
## 4 Feb 15 President's Day
## 5 Mar 17 St. Patrick's Day
## 6 Jul 4 Independence Day
## 7 Jul 14 Bastille Day
## 8 Oct 31 Halloween
## 9 Dec 25 ChristmasSection 3.3.1
high_school_data_clean <- high_school_data %>%
rename("Acad_Standing" = `Acad Standing After FA20 (Dean's List, Good Standing, Academic Warning, Academic Probation, Academic Dismissal, Readmission on Condition)`,
"hs_qual_acad" = `High School Quality Academic`,
"FA20_cr_hrs_att" = `FA20 Credit Hours Attempted`,
"FA20_cr_hrs_earned" = `FA20 Credit Hours Earned`)Section 3.4.1
The
NAs in the other columns should probably not be replaced. Doing so would require too much speculation on the analyst’s part.There is no
NAcount for the categorical variables.
sum(is.na(high_school_data_clean$`Bridge Program?`))## [1] 266sum(is.na(high_school_data_clean$Acad_Standing))## [1] 0sum(is.na(high_school_data_clean$Race))## [1] 21sum(is.na(high_school_data_clean$Ethnicity))## [1] 7high_school_data_clean <- high_school_data_clean %>%
mutate(converted_ACT = 41.6*`ACT Comp` + 102.4)high_school_data_clean <- high_school_data_clean %>%
mutate(best_score = pmax(`SAT Comp`, converted_ACT, na.rm = TRUE))Batting_PA <- Batting %>%
mutate(PA = AB + BB + IBB + HBP + SH + SF)b. There are several missing values in the `IBB`, `HBP`, `SH`, and `SF` columns. When one of these values is missing, any sum that contains it as a summand will be reported as `NA`.
c. sum(is.na(Batting$AB))## [1] 0sum(is.na(Batting$BB))## [1] 0sum(is.na(Batting$IBB))## [1] 36650sum(is.na(Batting$HBP))## [1] 2816sum(is.na(Batting$SH))## [1] 6068sum(is.na(Batting$SF))## [1] 36103d. Batting_PA <- Batting %>%
mutate(IBB = replace_na(IBB, 0),
HBP = replace_na(HBP, 0),
SH = replace_na(SH, 0),
SF = replace_na(SF, 0))e. Batting_PA <- Batting_PA %>%
mutate(PA = AB + BB + IBB + HBP + SH + SF)
sum(is.na(Batting_PA$PA))## [1] 0f. The single season record-holder for plate appearances is Pete Rose in 1974, with a total of 784.Batting_PA %>%
arrange(desc(PA))## playerID yearID stint teamID lgID G AB R H X2B X3B HR RBI SB CS BB SO IBB HBP SH SF GIDP PA
## 1 rosepe01 1974 1 CIN NL 163 652 110 185 45 7 3 51 2 4 106 54 14 5 1 6 9 784
## 2 rolliji01 2007 1 PHI NL 162 716 139 212 38 20 30 94 41 6 49 85 5 7 0 6 11 783
## 3 dykstle01 1993 1 PHI NL 161 637 143 194 44 6 19 66 37 12 129 64 9 2 0 5 8 782
## 4 suzukic01 2004 1 SEA AL 161 704 101 262 24 5 8 60 36 11 49 63 19 4 2 3 6 781
## 5 reyesjo01 2007 1 NYN NL 160 681 119 191 36 12 12 57 78 21 77 78 13 1 5 1 6 778
## 6 rosepe01 1975 1 CIN NL 162 662 112 210 47 4 7 74 0 1 89 50 8 11 1 1 13 772
## 7 cashda01 1975 1 PHI NL 162 699 111 213 40 3 4 57 13 6 56 34 5 4 0 7 8 771
## 8 vaughmo01 1996 1 BOS AL 161 635 118 207 29 1 44 143 2 0 95 154 19 14 0 8 17 771
## 9 reyesjo01 2008 1 NYN NL 159 688 113 204 37 19 16 68 56 15 66 82 8 1 5 3 9 771
## 10 suzukic01 2006 1 SEA AL 161 695 110 224 20 9 9 49 45 2 49 71 16 5 1 2 2 768
## 11 rosepe01 1976 1 CIN NL 162 665 130 215 42 6 10 63 9 5 86 54 7 6 0 2 17 766
## 12 morenom01 1979 1 PIT NL 162 695 110 196 21 12 8 69 77 21 51 104 9 3 6 2 7 766
## 13 molitpa01 1991 1 ML4 AL 158 665 133 216 32 13 17 75 19 8 77 62 16 6 0 1 11 765
## 14 boggswa01 1985 1 BOS AL 161 653 107 240 42 3 8 78 2 1 96 61 5 4 3 2 20 763
## 15 anderbr01 1992 1 BAL AL 159 623 100 169 28 10 21 80 53 16 98 98 14 9 10 9 2 763
## 16 suzukic01 2005 1 SEA AL 162 679 111 206 21 12 15 68 33 8 48 66 23 4 2 6 5 762
## 17 boggswa01 1989 1 BOS AL 156 621 113 205 51 7 3 54 2 6 107 51 19 7 0 7 19 761
## 18 suzukic01 2008 1 SEA AL 162 686 103 213 20 7 6 42 43 4 51 65 12 5 3 4 8 761
## 19 willsma01 1962 1 LAN NL 165 695 130 208 13 10 6 48 104 13 51 57 1 2 7 4 7 760
## 20 rolliji01 2006 1 PHI NL 158 689 127 191 45 9 25 83 36 4 57 80 2 5 0 7 12 760
## 21 rosepe01 1965 1 CIN NL 162 670 117 209 35 11 11 81 8 3 69 76 2 8 8 2 10 759
## 22 sizemgr01 2006 1 CLE AL 162 655 134 190 53 11 28 76 22 6 78 153 8 13 1 4 2 759
## 23 sizemgr01 2008 1 CLE AL 157 634 101 170 39 5 33 90 38 5 98 130 14 11 0 2 5 759
## 24 rosepe01 1973 1 CIN NL 160 680 115 230 36 8 5 64 10 7 65 42 6 6 1 0 14 758
## 25 biggicr01 1999 1 HOU NL 160 639 123 188 56 0 16 73 28 14 88 107 9 11 5 6 5 758
## 26 crosefr01 1938 1 NYA AL 157 631 113 166 35 3 9 55 27 12 106 97 0 15 5 0 NA 757
## 27 sizemgr01 2007 1 CLE AL 162 628 118 174 34 5 24 78 33 10 101 155 9 17 0 2 3 757
## 28 dimagdo01 1948 1 BOS AL 155 648 127 185 40 4 9 87 10 2 101 58 0 2 5 0 11 756
## 29 morenom01 1980 1 PIT NL 162 676 87 168 20 13 2 36 96 33 57 101 11 2 3 7 9 756
## 30 erstada01 2000 1 ANA AL 157 676 121 240 39 6 25 100 28 8 64 82 9 1 2 4 8 756
## 31 engliwo01 1930 1 CHN NL 156 638 152 214 36 17 14 59 3 NA 100 72 0 6 11 0 NA 755
## 32 richabo01 1962 1 NYA AL 161 692 99 209 38 5 8 59 11 9 37 24 1 1 20 4 13 755
## 33 alouma01 1969 1 PIT NL 162 698 105 231 41 6 1 48 22 8 42 35 9 2 1 3 5 755
## 34 suzukic01 2002 1 SEA AL 157 647 111 208 27 8 8 51 31 15 68 62 27 5 3 5 8 755
## 35 jeterde01 2005 1 NYA AL 159 654 122 202 25 5 19 70 14 5 77 117 3 11 7 3 15 755
## 36 weeksri01 2010 1 MIL NL 160 651 112 175 32 4 29 83 11 4 76 184 0 25 0 2 5 754
## 37 riceji01 1978 1 BOS AL 163 677 121 213 25 15 46 139 7 5 58 126 7 5 1 5 15 753
## 38 mattido01 1986 1 NYA AL 162 677 117 238 53 2 31 113 0 0 53 35 11 1 1 10 17 753
## 39 douthta01 1928 1 SLN NL 154 648 111 191 35 3 3 43 11 NA 84 36 0 10 10 0 NA 752
## 40 bondsbo01 1970 1 SFN NL 157 663 134 200 36 10 26 78 48 10 77 189 7 2 1 2 6 752
## 41 molitpa01 1982 1 ML4 AL 160 666 136 201 26 8 19 71 41 9 69 93 1 1 10 5 9 752
## 42 gonzalu01 2001 1 ARI NL 162 609 128 198 36 7 57 142 1 1 100 83 24 14 0 5 14 752
## 43 lindofr01 2018 1 CLE AL 158 661 129 183 42 2 38 92 25 10 70 107 7 8 3 3 5 752
## [ reached 'max' / getOption("max.print") -- omitted 110452 rows ]g. The career record-holder for plate appearances is also Pete Rose, with a total of 16,028.Batting_PA %>%
group_by(playerID) %>%
summarize(career_PA = sum(PA)) %>%
arrange(desc(career_PA))## # A tibble: 20,166 x 2
## playerID career_PA
## <chr> <dbl>
## 1 rosepe01 16028
## 2 aaronha01 14233
## 3 yastrca01 14181
## 4 henderi01 13407
## 5 bondsba01 13294
## 6 cobbty01 13071
## 7 murraed02 13039
## 8 pujolal01 13005
## 9 ripkeca01 12990
## 10 musiast01 12839
## # ... with 20,156 more rows
## # i Use `print(n = ...)` to see more rowsSection 3.5.1
- It looks like there are entry errors in the
SAT CompandACT Compcolumns. The minimumSAT Compscore is 100, which is below the lowest possible score on the SAT. The minimumACT Compscore is 4.05, which is not an integer and therefore can’t be an ACT score. We can see these from the summary statistics, histograms, and a sorted lists as follows:
# Part (a): summary statistics
summary(high_school_data)
# Part (b): histograms
ggplot(data = high_school_data) +
geom_histogram(mapping = aes(x = `SAT Comp`))
ggplot(data = high_school_data) +
geom_histogram(mapping = aes(x = `ACT Comp`))
# Part (c): sorted lists
head(arrange(high_school_data, `SAT Comp`))
head(arrange(high_school_data, `ACT Comp`))Since it’s not clear what numbers the 100 and 4.05 were supposed to be, it’s not appropriate to change them.
We first have to find the spot in the
SAT Compcolumn where the 100 is. Once we know this, we can change the value to 1000 as follows:
spot_100 <- which(high_school_data$`SAT Comp` == 100)
high_school_data$`SAT Comp`[spot_100] <- 1000t <- c("a", "b", "c", "d", "e")
t[c(1,4,5)]## [1] "a" "d" "e"- The idea for part (b) is to copy the technique from Exercise 4, but use the column
Batting$HRin place of the vectortand the vector from part (a) (namedHR50below) in place of the listc(1,4,5).
# Part (a):
which(Batting$HR >= 50 & Batting$HR <= 59)## [1] 18534 19054 22801 23980 24654 27739 27767 33002 33104 34154 37834 38467 41674 44657
## [15] 54762 67723 72980 74182 74879 75835 77079 77857 80418 81095 81689 83012 83167 86623
## [29] 87903 88247 89098 89784 92931 97328 103526 104124 105901# Part (b):
HR50 <- which(Batting$HR >= 50 & Batting$HR <= 59)
Batting$playerID[HR50]## [1] "ruthba01" "ruthba01" "ruthba01" "wilsoha01" "foxxji01" "foxxji01" "greenha01" "kinerra01"
## [9] "mizejo01" "kinerra01" "mayswi01" "mantlmi01" "mantlmi01" "mayswi01" "fostege01" "fieldce01"
## [17] "belleal01" "anderbr01" "mcgwima01" "griffke02" "griffke02" "vaughgr01" "sosasa01" "gonzalu01"
## [25] "rodrial01" "rodrial01" "thomeji01" "jonesan01" "howarry01" "ortizda01" "fieldpr01" "rodrial01"
## [33] "bautijo02" "davisch02" "judgeaa01" "stantmi03" "alonspe01"Section 3.6.1
flights %>%
unite("date", month, day, year, sep="-")## # A tibble: 336,776 x 17
## date dep_time sched_de~1 dep_d~2 arr_t~3 sched~4 arr_d~5 carrier flight tailnum origin dest air_t~6
## <chr> <int> <int> <dbl> <int> <int> <dbl> <chr> <int> <chr> <chr> <chr> <dbl>
## 1 1-1-2013 517 515 2 830 819 11 UA 1545 N14228 EWR IAH 227
## 2 1-1-2013 533 529 4 850 830 20 UA 1714 N24211 LGA IAH 227
## 3 1-1-2013 542 540 2 923 850 33 AA 1141 N619AA JFK MIA 160
## 4 1-1-2013 544 545 -1 1004 1022 -18 B6 725 N804JB JFK BQN 183
## 5 1-1-2013 554 600 -6 812 837 -25 DL 461 N668DN LGA ATL 116
## 6 1-1-2013 554 558 -4 740 728 12 UA 1696 N39463 EWR ORD 150
## 7 1-1-2013 555 600 -5 913 854 19 B6 507 N516JB EWR FLL 158
## 8 1-1-2013 557 600 -3 709 723 -14 EV 5708 N829AS LGA IAD 53
## 9 1-1-2013 557 600 -3 838 846 -8 B6 79 N593JB JFK MCO 140
## 10 1-1-2013 558 600 -2 753 745 8 AA 301 N3ALAA LGA ORD 138
## # ... with 336,766 more rows, 4 more variables: distance <dbl>, hour <dbl>, minute <dbl>,
## # time_hour <dttm>, and abbreviated variable names 1: sched_dep_time, 2: dep_delay, 3: arr_time,
## # 4: sched_arr_time, 5: arr_delay, 6: air_time
## # i Use `print(n = ...)` to see more rows, and `colnames()` to see all variable names# Part (a):
library(readxl)
hof <- read_excel("hof.xlsx")
# Part (b):
hof2 <- hof %>%
filter(!is.na(Name)) %>% #Filter out the NAs
select(Name) %>% #Remove the other columns
separate(Name, into = c("First", "Last")) %>% #Separate into columns for first and last name
arrange(Last) %>% #Sort by last name
unite("Name", "First", "Last", sep = " ") #Re-unite the first and last name columns- Many people in the list have more than two parts to their names, such as Grover Cleveland Alexander. When the
separatefunction operates on this value, it doesn’t know what to do with the “Alexander” and so just drops it, as indicated in the warning message. When theunitefunction does its job, it re-unites this name as just “Grover Cleveland” rather than “Grover Cleveland Alexander.” This situation also arises for people whose name contains a non-syntactic character; for example, Buck O’Neil becomes Buck O.
- Many people in the list have more than two parts to their names, such as Grover Cleveland Alexander. When the
patients <- tribble(
~name, ~attribute, ~value,
"Stephen Johnson", "age", 47,
"Stephen Johnson", "weight", 185,
"Ann Smith", "age", 39,
"Ann Smith", "weight", 125,
"Ann Smith", "height", 64,
"Mark Davidson", "age", 51,
"Mark Davidson", "weight", 210,
"Mark Davidson", "height", 73
)
pivot_wider(patients, names_from = "attribute", values_from = "value")## # A tibble: 3 x 4
## name age weight height
## <chr> <dbl> <dbl> <dbl>
## 1 Stephen Johnson 47 185 NA
## 2 Ann Smith 39 125 64
## 3 Mark Davidson 51 210 73- There are two Ann Smiths, each with different ages, weights, and heights, so in the pivoted table, Ann Smith’s values show up as 2-entry vectors rather than single values. This problem could have been avoided by entering middle initials or some other kind of distinguishing character, such as Ann Smith 1 and Ann Smith 2.
patients <- tribble(
~name, ~attribute, ~value,
"Stephen Johnson", "age", 47,
"Stephen Johnson", "weight", 185,
"Ann Smith", "age", 39,
"Ann Smith", "weight", 125,
"Ann Smith", "height", 64,
"Mark Davidson", "age", 51,
"Mark Davidson", "weight", 210,
"Mark Davidson", "height", 73,
"Ann Smith", "age", 43,
"Ann Smith", "weight", 140,
"Ann Smith", "height", 66
)
pivot_wider(patients, names_from = "attribute", values_from = "value")## # A tibble: 3 x 4
## name age weight height
## <chr> <list> <list> <list>
## 1 Stephen Johnson <dbl [1]> <dbl [1]> <NULL>
## 2 Ann Smith <dbl [2]> <dbl [2]> <dbl [2]>
## 3 Mark Davidson <dbl [1]> <dbl [1]> <dbl [1]>- “male” and “female” are not variables but rather values of a variable we could call “gender.” This is fixed with a
pivot_longer. Also, “yes” and “no” are not values but rather variables whose values are listed to the right. This could be fixed with apivot_wider. The “number pregnant” column in the code below does not show up in the final tidied data set – it’s only a temporary column to store the values of “yes” and “no” after the first pivot.
preg <- tribble(
~pregnant, ~male, ~female,
"yes", NA, 10,
"no", 20, 12
)
preg %>%
pivot_longer(cols = c("male", "female"), names_to = "gender", values_to = "number pregnant") %>%
pivot_wider(names_from = "pregnant", values_from = "number pregnant")## # A tibble: 2 x 3
## gender yes no
## <chr> <dbl> <dbl>
## 1 male NA 20
## 2 female 10 12Section 3.7.1
nameis a key formsleep.- The combination
playerID,yearID, andstintis a key forBatting. - The combination
month,day,dep_time,carrier, andflightis a key forflights.
mpgdoes not have a key. When a data set does not have a key, you can always use the row number as a key. We can add the row numbers as their own column as follows:
mpg_w_rows <- mpg %>%
mutate(row = row_number())inner_joincreates a merged data set that shows only the records corresponding to the key values that are in both data sets.left_joincreates a merged data set that shows only the records corresponding to the key values that are in the first listed data set.right_joincreates a merged data set that shows only the records corresponding to the key values that are in the second listed data set.full_joincreates a merged data set that shows all of the records corresponding to the key values that are in either of the data sets.
left_join(Pitching, Salaries, by = c("playerID", "yearID", "teamID", 'lgID'))- The
full_joinfunction merges theBattingandPitchingdata sets. This is the appropriate join because we don’t want to exclude any player from either data set. Theleft_joinfunction merges theSalariesdata set with the result of the previousfull_join.
full_join(Batting, Pitching) %>%
left_join(Salaries)Section 4.1.1
- It looks like a regression line might cross the y-axis around 35 and would have a rise of about -15 over a run of about 5, giving us a slope of -15/5, which equals -3.
ggplot(mpg) +
geom_point(aes(displ, hwy)) +
geom_abline(intercept = 35, slope = -3)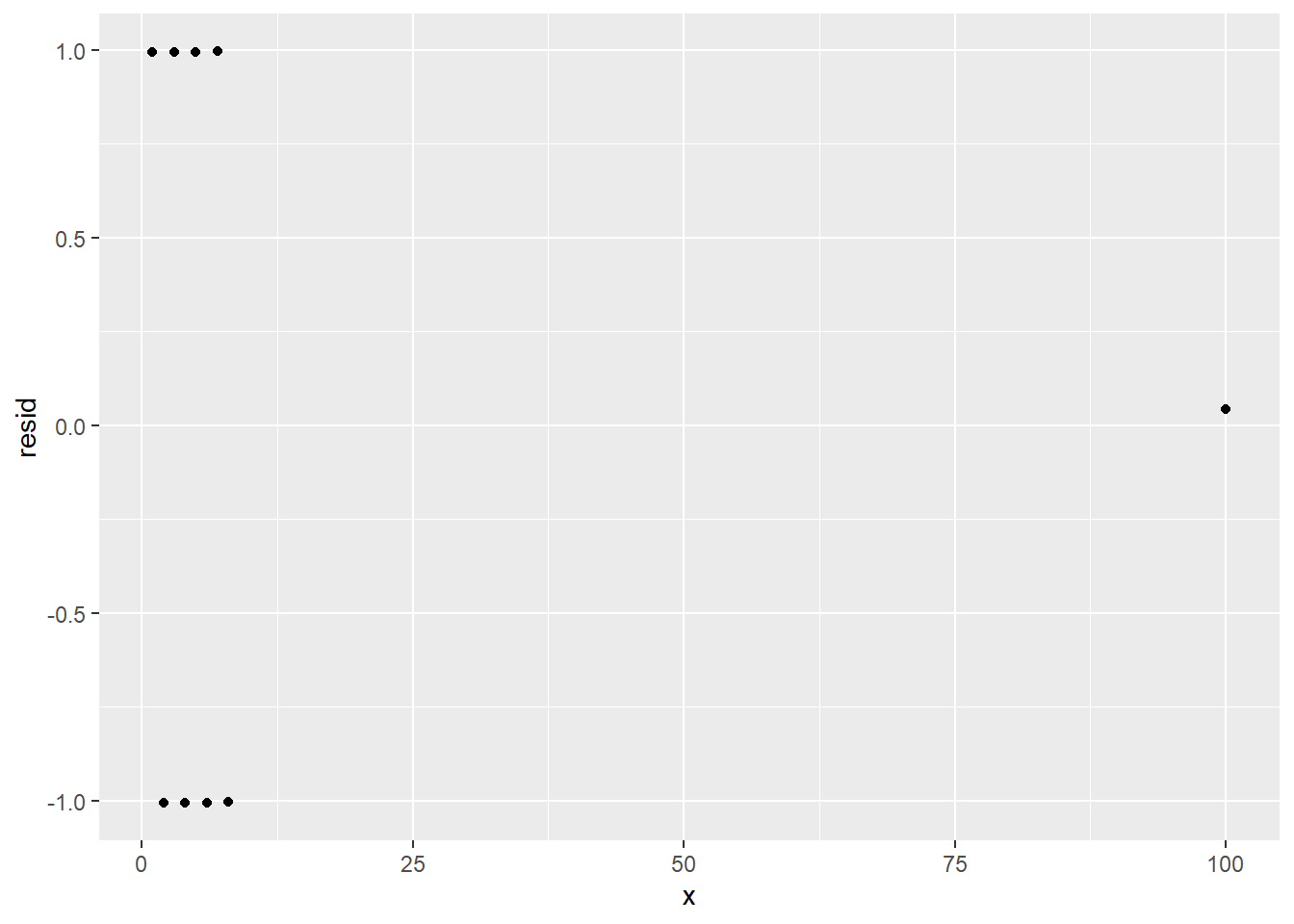
mpg_model <- lm(hwy ~ displ, data = mpg)
b <- coef(mpg_model)[1]
m <- coef(mpg_model)[2]
ggplot(mpg) +
geom_point(aes(displ, hwy)) +
geom_abline(intercept = 35, slope = -3) + # guessed regression line
geom_abline(intercept = b, slope = m, color = "red") # actual regression line
Section 4.2.1
X <- c(32.5, 29.1, 17.7, 25.1, 19.2, 26.8, 27.4, 21.1)
n <- length(X)
variance <- 1/(n - 1) * sum((X - mean(X))^2)
st_dev <- sqrt(variance)
variance## [1] 26.29411var(X)## [1] 26.29411st_dev## [1] 5.127778sd(X)## [1] 5.127778- To say that the variance of
Xis 26.29411 means that the average squared distance between the values ofXand the population mean is about 26.29411. To say that the standard deviation ofXis 5.127778 means that the average distance between the values ofXand the population mean is about 5.127778.
X <- rnorm(100, 50, 0.6)
X## [1] 49.89465 49.12142 49.64500 50.87677 49.59174 49.88390 51.28505 49.56135 50.47865 48.75779 50.52935
## [12] 49.20785 49.54296 49.95081 50.22636 49.50480 50.20584 50.21754 49.94540 48.98020 50.34305 50.91365
## [23] 49.84353 49.38462 51.34816 50.47381 50.43722 49.95626 49.78356 50.67631 49.74885 50.48481 50.38605
## [34] 49.67852 49.82027 49.27375 49.27381 50.54384 49.54337 50.85396 50.73716 50.39720 50.48076 49.01576
## [45] 50.45556 48.65065 49.63398 50.08084 49.46907 50.11526 51.11121 49.33433 49.24255 50.05063 50.10876
## [56] 49.81916 50.98534 49.57775 49.18555 49.67709 50.02491 49.60895 51.42723 50.36917 48.88077 49.61436
## [67] 50.11477 49.70123 49.01726 49.52215 50.29561 50.14371 51.37228 50.09824 49.14205 50.23992 50.77307
## [78] 50.91305 48.75136 49.71756 49.99803 49.65298 49.27288 49.99510 50.09797 49.41810 49.82271 50.27230
## [89] 49.62548 49.86603 50.43390 50.13443 48.90100 49.34616 50.52887 50.47013 49.39545 50.50800 50.69149
## [100] 50.77787a. We know that about 70% of the values should be within 1 standard deviation (0.6) of the mean (50). We can either make this count manually by examining the list above or have R do it for us as below. We should expect about 70 of the values to fall between 49.4 and 50.6.sum(X < 50.6 & X > 49.4)## [1] 65b. We know that about 95% of the values should be within 2 standard deviations (1.2) of the mean (50). We should thus expect about 95 of the values to fall between 48.8 and 51.2.sum(X < 51.2 & X > 48.8)## [1] 93- When \(n\) is large, the difference between \(\frac{1}{n}\) and \(\frac{1}{n-1}\) is very small. For example, suppose \(n=100000\). The values are very close, which means we’ll get almost the same number for the biased variance and standard deviation as we do for the unbiased versions.
n <- 100000
1/n## [1] 1e-051/(n - 1)## [1] 1.00001e-05Section 4.3.1
The model built in Section 5.1.1 which is needed for Exercises 1-5 is:
hwy = -3.53(displ) + 35.7.
The predicted value is -3.53(3.5) + 35.7 = 23.3. The actual value is 29, which gives a residual of 29 - 23.3 = 5.7.
mpg_model <- lm(hwy ~ displ, data = mpg)
mpg_w_pred_resid <- mpg %>%
add_predictions(mpg_model) %>%
add_residuals(mpg_model) %>%
select(-(cyl:cty), -fl, -class)
mpg_w_pred_resid## # A tibble: 234 x 7
## manufacturer model displ year hwy pred resid
## <chr> <chr> <dbl> <int> <int> <dbl> <dbl>
## 1 audi a4 1.8 1999 29 29.3 -0.343
## 2 audi a4 1.8 1999 29 29.3 -0.343
## 3 audi a4 2 2008 31 28.6 2.36
## 4 audi a4 2 2008 30 28.6 1.36
## 5 audi a4 2.8 1999 26 25.8 0.188
## 6 audi a4 2.8 1999 26 25.8 0.188
## 7 audi a4 3.1 2008 27 24.8 2.25
## 8 audi a4 quattro 1.8 1999 26 29.3 -3.34
## 9 audi a4 quattro 1.8 1999 25 29.3 -4.34
## 10 audi a4 quattro 2 2008 28 28.6 -0.636
## # ... with 224 more rows
## # i Use `print(n = ...)` to see more rowsggplot(mpg_w_pred_resid) +
geom_point(aes(displ, resid)) +
geom_hline(yintercept = 0)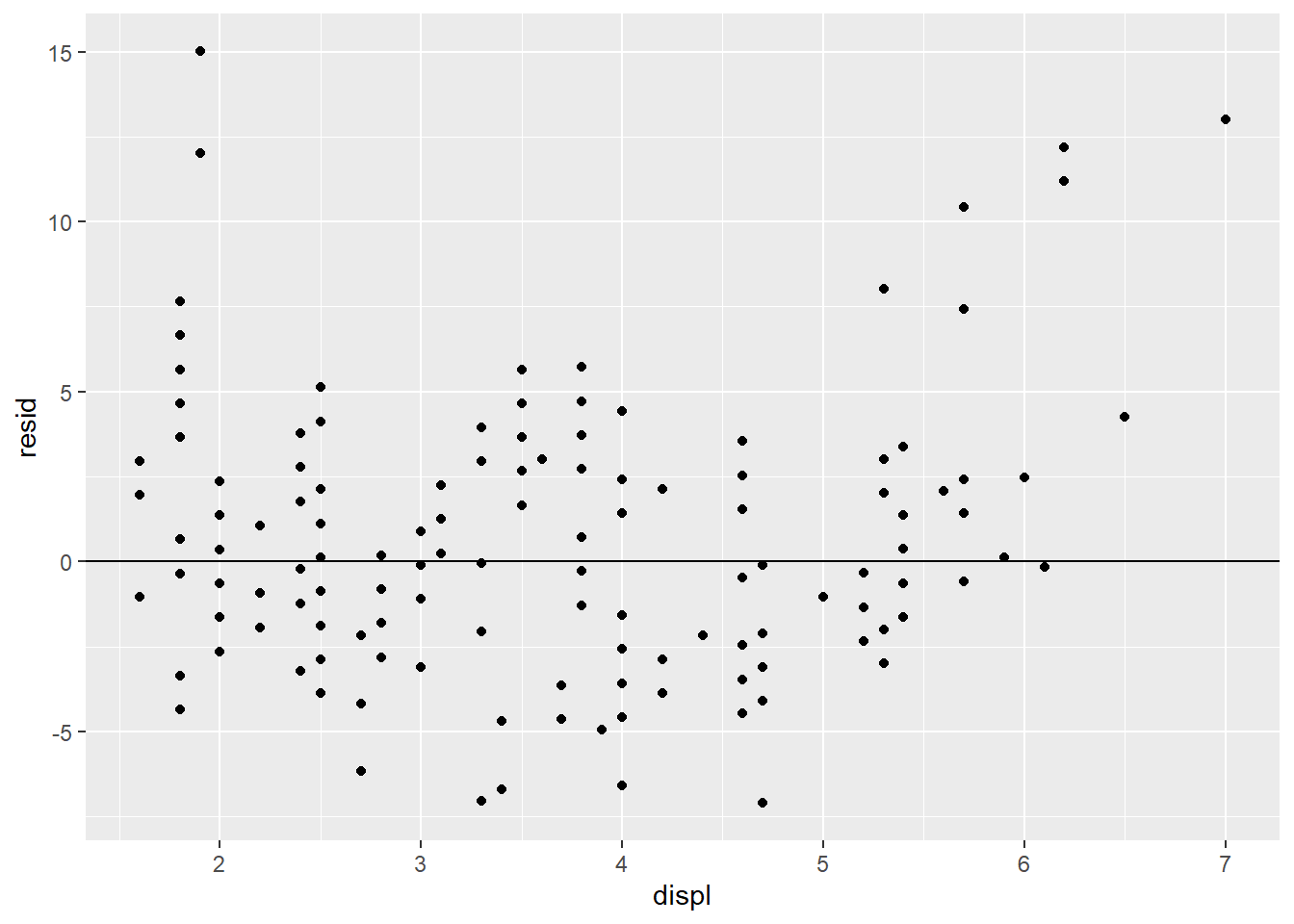
ggplot(mpg_w_pred_resid) +
geom_histogram(aes(resid), binwidth = 1)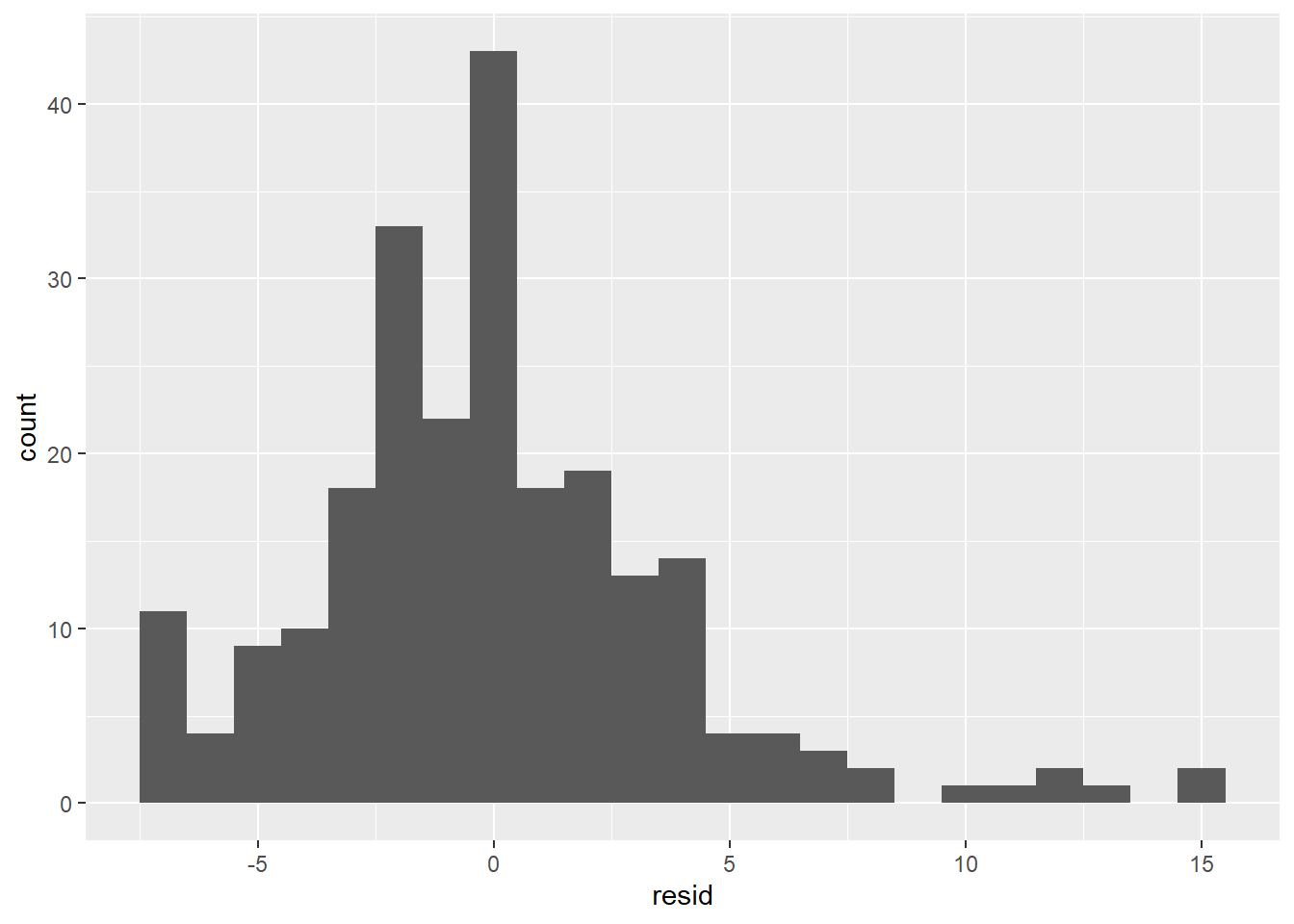
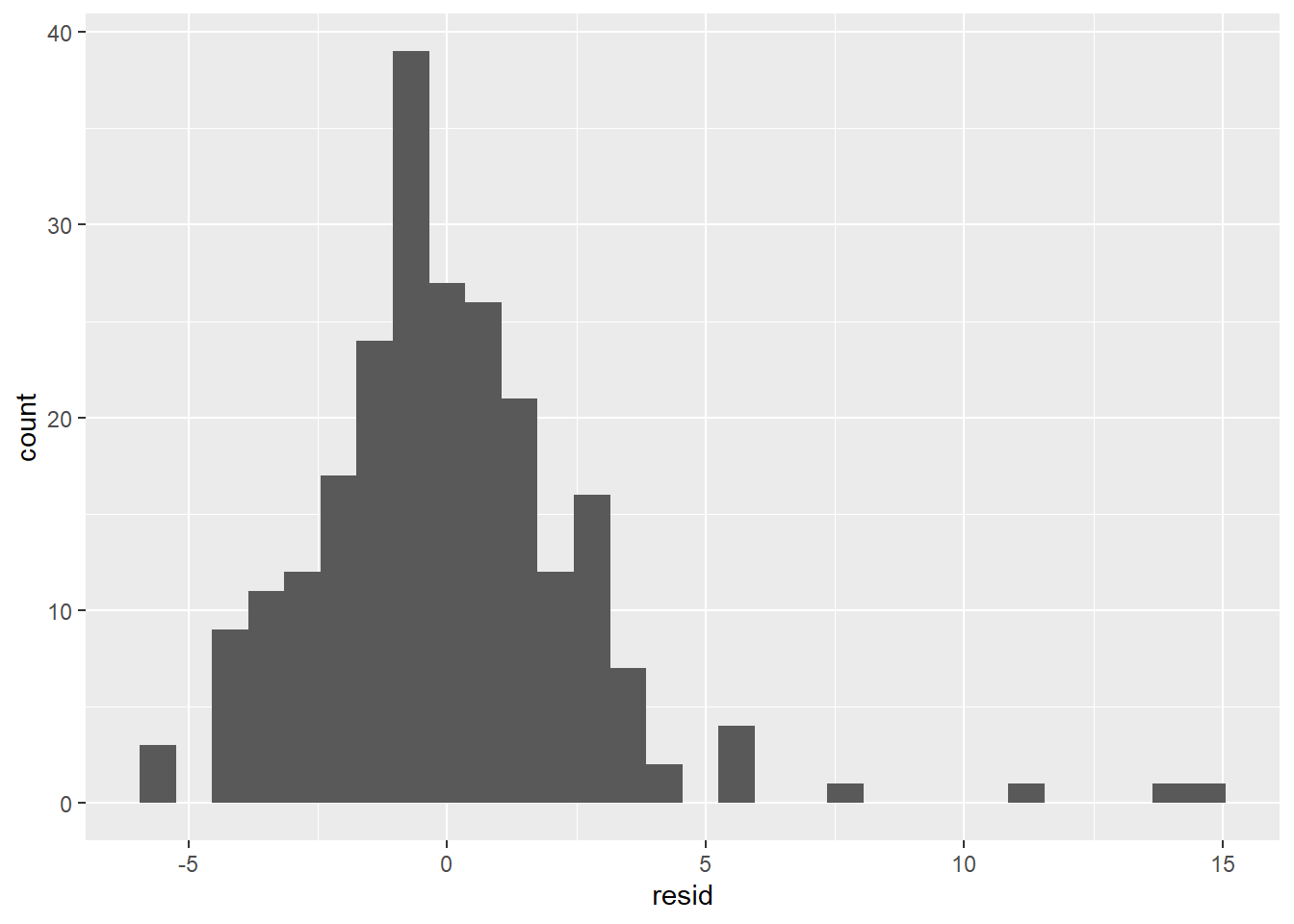
- The residuals don’t exhibit a very strong pattern. However, it does seem that the model significantly under-predicts for cars with very small or very large engines, which means that for these cars, engine size alone is not a good predictor of highway fuel efficiency. There must be other variables that explain why the
hwyvalues are so high. (Small-engine cars include hybrids, which get very high gas mileage. Among large-engine cars are two-seaters, which get surprisingly high gas mileage for their engine size because they are light weight and very aerodynamic.)
The residual distribution is somewhat normal, although it seems right-skewed rather than symmetrical. The reason is the same as given in the last paragraph: There are some large positive residuals, which are explained by cars for which the model significantly under-predicts the hwy value. There is also a spike at the far left, indicating that there are also cars for which the model significantly over-predicts.
Based on these residual visualizations, we can say that a linear regression model is appropriate, but it’s clear that there are other variables that would be needed to predict hwy better, such as class. We’ll see how to add extra predictors to a model in Section 4.6.
The residuals in (c) appear to exhibit less of a pattern than the others, which means that it is most indicative of a good linear model.
The residuals do not exhibit a pattern and the distribution is roughly normal, meaning that the data is appropriately modeled by a linear regression model.
library(readr)
height_weight <- read_csv("height-weight.csv")
height_model <- lm(`Weight(Pounds)` ~ `Height(Inches)`, data = height_weight)
height_weight_w_resid <- height_weight %>%
add_residuals(height_model)
ggplot(height_weight_w_resid) +
geom_point(aes(`Height(Inches)`, resid)) +
geom_hline(yintercept = 0)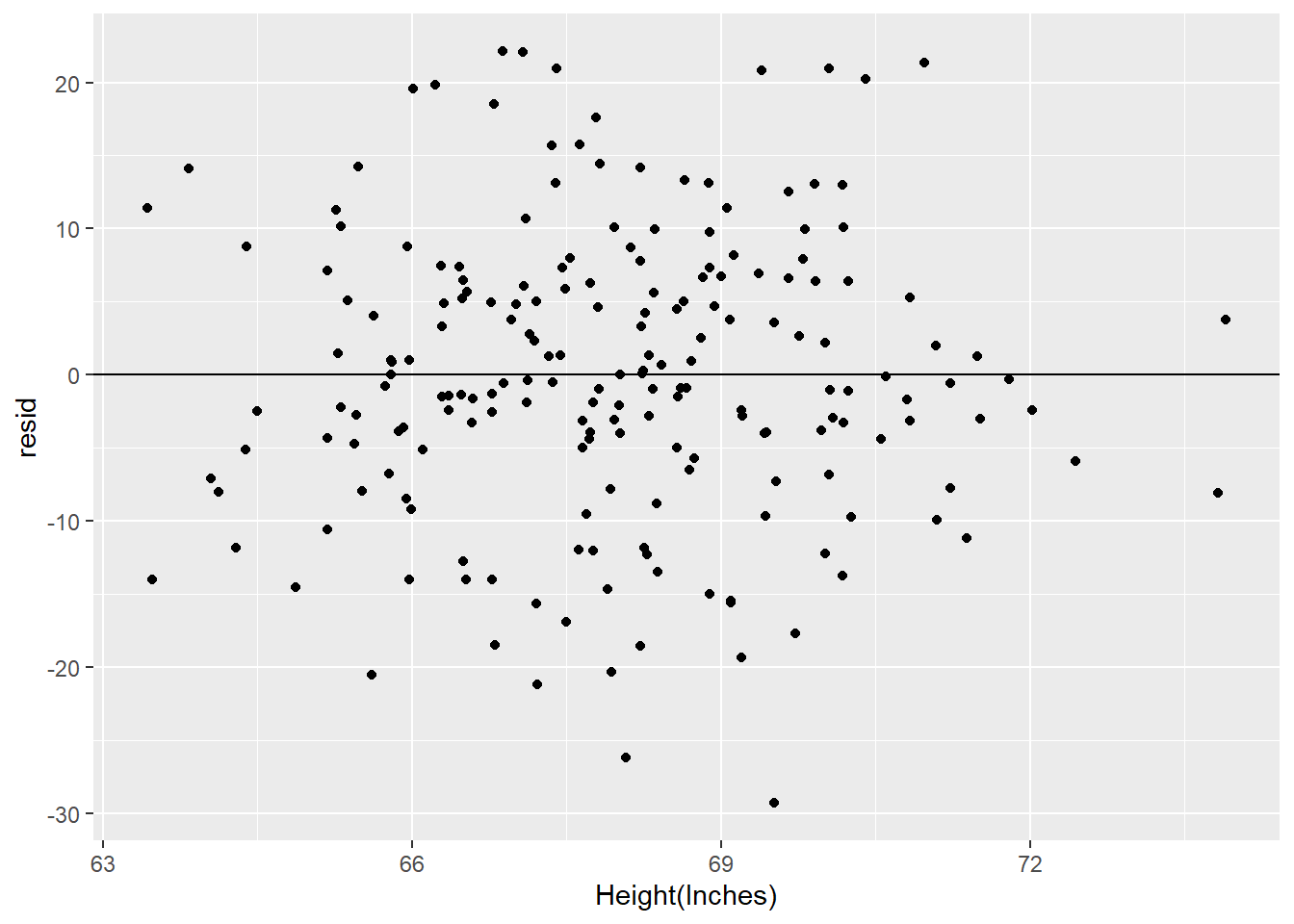
ggplot(height_weight_w_resid) +
geom_histogram(aes(resid), binwidth = 1.5)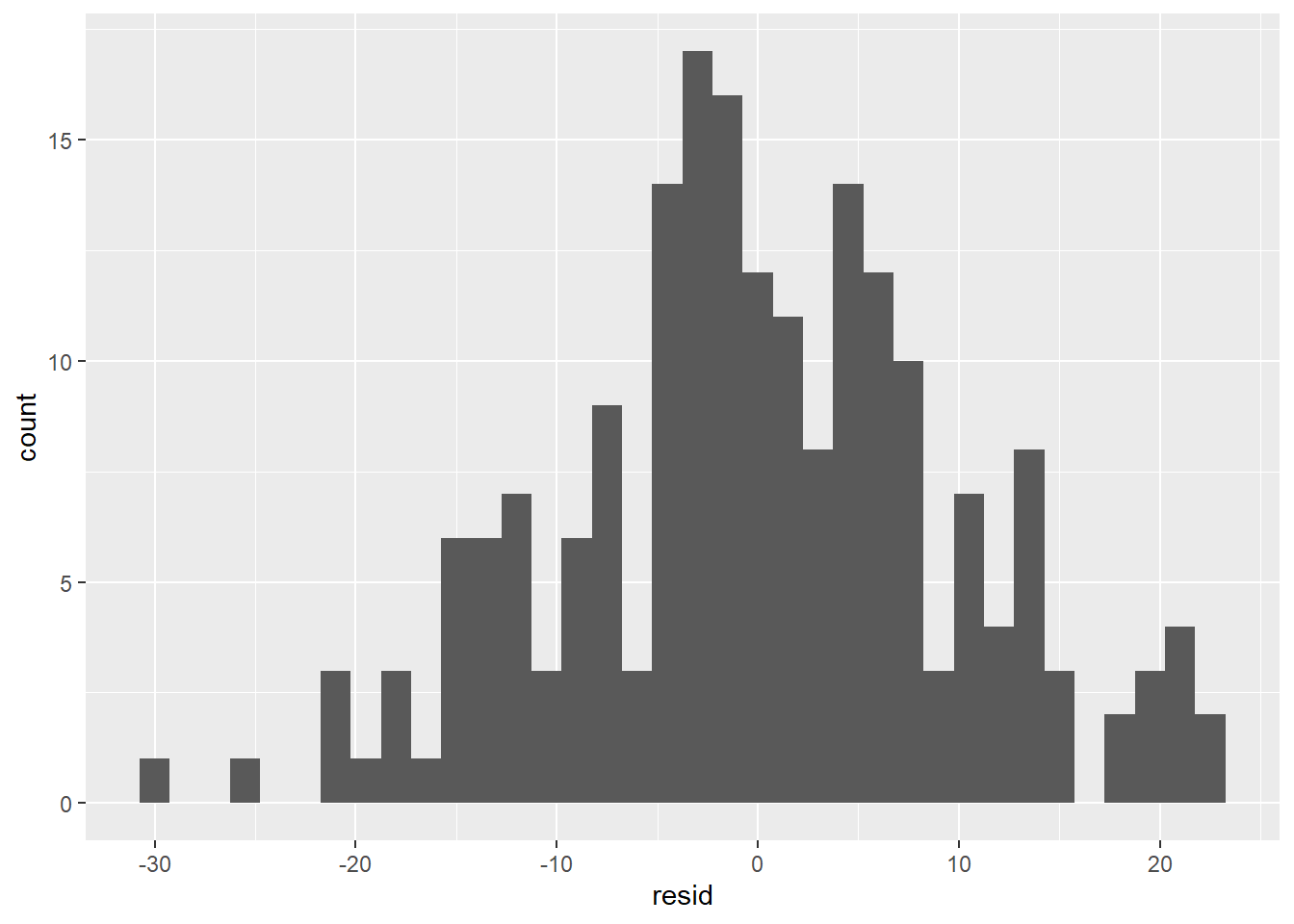
Section 4.4.1
The model we need for Exercises 1-2 is:
mpg_model <- lm(hwy ~ displ, data = mpg)- We can run the model’s summary to see that the RSE is 3.836.
summary(mpg_model)##
## Call:
## lm(formula = hwy ~ displ, data = mpg)
##
## Residuals:
## Min 1Q Median 3Q Max
## -7.1039 -2.1646 -0.2242 2.0589 15.0105
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 35.6977 0.7204 49.55 <2e-16 ***
## displ -3.5306 0.1945 -18.15 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.836 on 232 degrees of freedom
## Multiple R-squared: 0.5868, Adjusted R-squared: 0.585
## F-statistic: 329.5 on 1 and 232 DF, p-value: < 2.2e-16a. 3.836
b. 7.672 (which equals twice the RSE)- By examining the plot below, we see there are 8 points outside the blue lines which make up the 95% confidence band. We also know there are 234 data points total in
mpg. Thus, the band contains 226 out of 234 data points, which is about 96.6%. This is close to the expected 95%.
mpg_coefs <- coef(mpg_model)
b <- mpg_coefs[1]
m <- mpg_coefs[2]
ggplot(mpg) +
geom_point(aes(displ, hwy)) +
geom_abline(slope = m, intercept = b) +
geom_abline(slope = m, intercept = b + 7.672, color = "blue") +
geom_abline(slope = m, intercept = b - 7.672, color = "blue")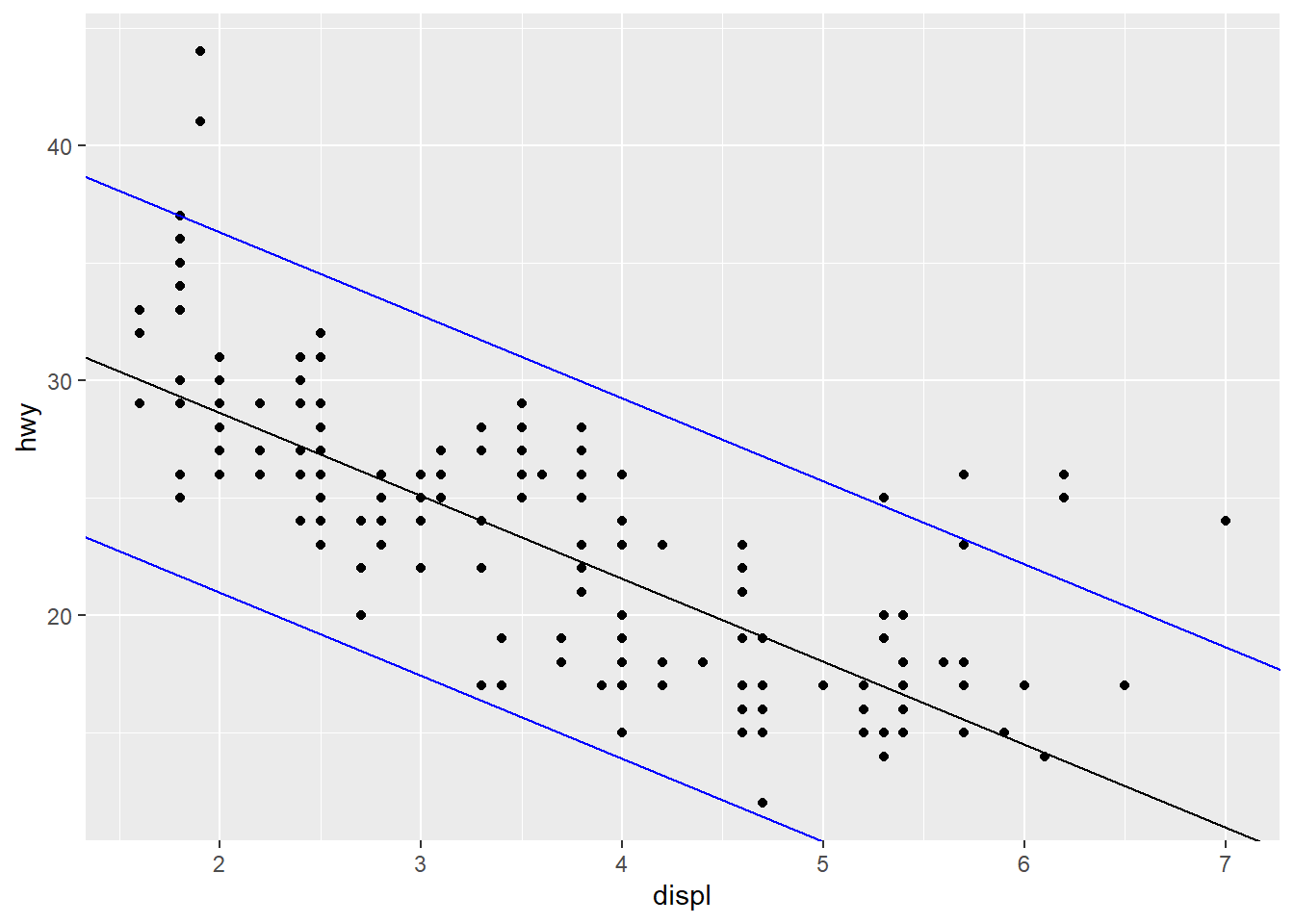
- The RSE of the model is 0.7634, which means that we can be 95% confident that the prediction is within 1.5268 of the true value. However, the residual distribution was not very normal-looking, and there are only 10 data points. Further, the RSE was computed with only 8 degrees of freedom, which is extremely low. Therefore, we should not put too much trust in this confidence interval.
sim1a <- readr::read_csv("https://raw.githubusercontent.com/jafox11/MS282/main/sim1_a.csv")
sim1a_model <- lm(y ~ x, data = sim1a)
summary(sim1a_model)##
## Call:
## lm(formula = y ~ x, data = sim1a)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.1206 -0.4750 0.1561 0.5620 0.9511
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 6.36737 0.52153 12.21 1.88e-06 ***
## x 1.45886 0.08405 17.36 1.24e-07 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.7634 on 8 degrees of freedom
## Multiple R-squared: 0.9741, Adjusted R-squared: 0.9709
## F-statistic: 301.3 on 1 and 8 DF, p-value: 1.237e-07Section 4.5.1
- The multiple R2 value is 0.5868, which means that the model can explain about 58.68% of the variance in the
hwyvariable. The rest of the variance is explained by randomness or other variables not used by the model. The adjusted R2 value is 0.585, which can be interpreted in the same way: The model explains about 58.5% of the variance inhwy. The difference is that multiple R2 values are biased, whereas adjust R2 values are not.
mpg_model <- lm(hwy ~ displ, data = mpg)
summary(mpg_model)##
## Call:
## lm(formula = hwy ~ displ, data = mpg)
##
## Residuals:
## Min 1Q Median 3Q Max
## -7.1039 -2.1646 -0.2242 2.0589 15.0105
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 35.6977 0.7204 49.55 <2e-16 ***
## displ -3.5306 0.1945 -18.15 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.836 on 232 degrees of freedom
## Multiple R-squared: 0.5868, Adjusted R-squared: 0.585
## F-statistic: 329.5 on 1 and 232 DF, p-value: < 2.2e-16- The R2 value is 0.999, which would indicate that the model can explain almost 100% of the variance in the
yvariable. However, the residuals exhibit a very obvious pattern, which means that the model is still missing an important relationship betweenxandy, despite the very high R2 value.
df <- tibble(
x = c(1, 2, 3, 4, 5, 6, 7, 8, 100),
y = c(2, 1, 4, 3, 6, 5, 8, 7, 100)
)
df_model <- lm(y ~ x, data = df)
summary(df_model)##
## Call:
## lm(formula = y ~ x, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.00644 -1.00447 0.04167 0.99406 0.99602
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 0.007418 0.398710 0.019 0.986
## x 0.999509 0.011841 84.410 8.62e-12 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1.069 on 7 degrees of freedom
## Multiple R-squared: 0.999, Adjusted R-squared: 0.9989
## F-statistic: 7125 on 1 and 7 DF, p-value: 8.624e-12df_w_resid <- df %>%
add_residuals(df_model)
ggplot(df_w_resid) +
geom_point(aes(x, resid))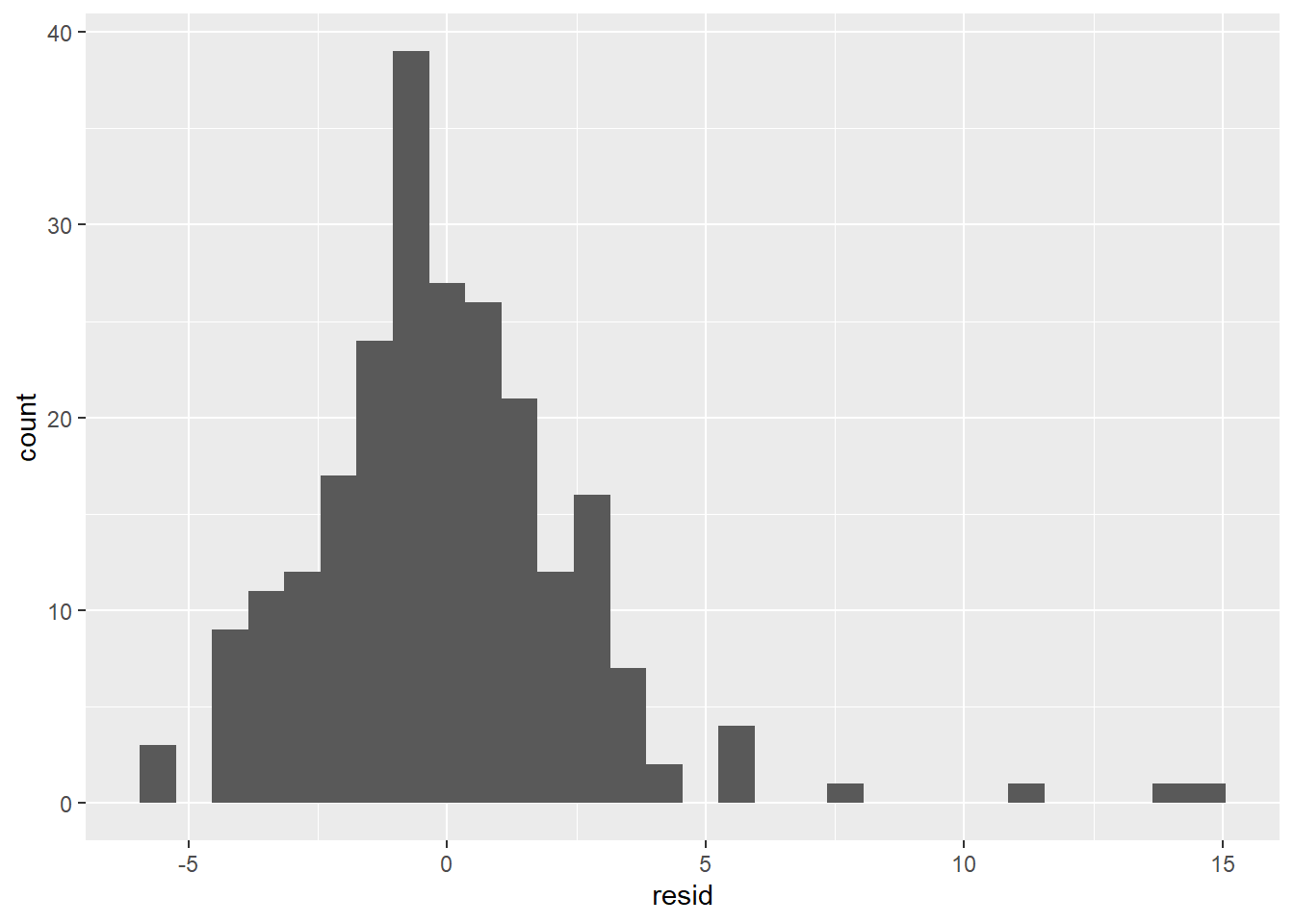
For an R2 value to be negative, the ratio \(\frac{\textrm{MSR}}{\textrm{variance}}\) would have to be greater than 1. This happens when the MSR is greater than the variance. When this is the case, the actual values of the response variable are, on average, closer to the mean response value than they are to the model’s predicted response values. In other words, the R2 value will be negative when the model does a worse job making predictions than one which would just predict the mean.
Statement (2) is more meaningful. Knowing the RSE without knowing anything about the magnitude of the response values doesn’t tell us anything; we would have no frame of reference to know whether an RSE of 2.25 should be considered high or low. The R2 value, on the other hand, does not need a frame of reference. To say that the model can explain 78% of the variation is meaningful even if we don’t know anything about the data set.
Section 4.6.1
- The coefficient (slope) is -3.5306, the standard error is 0.1945, the t-statistic is -18.15, and the p-value is \(2\times 10^{-16}\) (which is the smallest non-zero number R can display).
mpg_model <- lm(hwy ~ displ, data = mpg)
summary(mpg_model)##
## Call:
## lm(formula = hwy ~ displ, data = mpg)
##
## Residuals:
## Min 1Q Median 3Q Max
## -7.1039 -2.1646 -0.2242 2.0589 15.0105
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 35.6977 0.7204 49.55 <2e-16 ***
## displ -3.5306 0.1945 -18.15 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.836 on 232 degrees of freedom
## Multiple R-squared: 0.5868, Adjusted R-squared: 0.585
## F-statistic: 329.5 on 1 and 232 DF, p-value: < 2.2e-16- To say that the coefficient is -3.5306 means that for every increase of 1 liter in the
displvariable, the highway fuel efficiency drops by about 3.5306 miles per gallon.
To say that the standard error is 0.1945 means that on average, we would estimate a deviation of about 0.1945 from the true coefficient value when estimating the value of the coefficient.
To say that the t-statistic is -18.15 means that the value of our coefficient is about 18.15 standard errors below 0.
To say that the p-value is \(2\times 10^{-16}\) means that assuming the true value of our coefficient is 0, the probability of getting a value of -3.5306 or less as the result of statistical randomness is \(2\times 10^{-16}\).
Yes, it does since the p-value is less than 0.05.
The intercept is about 35.7 and it has a very small p-value, so it’s statistically significant. However, it does not have a meaningful interpretation in this context because it would be the highway fuel efficiency of a car with a 0-liter engine.
For part (a), we should make it so that the coefficient of
wis a large positive number and/or the standard error forwis small. We can guarantee this by making the relationship betweenwandzhighly linear with a large positive slope.
For part (b), we can replicate what we did for part (a), but make the slope a large negative number.
For part (c), we should make it so that the coefficient of y is near 0 and/or there is no linear relationship between y and z.
The following data set is one such example.
df <- tibble(
w = c(1, 2, 3, 4, 5, 6, 7, 8, 9, 10),
x = c(10, 9, 8, 7, 6, 5, 4, 3, 2, 1),
y = c(1, 2, 3, 4, 5, 5, 4, 3, 2, 1),
z = c(5, 10, 15, 20, 25, 30, 35, 40, 45, 50)
)
df_model_w <- lm(z ~ w, data = df)
summary(df_model_w)##
## Call:
## lm(formula = z ~ w, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -4.756e-15 -1.193e-15 -2.021e-16 1.623e-15 3.315e-15
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -4.494e-15 1.736e-15 -2.588e+00 0.0322 *
## w 5.000e+00 2.798e-16 1.787e+16 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 2.541e-15 on 8 degrees of freedom
## Multiple R-squared: 1, Adjusted R-squared: 1
## F-statistic: 3.193e+32 on 1 and 8 DF, p-value: < 2.2e-16df_model_x <- lm(z ~ x, data = df)
summary(df_model_x)##
## Call:
## lm(formula = z ~ x, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -9.425e-15 -2.428e-16 7.752e-16 1.120e-15 5.908e-15
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 5.500e+01 2.766e-15 1.988e+16 <2e-16 ***
## x -5.000e+00 4.458e-16 -1.122e+16 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 4.049e-15 on 8 degrees of freedom
## Multiple R-squared: 1, Adjusted R-squared: 1
## F-statistic: 1.258e+32 on 1 and 8 DF, p-value: < 2.2e-16df_model_y <- lm(z ~ y, data = df)
summary(df_model_y)##
## Call:
## lm(formula = z ~ y, data = df)
##
## Residuals:
## Min 1Q Median 3Q Max
## -22.50 -11.25 0.00 11.25 22.50
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 2.750e+01 1.191e+01 2.309 0.0497 *
## y 1.589e-15 3.590e+00 0.000 1.0000
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 16.06 on 8 degrees of freedom
## Multiple R-squared: 4.284e-32, Adjusted R-squared: -0.125
## F-statistic: 3.427e-31 on 1 and 8 DF, p-value: 1- There are two things wrong with this statement. First, the p-value tells you the probability of obtaining a slope whose absolute value is 3.1 or greater. Second, the p-value is a probability calculated under the assumption that the null hypothesis is true. The correct way to word this statement is, “Assuming that E has no significant impact on R, the probability of obtaining a slope whose absolute value is 3.1 or greater is 23%.”
- Getting a p-value above the 0.05 threshold means that there is not enough evidence to reject the null hypothesis, which is not the same as accepting it. (By analogy, you might not believe there’s enough evidence to disbelieve is Santa Claus, but that doesn’t necessarily mean you have to believe in him.) The correct way to word the statement is, “Since the p-value is above the 0.05 threshold, we cannot reject the null hypothesis, which means we don’t have enough evidence to conclude that E has a significant impact on R.”
- To reject the null hypothesis is to say that we have enough evidence to conclude that the intercept is not 0. It doesn’t necessarily mean that the true value is 77.09, though.
- The multiple R2 is 0.0003758, which means our model can only explain about 0.038% of the variation. Even worse, the adjusted R2 value is -0.05516. A negative R2 value means that we’d be better off just using the mean of the exam scores as our prediction rather than using the model to make a prediction. In other words, our model is useless, so despite their low p-values, we shouldn’t attribute any real meaning to the coefficients. This model would not make it past the post-assessment stage, so we shouldn’t be trying to interpret anything with it.
- Since the RSE is 10.34, the endpoints of our 95% CI are +/- 20.68. This means that if our model predicts an exam score of 60%, then there’s a 95% chance that the actual score is somewhere between about 40% and 80%. That’s not very impressive! The large RSE is more reason to not move on to the interpretation stage.
Section 4.7.1
library(readr)
car_prices <- read_csv("car_prices.csv")car_prices_model <- lm(price ~ `highway-mpg`, data = car_prices)- The coefficient is -821.50, which means that for each additional mile per gallon of highway fuel efficiency, the selling price drops by about $821.50. The p-value is extremely small, meaning that this coefficient is statistically significant. The conclusion we can draw is that cars with better fuel efficiency are cheaper. This might seem surprising since it should be the case that high fuel efficiency adds value to a car rather than subtracts from it.
summary(car_prices_model)##
## Call:
## lm(formula = price ~ `highway-mpg`, data = car_prices)
##
## Residuals:
## Min 1Q Median 3Q Max
## -8679 -3418 -1136 1063 20938
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 38450.00 1849.66 20.79 <2e-16 ***
## `highway-mpg` -821.50 58.84 -13.96 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 5671 on 197 degrees of freedom
## Multiple R-squared: 0.4973, Adjusted R-squared: 0.4948
## F-statistic: 194.9 on 1 and 197 DF, p-value: < 2.2e-16Both
curb-weightandhorsepowermight be confounding variables since they are correlated with bothpriceandhighway-mpg. Ascurb-weightandhorsepowerincrease,pricegenerally increases, buthighway-mpggenerally decreases.
car_prices_model2 <- lm(price ~ `highway-mpg` + `curb-weight` + horsepower, data = car_prices)- The new coefficient of
highway-mpgis about 183.6, meaning that for a given curb weight and horsepower, each additional mpg of highway fuel efficiency adds about $183.60 to the car’s value. The p-value is 0.0157, which is below the 0.05 cutoff, meaning that the coefficient is statistically significant.
summary(car_prices_model2)##
## Call:
## lm(formula = price ~ `highway-mpg` + `curb-weight` + horsepower,
## data = car_prices)
##
## Residuals:
## Min 1Q Median 3Q Max
## -9002.2 -2021.6 53.3 1617.2 15977.7
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -2.624e+04 4.324e+03 -6.069 6.59e-09 ***
## `highway-mpg` 1.836e+02 7.531e+01 2.438 0.0157 *
## `curb-weight` 9.014e+00 9.034e-01 9.978 < 2e-16 ***
## horsepower 1.046e+02 1.278e+01 8.183 3.52e-14 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3786 on 195 degrees of freedom
## Multiple R-squared: 0.7782, Adjusted R-squared: 0.7748
## F-statistic: 228.1 on 3 and 195 DF, p-value: < 2.2e-16The conclusion from Exercise 3 is an example of omitted variable bias because it is a misleading statement that is due to the omission of relevant variables as predictors.
One example of omitted variable bias might be to say that since the number of reported sunburns seems to increase when an ice cream stand sells more ice cream, that it must be the case that ice cream causes sun burns. The omitted variable here, though, would be the air temperature. As temperature goes up, so does the amount of ice cream sold and the number of reported sun burns.
Section 4.8.1
- The model is given by:
diamonds_model_4 <- lm(price ~ carat + depth + table, data = diamonds)- The summary below shows that the multiple and adusted R2 values are both 0.8537. If more decimal places could be displayed, we’d eventually see a difference, but they’re the same through four decimal places. This is because
diamondshas 53,940 observations, whereas the number of degrees of freedom is 53,936. The values of the fractions \(\frac{1}{53940}\) and \(\frac{1}{53936}\) are so close, the amount of bias built into the multiple R2 won’t be noticeable, and this leads to nearly equal multiple and adjusted R2 values.
summary(diamonds_model_4)##
## Call:
## lm(formula = price ~ carat + depth + table, data = diamonds)
##
## Residuals:
## Min 1Q Median 3Q Max
## -18288.0 -785.9 -33.2 527.2 12486.7
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 13003.441 390.918 33.26 <2e-16 ***
## carat 7858.771 14.151 555.36 <2e-16 ***
## depth -151.236 4.820 -31.38 <2e-16 ***
## table -104.473 3.141 -33.26 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1526 on 53936 degrees of freedom
## Multiple R-squared: 0.8537, Adjusted R-squared: 0.8537
## F-statistic: 1.049e+05 on 3 and 53936 DF, p-value: < 2.2e-16The coefficient of
caratmeans that with each additioncaratadded to the weight of a diamond, the price goes up by about $7,858. The coefficient ofdepthmeans that with each additional percentage point added to thedepth, the price drops by about $151. The coefficient oftablemeans that with each unit added to thetablevalue, the price drops by about $104.The p-value of each coefficient is extremely low, meaning that the coefficient values are statistically significant.
The residuals appear to be approximately normal.
diamonds_w_resid <- diamonds %>%
add_residuals(diamonds_model_4)
ggplot(diamonds_w_resid) +
geom_histogram(aes(resid), binwidth = 750)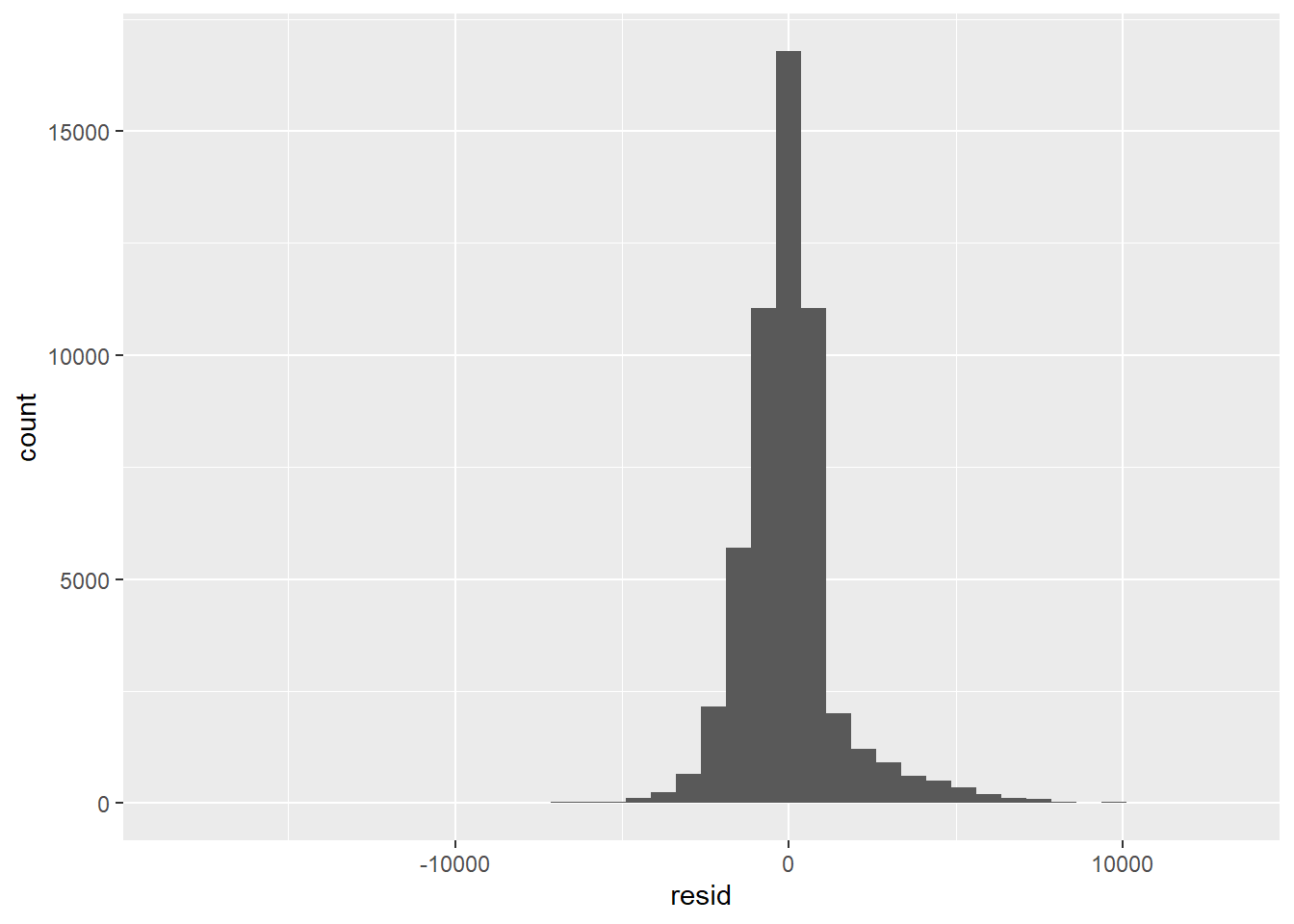
- Since the residuals are normally distributed, we can state the 95% CI. The summary above shows that the RSE of the model is 1526. This means that we can be 95% confident that a given prediction is within $3,052 of the actual price.
delay_model <- lm(arr_delay ~ dep_delay + air_time + distance, data = flights)
delay_model##
## Call:
## lm(formula = arr_delay ~ dep_delay + air_time + distance, data = flights)
##
## Coefficients:
## (Intercept) dep_delay air_time distance
## -15.91942 1.01957 0.68698 -0.08919The
air_timeanddistancecoefficients seem contradictory. You wouldn’t expect them to affect the arrival delay in opposite ways, but the reason is that these two variables are likely highly correlated. So the counter-intuitive coefficients are probably side-effects of collinearity.The correlation coefficient between
air_timeanddistanceis almost 1, which means these two variables are highly correlated.
arr_delay_predictors <- flights %>%
select(dep_delay, air_time, distance) %>%
filter(!is.na(dep_delay) & !is.na(air_time) & !is.na(distance))
cor(arr_delay_predictors)## dep_delay air_time distance
## dep_delay 1.00000000 -0.02240508 -0.0216809
## air_time -0.02240508 1.00000000 0.9906496
## distance -0.02168090 0.99064965 1.0000000- We’ll keep
air_timeand deletedistance. Previously,air_timehad a positive coefficient, but in the new non-collinear model,air_timehas a negative coefficient. A negative coefficient makes more sense because the longer the flight is, the more time you have to make up for a departure delay, thus decreasing the arrival delay.
delay_model2 <- lm(arr_delay ~ dep_delay + air_time, data = flights)
delay_model2##
## Call:
## lm(formula = arr_delay ~ dep_delay + air_time, data = flights)
##
## Coefficients:
## (Intercept) dep_delay air_time
## -4.831805 1.018723 -0.007055- Comparing the summaries below, we see that the R2 value increases a very little bit after the outliers are removed. Also, the coefficient of
xincreases slightly. In this case, removing the outliers didn’t make much of a difference, probably because there are very few of them compared to the size of the data set.
outliers_present <- lm(price ~ x, data = diamonds)
summary(outliers_present)##
## Call:
## lm(formula = price ~ x, data = diamonds)
##
## Residuals:
## Min 1Q Median 3Q Max
## -8426 -1264 -185 973 32128
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -14094.056 41.732 -337.7 <2e-16 ***
## x 3145.413 7.146 440.2 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1862 on 53938 degrees of freedom
## Multiple R-squared: 0.7822, Adjusted R-squared: 0.7822
## F-statistic: 1.937e+05 on 1 and 53938 DF, p-value: < 2.2e-16diamonds2 <- diamonds %>%
filter(x > 3)
outliers_removed <- lm(price ~ x, diamonds2)
summary(outliers_removed)##
## Call:
## lm(formula = price ~ x, data = diamonds2)
##
## Residuals:
## Min 1Q Median 3Q Max
## -8476.1 -1265.4 -178.1 990.5 12087.7
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) -14184.869 41.332 -343.2 <2e-16 ***
## x 3160.674 7.077 446.6 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 1840 on 53930 degrees of freedom
## Multiple R-squared: 0.7872, Adjusted R-squared: 0.7872
## F-statistic: 1.995e+05 on 1 and 53930 DF, p-value: < 2.2e-16- In this case, it would not make sense to impute the mean value into the missing values of
dep_delaysince theNAs in this column are meaningful; they indicate a cancelled flight. To give them a value would cause us to pretend that these flights weren’t cancelled. In this case, it would be better to filter our the rows with missing values in thedep_delaycolumn.
Section 4.9.1
- The model is
hwy= 24.8 + 3.498(classcompact) + 2.493(classmidsize) - 2.436(classminivan) - 7.921(classpickup) + 3.343(classsubcompact) - 6.671(classsuv)
class_model <- lm(hwy ~ class, data = mpg)
class_model##
## Call:
## lm(formula = hwy ~ class, data = mpg)
##
## Coefficients:
## (Intercept) classcompact classmidsize classminivan classpickup classsubcompact
## 24.800 3.498 2.493 -2.436 -7.921 3.343
## classsuv
## -6.671- Use 1 for
classsuvand 0 for the others. This gives 18.129. - Use 1 for
classsubcompactand 0 for the others. This gives 28.143. - Since there is no
class2seatervariable, we use 0 for all the variables. This gives 24.8.
- Use 1 for
- The R2 value is 0.6879, which means the model can explain about 69% of the variation in the
hwyvariable. This is pretty good. The RSE is 3.37, which means we can be about 70% confident that any prediction is within 3.37 of the actualhwyvalue and 95% confident that any prediction is within 6.74 of the actualhwyvalue. This is also moderately good, given the sizes of thehwyvalues.
summary(class_model)##
## Call:
## lm(formula = hwy ~ class, data = mpg)
##
## Residuals:
## Min 1Q Median 3Q Max
## -8.1429 -1.8788 -0.2927 1.1803 15.8571
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 24.800 1.507 16.454 < 2e-16 ***
## classcompact 3.498 1.585 2.206 0.0284 *
## classmidsize 2.493 1.596 1.561 0.1198
## classminivan -2.436 1.818 -1.340 0.1815
## classpickup -7.921 1.617 -4.898 1.84e-06 ***
## classsubcompact 3.343 1.611 2.075 0.0391 *
## classsuv -6.671 1.567 -4.258 3.03e-05 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 3.37 on 227 degrees of freedom
## Multiple R-squared: 0.6879, Adjusted R-squared: 0.6797
## F-statistic: 83.39 on 6 and 227 DF, p-value: < 2.2e-16- First, the R2 value is 0.4836. This isn’t great, but it’s not terrible. We can’t expect big R2 values for this simple regression model with a binary predictor, since something as unpredictable as life expectancy can’t be explained by a single yes/no variable. Regardless, the coefficient of the Western Europe value is -1.1235, which means that the average life expectancy in Western Europe is about 1.1235 years less than the average life expectancy in Northern Europe. The p-value of this coefficient is 0.00831, which is less than the 0.05 cutoff. This indicates that the 1.1235-year life expectancy difference is significant.
library(readr)
Europe <- read_csv("Europe.csv")
Europe_model <- lm(Life_Expectancy ~ Region, data = Europe)
summary(Europe_model)##
## Call:
## lm(formula = Life_Expectancy ~ Region, data = Europe)
##
## Residuals:
## Min 1Q Median 3Q Max
## -1.02520 -0.51150 -0.04321 0.51626 0.75192
##
## Coefficients:
## Estimate Std. Error t value Pr(>|t|)
## (Intercept) 82.5764 0.2569 321.475 < 2e-16 ***
## RegionWestern Europe -1.1235 0.3501 -3.209 0.00831 **
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## Residual standard error: 0.6292 on 11 degrees of freedom
## Multiple R-squared: 0.4836, Adjusted R-squared: 0.4366
## F-statistic: 10.3 on 1 and 11 DF, p-value: 0.008314- The long right tail of the histogram means that there are a significant number of cars with large positive residuals, which means that the model underpredicts the value of
hwy. As we have seen, this must be due to the2seaterandcompactclasses, which have cars that get much better gas mileage than would be predicted.
- The long right tail of the histogram means that there are a significant number of cars with large positive residuals, which means that the model underpredicts the value of
mpg_model <- lm(hwy ~ displ, data = mpg)
mpg_model_w_resid <- mpg %>%
add_residuals(mpg_model)
ggplot(mpg_model_w_resid) +
geom_histogram(aes(resid))
b. mpg_model2 <- lm(hwy ~ displ + class, data = mpg)c. The new histogram looks a little more normal. We see few cars in the right tail, indicating that by controlling for `class`, we were able to partly explain the under-predictions the original model had made.mpg_w_resid2 <- mpg %>%
add_residuals(mpg_model2)
ggplot(mpg_w_resid2) +
geom_histogram(aes(resid))